10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2007; 3(7):486-492. doi:10.7150/ijbs.3.486 This issue Cite
Short Research Communication
Concerted Transcriptional Regulation by BRCA1 and COBRA1 in Breast Cancer Cells
1. Department of Biochemistry and Molecular Genetics, University of Virginia, Charlottesville, VA 22908, USA
2. Public Health Sciences Administration, School of Medicine, University of Virginia, Charlottesville, VA 22908, USA
3. Department of Molecular Medicine/Institute of Biotechnology, University of Texas Health Science Center at San Antonio, San Antonio, TX 78245, USA
Received 2007-11-6; Accepted 2007-11-22; Published 2007-11-26
Abstract
Cofactor of BRCA1 (COBRA1) was first identified as a protein that binds to the breast cancer susceptibility gene product BRCA1. COBRA1 modulates estrogen-dependent and independent transcription and suppresses the growth of breast cancer cells. Its expression is significantly reduced in metastatic and recurrent breast cancer, pointing to a tumor suppressor function in breast cancer development. In light of these initial implications of COBRA1 in human breast cancer, the current investigation sought to obtain more direct functional evidence that links COBRA1 with BRCA1 in transcriptional regulation in breast cancer cells. Small hairpin RNA (shRNA)-mediated gene knockdown and gene expression microarray were used to study the impact of COBRA1 and BRCA1 on global transcription in the same breast cancer cell background. The gene expression profiling study in tissue culture cells uncovers a significant overlap of COBRA1- and BRCA1-regulated genes, many of which have been previously implicated in breast cancer progression. The data shown herein support the notion that COBRA1 and BRCA1 may engage in common gene regulatory pathways to suppress breast cancer progression.
Keywords: COBRA1, BRCA1, transcription regulation, shRNA knockdown, breast cancer.
1. Introduction
Cofactor of BRCA1 (COBRA1) was first identified through a yeast two-hybrid screen as a protein that binds to the BRCT domain of the tumor suppressor BRCA1 [1]. In support of its role in transcriptional regulation, COBRA1 is found in the negative elongation factor (NELF) complex, which stalls RNA polymerase II (RNAPII) during transcription elongation in vitro [24]. Furthermore, in vivo studies also demonstrate that COBRA1 and the rest of the NELF complex are recruited to distinct regions of the genome and modulate both estrogen-dependent and -independent expression of cancer-associated genes in breast cancer cell lines [5,6]. Consistent with the in vitro biochemical finding, COBRA1 modulates the movement of the transcription elongation complex at several estrogen-responsive transcription promoters in vivo, thus distinguishing itself from most of the known transcription coregulators that act at the transcription initiation stage [5].
COBRA1 is enriched in the luminal epithelial cells of normal mammary gland [5], and COBRA1 mRNA level has been reported to fluctuate in various breast cancer cell lines [7]. In addition, ectopic expression of COBRA1 in breast cancer cells suppresses tumor cell proliferation, whereas partial depletion of COBRA1 or another NELF subunit (NELF-E) results in accelerated cell colony growth [5]. In addition, COBRA1 expression was inversely correlated with breast cancer progression, with low expression in primary tumors from those patients who had distant metastasis and local recurrence [29]. These observations are consistent with a potential role of COBRA1 in tumor suppression.
Several lines of evidence suggest that COBRA1 and BRCA1 may share a similar functional pathway(s) in transcriptional regulation. First, like COBRA1, BRCA1 modulates ligand-dependent and independent transcription in ERα-positive breast cancer cells [8,9]. Second, both COBRA1 and BRCA1 are physically associated with the promoter region of an estrogen-responsive gene (pS2/TFF1) [5,9]. Third, both proteins interact with the C-terminal domain (CTD) of RNAPII as well as several transcription elongation factors [10, 13]. Lastly, the two proteins also share the property of inducing large-scale chromatin reorganization [1]. Therefore, it is conceivable that the physical interaction between COBRA1 and BRCA1 may facilitate communication between the two proteins and thus mutually coordinate their regulation of common regulatory targets in the genome. In the current study, we took a genome-wide approach to compare the impact of COBRA1 and BRCA1 on global transcription in the same breast cancer cell background. Our work led to the intriguing finding that COBRA1 and BRCA1 knockdown resulted in substantially overlapping gene expression profiles. Furthermore, a number of COBRA1/BRCA1-regulated genes are associated with breast cancer progression.
