10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2008; 4(2):96-102. doi:10.7150/ijbs.4.96 This issue Cite
Research Paper
Comparative understanding of UTS2 and UTS2R genes for their involvement in type 2 diabetes mellitus
1. Department of Animal Sciences, Washington State University, Pullman, WA 99164–6351, USA
2. School of Molecular Biosciences, Washington State University, Pullman, WA 99164-4660, USA
3. USDA Agricultural Research Service Fort Keogh Livestock and Range Research Laboratory, Miles City, MT 59301, USA
Received 2008-3-17; Accepted 2008-4-22; Published 2008-4-23
Abstract
Several reports have shown that urotensin 2 (UTS2) and its receptor (UTS2R) are involved in glucose metabolism and insulin resistance, which lead to development of type 2 diabetes mellitus (T2DM) in humans. In the present study, we annotated both bovine UTS2 and UTS2R genes and identified 5 single nucleotide polymorphisms (SNPs) for the former gene and 14 mutations for the latter gene. Four mutations were genotyped on a Wagyu x Limousin reference population, including 6 F1 bulls, 113 F1 dams and ~250 F2 progeny. Among 12 phenotypes related to fat deposition and fatty acid composition, we observed that the UTS2 gene was significantly associated with the amount of skeletal saturated fatty acids, while its receptor (UTS2R) gene had significant effects on amounts of saturated and monounsaturated fatty acids, Δ9 desaturase activity for converting 16:0 into 16:1, muscle fat (marbling) score and Longissimus Dorsi muscle area. However, in this population, these markers were not associated with subcutaneous fat depth or percent kidney, pelvic and heart fat. We also found that mutations in the promoter regions altered the promoter activities in both genes and coding SNPs might affect the mRNA stability in the UTS2R gene. Overall, our present study provides the first evidence that both UTS2 and UTS2R genes regulate skeletal muscle fat accumulation and fatty acid metabolism, thus indicating their potential pathological functions related to obesity and T2DM in humans.
Keywords: Type 2 diabetes mellitus, UTS2, UTS2R, skeletal fat accumulation, fatty acid composition.
1. Introduction
Three members of the urotensin II family of peptides have been discovered in mammals, including urotensin 2 (UTS2), urotensin 2 receptor (UTS2R) and urotensin 2 domain containing (UTS2D). UTS2 is cyclic peptide with an 11 amino acid mature peptide in human, but it has been recognized as a hormone in the neurosecretory system of teleost fish for over 30 years [1]. In addition, UTS2 is a potent vasoconstrictive peptide that regulates both endothelium-dependent and independent vasodilation [2]. The human orphan G protein-coupled receptor 14 (GPR14) was cloned in 1995 [3] and then confirmed to function as the UTS2 receptor (UTS2R) [4]. Interestingly, the UTS2R shares highest identity with the somatostatin receptor SSTR4 [3]. A novel urotensin II-related peptide, now named the urotensin 2 domain containing (UTS2D) was also isolated in human, mouse and rat [5]. UTS2D binds and activates the urotensin 2 receptor, suggesting that it is the endogenous and functional ligand for UTS2R.
Among these three genes, both UTS2 and UTS2R have been reported to affect glucose metabolism and insulin resistance, a core pathological characteristic of patients with type 2 diabetes mellitus (T2DM). In Hong Kong Chinese, the GGT haplotype (-605G, 143G and 3836T) in the UTS2 gene is associated with higher plasma level of urotensin 2 and insulin, and higher homeostasis model assessment of insulin resistance index and beta-cell function, while the AC haplotype (-11640A and -8515C) in the UTS2R gene has a higher amount of plasma glucose 2 h after a 75 g oral glucose load [6]. In human diabetic tissue, Langham and colleagues [7] found that expression of both UTS2 and UTS2R are increased 45- and almost 2,000-fold in comparison to control nephrectomy tissue, respectively (P<0.0001) using quantitative real-time polymerase chain reaction. In the healthy rat, infusion of synthetic rat urotensin 2 inhibits both insulin release induced by glucose and insulin responses induced by carbachol, glucagon-like peptide-1, and a calcium channel agonist [8]. However, in streptozotocin-induced diabetic rats, long-term treatment with palosuran, a UTS2R antagonist, improved survival, increased insulin, and slowed the increase in glycemia, glycosylated hemoglobin, and serum lipids [9]. Therefore, the urotensin 2 system plays a unique role both in insulin secretion and in the renal complications of diabetes.
Studies have shown that the fat droplets accumulated in human skeletal muscle are a major contributor to insulin resistance [10]. For example, in male Pima Indians, negative relationships were found between amounts of triglyceride in skeletal muscle and physiological and supraphysiological insulin levels, and nonoxidative glucose disposal (r = -0.44 – -0.53, P < 0.01) [11]. In a European population, muscle lipid was correlated with percent body fat (r = 0.50, p = 0.028), waist:hip ratio (r = 0.74, p < 0.001), visceral fat (r = 0.62, p = 0.004) and insulin sensitivity (r = -0.53, p = 0.016) [12]. Just recently, Suzuki and colleagues [13] reported that enhanced muscle mass might have positive effects on fat utilization, thus reducing the metabolic problems and insulin resistance associated with obesity and T2DM. In addition, both UTS2 and UTS2R mRNA are expressed in skeletal muscle as well as in lung, pancreas, kidney and liver [14]. Therefore, the objective of the present study was to determine whether both UTS2 and UTS2R genes contribute to muscle lipid metabolism using cattle as a model organism.
2. Materials and Methods
Animals, body fat deposition and fatty acid composition
A Wagyu–Limousin F2 reference population was used in the present study, including 6 F1 bulls, 113 F1 dams and ~250 F2 progeny. The entire core of the longissimus dorsi was sampled from these F2 progeny and relative amounts of saturated (SFA), monounsaturated (MUFA) and polyunsaturated fatty acids (PUFA), three indexes of Δ9 desaturase activity as R1 = (14:1/14:0) x 100%; R2 = (16:1/16:0) x 100% and R3 = (18:1/18:0) x 100%, conjugated linoleic acid mg/100 g dry meat (CLA), cholesterol mg/100 g dry meat (CHOL), Longissimus Dorsi area (LDA in in2) and muscle fat (marbling) score (MFMS) were measured. In addition, subcutaneous fat depth (SFD) and percent kidney-pelvic-heart fat (KPH) were also recorded in the population. Muscle fat (marbling