2. Materials and Methods
Cell lines and culture
T47D cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and grown in Dulbecco's Eagle's medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 μg/ml). Phoenix packaging cells were a generous gift from Dr. David Rekosh at the University of Virginia. The cell lines were tested to be mycoplasma free.
shRNA constructs
To generate the stable knockdown cell lines, BRCA1 and COBRA1 shRNAs were cloned between the BglII and HindIII sites of the pSUPER.retro/puromycin and pSUPER.retro/neomycin vectors, respectively (Oligoengine). The following sequences directed against BRCA1 and COBRA1 were used: shBRCA1-03: 5'-GAAGCCAGCTCAAGCAATA-3'; shCOBRA1-2A: 5'-GACCTTCTGGAGAAGAGCT-3'. For the negative control, the EGFP-targeted shRNA sequence (5'-GAACGGCATCAAGGTGAAC-3') was cloned into both the puromycin- and neomycin-resistant pSUPER.retro constructs. For verification of the BRCA1 knockdown effect, the following siRNA oligos were used in a transient transfection experiment: BRCA1-DO4 (5'-GCAGATAGTTCTACCAGTA-3'); BRCA1-DO7 (5'-GAAGGAGCTTTCATCATTC-3').
Viral supernatants were prepared and used to infect T47D cells as described previously [5]. Stable pools were selected with either 10 μg/ml puromycin for one week, or 800 μg/ml neomycin for two weeks. To generate the COBRA1/BRCA1 double knockdown cells, the BRCA1 single knockdown pool was first generated and selected in puromycin-containing medium. The selected cell pool was further infected with the shCOBRA1-expressing virus, and a second selection was carried out for two weeks in the presence of both puromycin and neomycin.
Immunoblotting
To evaluate the expression of BRCA1 and COBRA1, whole cell extracts were prepared in SDS sample buffer containing a cocktail of protease inhibitors. Protein concentrations were determined by the BCA protein assay reagent (Pierce). Approximately 30 μg of total protein was used for 5% (for BRCA1) or 10% (for COBRA1) SDS-PAGE. The proteins were detected by incubation with antibody directed against: anti-BRCA1 (Ab1 from Calbiochem), anti-tubulin (Calbiochem), or anti-COBRA1 [5]. Antibody binding was visualized using chemiluminescence (SuperSignal West Pico, Pierce) according to the manufacturer's directions.
Microarray and data analysis
shRNA-expressing T47D cells were grown to 70% confluency and harvested for total RNA. Duplicates of the following cell lines were subjected to microarray study using the human gene array chips (HgU-133A) from Affymetrix: shEGFPpuro+shEGFPneo, shBRCA1puro+shEGFPneo, shEGFPpuro+shCOBRA1neo, shBRCA1puro+shCOBRA1neo.
A total of 287 genes were found to be differentially regulated by BRCA1 and COBRA1, using the LPE test with statistical significance of p-value 0.003 [14]. The LPE test is specifically designed for small-sample microarray data analysis by significantly strengthening statistical power based on an advanced error pooling method at each local intensity range where signal-to-noise ratio is similar among different genes. Thus, by employing LPE, we were able to identify differentially regulated genes on our duplicated microarray data with statistical rigor.
Hierarchical clustering analysis was further performed with the 287 genes selected in the previous analysis, using Pearson correlation as a measure of similarity between genes and the average linkage (average distance) as a clustering allocation algorithm. In this clustering analysis, genes with low-intensity expression patterns were found to be closely clustered at the left-hand side of the heat map in Fig. 1B. Note that high expression intensities are represented by red, low expression intensities by green, and medium intensities by black.
RNA isolation and cDNA synthesis
T47D cells were grown to 70% confluency and total RNA was extracted with TRIzol Reagent according to the manufacturer's instructions (Invitrogen, Carlsbad, CA, USA). The concentration and purity of the RNA were determined spectrophotometrically at 260 and 280nm. Approximately 1 μg of RNA was reverse-transcribed using ImPromp-II reverse transcription system (Promega, Madison, WI, USA). The RNA extraction and reverse transcription kits used for RNA/cDNA preparation for the clinical samples were purchased from AbGene and Sigma, respectively.
Quantitative real-time PCR
The Affymetrix microarray results were verified by real-time RT-PCR for those genes shown in Fig. 3. Primer Express software (Applied Biosystems, Foster City, CA, USA) was used to design primers that covered two neighboring exons. Primer sequences are provided in the Table 1. An ABI 7300 real-time PCR machine was used for the quantitative real-time PCR analysis. Standard curves were generated by a serial dilution of cDNA and relative mRNA levels were measured. Expression levels were normalized against β-actin. Results shown were representatives of at least three independent experiments.
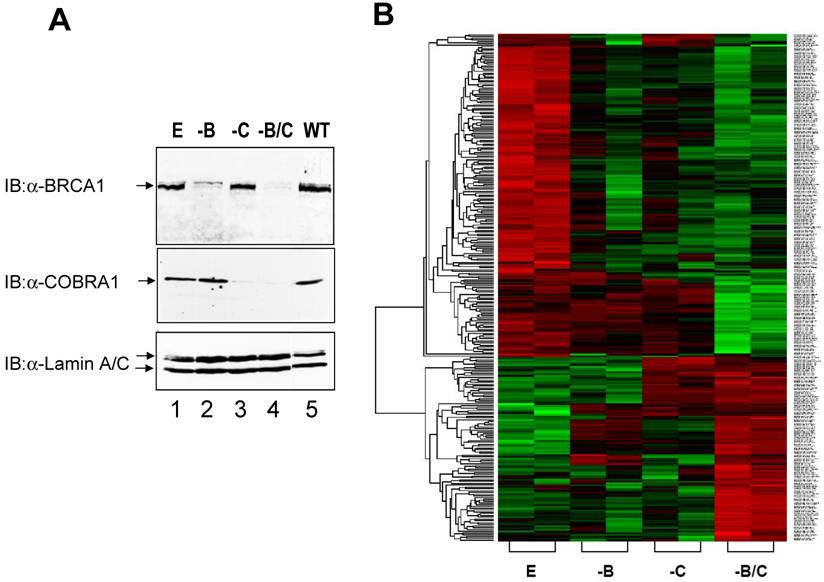
Microarray analysis of gene expression profiles in COBRA1 and/or BRCA1-depleted breast cancer cell lines. A. Immunoblot analysis showing the decreased expression of BRCA1 and COBRA1 protein in the shRNA-expressing T47D cells lines. Whole cell extracts were prepared from the following cell lines: shEGFPpuromycin+shEGFPneomycin (E); shBRCA1puromycin+shEGFPneomycin (-B); shEGFPpuromycin+shCOBRA1neomycin (-C); and shBRCA1puromycin+shCOBRA1neomycin (-B/C). Parental T47D cells (WT) were also included for comparison. Protein expression levels were analyzed using antibodies against BRCA1(top panel), COBRA1(middle panel), lamin A/C (bottom panel). Lamin was used as a loading control. B. Hierarchical clustering analysis of the 278 COBRA1 and/or BRCA1-regulated genes. Data from the duplicates of four knockdown cell lines are shown. High expression intensities are represented by red, while low expression intensities are represented by green. Black indicates medium intensities. In the tree dendogram on the left side of the plot, genes with similar expression patterns are closely linked in lower levels. On the right side, annotations of corresponding genes were added in the order of Affymetrix probe ID, locus ID, gene symbol, and chromosome location.
Primers used in the qRT-PCR for verification of COBRA1/BRCA1-regulated genes.
| Genbank Accession No. | Gene name | Symbol | Primer Forward (5'_3') Primer Reverse (5'_3') |
|---|---|---|---|
| NM_005980 | S100 calcium binding protein P | S100P | F 5'-GGAGAAGGAGCTACCAGGC R 5'-ATCCACGGCATCCTTGTCTT |
| NM_003254 | Tissue inhibitor of metalloproteinase1 | TIMP1 | F 5'-ATT CCG ACC TCG TCA TCA GG R 5'-CTG GTT GAC TTC TGG TGT CCC |
| NM_006301 | Mitogen-activated protein kinase kinase kinase 12 | MAP3K12 | F 5'-TTG GCA AAG CCT ACT CCA CTG R 5'-GGA CCT CCC AAA GGT CTT CC |
| NM_021903 | Gamma-aminobutyric acid (GABA) B receptor, 1 | GABBR1 | F 5'-CCT CAG AAG GTT GCC AGA TCA R 5'-TGA TAG CCT TCA CCT GGT CCC |
| NM_001547 | Apolipoprotein D | APOD | F 5'-CCA GGA GTT GAG AGC TGA TGG R 5'-GTG GCT TCA CCT TCG ATT TGA |
| NM_001547 | Apolipoprotein D | APOD | F 5'-CCA GGA GTT GAG AGC TGA TGG R 5'-GTG GCT TCA CCT TCG ATT TGA |
| NM_006846 | Serine protease inhibitor, Kazal type, 5 | SPINK5 | F 5'-GCC ACA GTG TCA GTG CTT CTG R 5'-GGC AGC ATC TTG TAT GAG GCA |
| D13889 | Inhibitor of DNA binding 1 | ID1 | F 5'- GAC GAG CAG CAG GTA AAC GTG R 5'- GGC GTG AGT AAC AGC CGT TC |
| NM_005380 | Neuroblastoma, suppression of tumorigenicity 1 | NBL1 | F 5'-TCC CTG GTT CAC TGT GAC TCC R 5'-AAT CTC CCA CAT GGA CTG GG |
3. Results and Discussion
Given the known function of COBRA1 in transcriptional regulation, it was reasonable to speculate that reduced COBRA1 expression might lead to aberrant gene expression, which in turn could promote tumor recurrence and metastasis. Decreased COBRA1 expression may also compromise the potential functional interaction between COBRA1 and BRCA1. To test these intriguing possibilities, we stably knocked down COBRA1 and BRCA1 either separately or together in the ER-positive T47D breast cancer cell line. As a negative control, we also expressed small hairpin RNA (shRNA) specifically targeting at enhanced green fluorescent protein (EGFP). Immunoblot analysis was carried out to examine the expression levels of BRCA1 (Figure 1A, top panel) and COBRA1 (middle panel) in these cell pools. When compared with the parental T47D cells (lane 5) and the EGFP-shRNA control cells (lane 1), both COBRA1 and BRCA1 shRNA sequences significantly reduced the expression levels of the corresponding proteins in the single (lane 2 and 3) and double knockdown cells (lane 4). Importantly, the BRCA1 level was not affected by the expression of COBRA1-specific shRNA, and vice versa. Thus, expression of BRCA1 and COBRA1 is not dependent of each other.
Total RNA samples from biologically duplicated cell lines of control (E), BRCA1 knockdown (-B), COBRA1 knockdown (-C), and BRCA1-COBRA1 double knockdown (-B/C) were isolated and subjected to microarray analyses using Affymetrix HG-U133A GeneChipsTM with ~23K human gene transcripts. The LPE test was applied to identify the differentially expressed genes between the control and each of COBRA1 and/or BRCA1 knockdown cells. The program revealed 287 genes that were significantly affected by either BRCA1/COBRA1 alone or double knockdown, respectively (p<0.003; Supplemental Material).
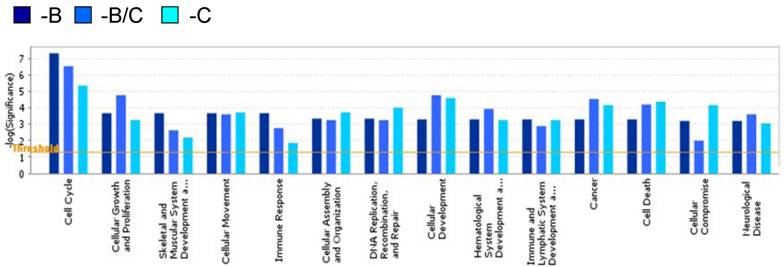
A hierarchical clustering analysis of the 287 genes was carried out to further interrogate the relationship between BRCA1- and COBRA1-mediated gene regulation (Fig. 1B). A careful examination of the heat map revealed some interesting expression patterns among the 287 genes. In particular, depletion of BRCA1 or COBRA1 alone affected a large number of commonly regulated genes (a total of 151 genes), strongly suggesting a significant degree of functional overlap between the two proteins. Gene Ontology Analysis indicates that the BRCA1 and COBRA1-regulated genes are enriched with those involved in cell cycle control, cell proliferation and development, cell death, and cancer (Fig. 2).
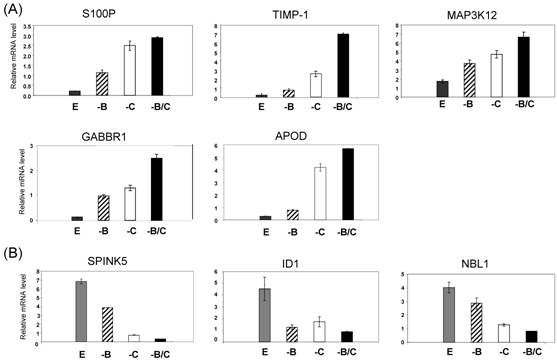
Following the microarray study, we performed quantitative RT-PCR (qRT-PCR) analysis to validate the effects of BRCA1 and COBRA1 on gene expression. The genes that were chosen for further verification include those that have been previously implicated in breast cancer, such as S100P [15] and TIMP-1 [16]. We first confirmed the changes in gene expression by using the same RNA prepared for the microarray analysis. As shown in Fig. 3, the expression level of all eight genes was altered by BRCA1/COBRA1 knockdown in a manner that was consistent with the microarray data (Supplemental Material). For example, expression of S100P was largely affected by COBRA1 or BRCA1 knockdown alone, respectively. Depletion of BRCA1 on top of COBRA1 did not lead to additional changes in mRNA abundance (compare “-C” with “-B/C” for S100P). On the other hand, the mRNA level of TIMP1 increased most substantially by the simultaneous knockdown of COBRA1 and BRCA1 (e.g. compare column “–B” and “–C” with “–B/C” for TIMP1), suggesting the involvement of both proteins in gene regulation at these loci. We were also able to verify the effect of COBRA1/BRCA1 knockdown on several down-regulated genes including SPINK5, ID1, and NBL1 (Fig. 3B).
Among the BRCA1/COBRA1-coregulated genes, TIMP1 has been reported to be over-expressed in breast cancer [16,17] and especially in patients with advanced breast cancer [18], thus serving as a potential predictor for poor prognosis [19]. A recent study has also correlated TIMP1 expression with breast cancer that is metastasized to liver [20]. To confirm the TIMP1 results obtained from the stable knockdown cells, we used two different BRCA1-specific siRNA oligos (DO4 and DO7) to transiently knock down BRCA1 in COBRA1-depleted T47D cells. As shown in Fig. 4A and 4B, BRCA1 knockdown by either oligo significantly increased TIMP1 expression. This result alleviates the concern over the possible secondary genetic changes that might have occurred in the stable BRCA1 knockdown cells due to the impaired DNA checkpoint function. It has been reported that TIMP1 expression is estrogen-stimulated [21]. Consistent with the published finding, we observed a modest (2-3 fold) stimulation of TIMP1 by estrogen treatment in both control and BRCA1/COBRA1-knockdown cells. Intriguingly, both estrogen-dependent and independent transcription of the TIMP1 gene was markedly elevated by the COBRA1/BRCA1 knockdown (Fig. 4C).
Gene ontology of the COBRA1/BRCA1-regulated genes. The Ingenuity Gene Ontology Analysis tool is used to identify several biological pathways that are relatively enriched with COBRA1/BRCA1-regulated genes.
Verification of the microarray results by real-time RT-PCR. Four different cell lines are compared: shEGFP (E; dark gray bars), shBRCA1 (-B; black stripe bars), shCOBRA1 (-C; white bars), and shBRCA1+shCOBRA1 (-B/C; black bars). The selected genes for verification include those that are up- (A) and down-regulated (B) by the depletion of BRCA1 and COBRA1.
Further verification of the effect of BRCA1 knockdown on TIMP1 transcription. (A-B). Transient knockdown of BRCA1 using BRCA1-specific siRNA oligo (DO4 in A and DO7 in B). Mock transfection and a luciferase-specific siRNA oligo were included as controls. siRNA oligos were transfected into EGFP(E) or COBRA1(-C) stable knockdown T47D cells. The immunoblots in Fig. 4A show the extent of BRCA1 depletion in the transient transfection experiment. (B). Control or BRCA1/COBRA1 knockdown cells were starved in medium containing charcoal-stripped serum. Vehicle or estradiol (E2; 10nM final concentration) was added for 24 hr, and RNA was harvested for qRT-PCR analysis of TIMP1 mRNA abundance.

The effect of BRCA1 knockdown on TIMP1 expression is consistent with the previously published work that indicates elevated TIMP1 expression in BRCA1 mutation-associated ovarian tumors and “BRCA1-like” sporadic tumors [22,23]. The same group also showed that ectopic expression of BRCA1 in a BRCA1-deficient breast cancer cell line resulted in reduced TIMP-1 expression. In an independent study, ribozyme-mediated down-regulation of BRCA1 in a melanoma cell line also resulted in increased TIMP1 expression [24]. Taken together, TIMP1 may serve as one of the molecular links between BRCA1-mediated transcriptional regulation and tumor metastasis.
Many of the COBRA1-regulated genes identified in our previous and current work have been well documented for their roles in advanced breast cancer. For example, we recently demonstrated that COBRA1 is recruited to, and regulates the activity of, the promoters of the trefoil factor (TFF) gene family (TFF1-3) [6]. TFF3 was also identified as a COBRA1-regulated gene in the current study (Supplemental Material). It is worth noting that both TFF1 and TFF3 are associated with breast cancer metastasis to bone [25] and advanced prostate cancer [26]. Similar association with breast cancer metastasis has also been reported for a number of COBRA1-regulated genes identified in the current microarray study, including S100P [27], MUC1 [28], and TIMP1. Combined with the finding that reduced COBRA1 expression is associated with metastatic and recurrent breast cancer [29], the effect of COBRA1 knockdown on gene expression lends further support to the notion that COBRA1 plays an important role in suppressing breast cancer development.
One of the most intriguing findings of the current work is the significant overlap of gene expression profiles between the BRCA1 and COBRA1-depleted cells. The data in the current study strongly suggest a joint action of COBRA1 and BRCA1 in modulating transcription of a group of common target genes. Both COBRA1 and BRCA1 are known to be associated with the RNA polymerase II (RNAPII) holoenzyme [2,10,13,30,31] as well as other factors involved transcriptional elongation and RNA processing [3,4,12,32,33]. In light of the physical association and functional commonality between COBRA1 and BRCA1, we propose that these two proteins may intimately communicate with each other in modulating the movement of the transcription elongation complex for a common set of target genes.
Given the large overlap of the gene expression profiles in the COBRA1 and BRCA1-depleted cells, it is interesting to note that BRCA1-associated tumors are usually diagnosed as high-grade infiltrating ductal carcinoma, and that patients with BRCA1-associated breast tumors tend to have a poor prognosis [34]. Moreover, a recent report suggests a high incidence of brain metastasis in BRCA1-associated cancer cases [35]. These clinical features of BRCA1-associated tumors are reminiscent of the association of low COBRA1 expression with metastatic breast cancer [29]. It is conceivable that loss of COBRA1 and/or BRCA1 may result in aberrant expression of a common group of cancer metastasis-associated genes, which could account for the clinical outcomes in both cases.
A significant proportion of the genes in the current microaray study are down-regulated in the COBRA1 and BRCA1 knockdown cells. This is somewhat unexpected as COBRA1 is best known for its role in transcriptional repression. It is possible that expression of this group of genes is indirectly affected by COBRA1 knockdown. Alternatively, COBRA1 might act as a bi-functional protein to both repress and activate transcription. While more mechanistic studies may shed light on this issue, it is worth pointing out that COBRA1 is capable of inducing large-scale chromatin unfolding when targeted to specific regions of the mammalian genome [1]. Conceivably, the activity of COBRA1 in chromatin reorganization could account for a more direct role of COBRA1 in gene activation.
In conclusion, combination of shRNA-mediated knockdown and gene expression profiling points to a large degree of functional commonality between COBRA1 and BRCA1 in transcriptional regulation. This finding provides functional relevance to the previously identified physical association between the two proteins. Furthermore, altered expression of multiple breast cancer-associated genes in the COBRA1/BRCA1 knockdown cells offers molecular insight into the roles of COBRA1 and BRCA1 in progression of breast cancer.
Supplementary Material
Primers used in the qRT-PCR for verification of COBRA1/BRCA1-regulated genes.
Acknowledgements
The work was supported by National Institutes of Health grant (DK064604 and CA93506), and DOD Breast Cancer Research Program Award W81XWH-04-1-0519 (S.E.A).
Conflict of interest
The authors have declared that no conflict of interest exists.
References
1. Ye Q, Hu Y-F, Zhong H, Nye AC, Belmont AS, Li R. BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J. Cell Biol. 2001;155:911-921
2. Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim D-K, Hasegawa J, Omori M, Inukai N, Endoh M, Yamada T, Handa H. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 2003;23:1863-1873
3. Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose H, Sugimoto S, Hartzog GA, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymeerase II processitivity, is composed of human Spt4 and Spt5 homologs. Genes & Dev. 1998;12:343-356
4. Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41-51
5. Aiyar SE, Sun J-L, Blair AL, Moskaluk CA, Lv Y, Ye Q-N, Yamaguchi Y, Mukherjee A, Ren D-M, Handa H, Li R. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes & Dev. 2004;18:2134-2146
6. Aiyar SE, Blair AL, Hopkinson DA, Bekiranov S, Li R. Regulation of clustered gene expression by cofactor of BRCA1 (COBRA1) in breast cancer cells. Oncogene. 2007;26:2543-2553
7. Zhu J, Song S, Jiang Z, Yan J, Lu Q, Huang C, Ye Q. Characterization of COBRA1 in human breast cancer cell lines using a new polyclonal antibody against COBRA1. IUBMB Life. 2004;56:161-166
8. Fan S, Wang J, Yuan R, Ma Y, Meng Q, Erdos MR, Pestell RG, Yuan F, Auborn KJ, Goldberg ID, Rosen EM. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284(5418):1354-6
9. Zheng L, Annab LA, Afshari CA, Lee W-H, Boyer TG. BRCA1 mediates ligand-independent transcriptional repression of the estrogen receptor. Proc. Natl. Acad. Sci. USA. 2001;98:9587-9592
10. Neish AS, Anderson SF, Schlegel BP, Wei W, Parvin JD. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26(3):847-53
11. Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 2002;22:2918-2927
12. Cabart P, Chew HK, Murphy S. BRCA1 cooperates with NUFIP and P-TEFb to activate transcription by RNA polymerase II. Oncogene. 2004;23:5316-5329
13. Krum SA, Miranda GA, Lin C, Lane TF. BRCA1 associates with processive RNA polymerase II. J. Biol. Chem. 2003;278:52012-52020
14. Jain N, Thatte J, Braciale T, Ley K, O'Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003;19:1945-1951
15. Mackay A, Jones C, Dexter T, Silva RL, Bulmer K, Jones A, Simpson P, Harris RA, Jat PS, Neville AM, Reis LF, Lakhani SR, O'Hare MJ. cDNA microarray analysis of genes associated with ERBB2 (HER2/neu) overexpression in human mammary luminal epithelial cells. Oncogene. 2003;22:2680-2688
16. Nakopoulou L, Giannopoulou I, Lazaris AC, Alexandrou P, Tsirmpa I, Markaki S, Panayotopoulou E, Keramopoulos A. The favorable prognostic impact of tissue inhibitor of matrix metalloproteinases-1 protein overexpression in breast cancer cells. APMIS. 2003;111:1027-1036
17. Clavel C, Polette M, Doco M, Binninge I, Birembaut P. Immunolocalization of matrix metallo-proteinases and their tissue inhibitor in human mammary pathology. Bull Cancer. 1992;79:261-270
18. Holten-Andersen MN, Murphy G, Nielsen HJ, Pedersen AN, Christensen IJ, Hoyer-Hansen G, Brunner N, Stephens RW. Quantitation of TIMP-1 in plasma of healthy blood donors and patients with advanced cancer. Br J Cancer. 1999;80:495-503
19. McCarthy K, Maguire T, McGreal G, McDermott E, O'Higgins N, Duffy MJ. High levels of tissue inhibitor of metalloproteinase-1 predict poor outcome in patients with breast cancer. Int J Cancer. 1999;84:44-48
20. Van den Eynden GG, Van Laere SJ, Van der Auwera I, Gilles L, Burn JL, Colpaert C, van Dam P, Van Marck EA, Dirix LY, Vermeulen PB. Differential expression of hypoxia and (lymph)angiogenesis-related genes at different metastatic sites in breast cancer. Clin Exp Metastasis. 2007 [Epub ahead of print]
21. Chen B, Wen Y, Wang H, Polan ML. Differences in estrogen modulation of tissue inhibitor of matrix metalloproteinase-1 and matrix metalloproteinase-1 expression in cultured fibroblasts from continent and incontinent women. Am J Obstet Gynecol. 2003;189:59-65
22. Jazaeri AA, Yee CJ, Sotiriou C, Brantley KR, Boyd J, Liu ET. Gene expression profiles of BRCA1-linked, BRCA2-linked, and sporadic ovarian cancers. J. Natl. Cancer Inst. 2002;94:990-1000
23. Jazaeri AA, Chandramouli GVR, Aprelikova O, Nuber UA, Sotiriou C, Liu ET, Ropers HH, Yee CJ, Boyd J, Barrett JC. BRCA1-mediated repression of select X chromosome genes. J. Translational Med. 2004;2:32-39
24. Hesling C, D'Incan M, D'Incan C, Souteyrand P, Monboisse JC, Pasco S, Madelmont JC, Bignon YJ. Downregulation of BRCA1 in A375 melanoma cell line increases radio-sensitivity and modifies metastatic and angiogenic gene expression. J Invest Dermatol. 2004;122:369-380
25. Smid M, Wang Y, Klijn JG, Sieuwerts AM, Zhang Y, Atkins D, Martens JW, Foekens JA. Genes Associated With Breast Cancer Metastatic to Bone. J Clin Oncol. 2006;24:2261-2267
26. Vestergaard EM, Borre M, Poulsen SS, Nexo E, Torring N. Plasma levels of trefoil factors are increased in patients with advanced prostate cancer. Clin Cancer Res. 2006;12:807-812
27. Wang G, Platt-Higgins A, Carroll J, de Silva Rudland S, Winstanley J, Barraclough R, Rudland PS. Induction of metastasis by S100P in a rat mammary model and its association with poor survival of breast cancer patients. Cancer Res. 2006;66:1199-1207
28. Rakha EA, Boyce RW, Abd El-Rehim D, Kurien T, Green AR, Paish EC, Robertson JF, Ellis IO. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod Pathol. 2005;18:195-304
29. Sun J, Watkins G, Blair AL, Moskaluk C, Ghosh S, Jiang WG, Li R. Deregulation of cofactor of BRCA1 expression in breast cancer cells. J Cell Biochem. 2007 [Epub ahead of print]
30. Scully R, Anderson SF, Chao DM, Wei W, Ye L, Young RA, Livingston DM, Parvin JD. BRCA1 is a component of the RNA polymerase II holoenzyme. Proc Natl Acad Sci U S A. 1997;94(11):5605-10
31. Anderson SF, Schlegel BP, Nakajima T, Wolpin ES, Parvin JD. BRCA1 protein is linked to the RNA polymerase II holoenzyme complex via RNA helicase A. Nat. Genet. 1998;19:254-256
32. Kleiman FE, Manley JL. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science. 1999;285:1329-1339
33. Kleiman FE, Manley JL. The BARD1-CstF-50 interaction links mRNA 3' end formation to DNA damage and tumor suppression. Cell. 2001;104:743-753
34. Narod SA, Brunet JS, Ghadirian P, Robson M, Heimdal K, Neuhausen SL, Stoppa-Lyonnet D, Lerman C, Pasini B, de los Rios P, Weber B, Lynch H. HBCCS. Tamoxifen and risk of contralateral breast cancer in BRCA1 and BRCA2 mutation carriers: a case-control study. Hereditary Breast Cancer Clinical Study Group. Lancet. 2000;356:1876-1881
35. Albiges L, Andre F, Balleyguier C, Gomez-Abuin G, Chompret A, Delaloge S. Spectrum of breast cancer metastasis in BRCA1 mutation carriers: highly increased incidence of brain metastases. Ann Oncol. 2005;16:1846-1847
Author contact
![]() Correspondence to: Rong Li, PhD, Professor, Department of Molecular Medicine, Institute of Biotechnology, 15355 Lambda Drive, University of Texas, Health Science Center at San Antonio, San Antonio, TX 78245. Tel: 210-567-7215; Fax: 210-567-7324; Email: lir3edu
Correspondence to: Rong Li, PhD, Professor, Department of Molecular Medicine, Institute of Biotechnology, 15355 Lambda Drive, University of Texas, Health Science Center at San Antonio, San Antonio, TX 78245. Tel: 210-567-7215; Fax: 210-567-7324; Email: lir3edu

 Global reach, higher impact
Global reach, higher impact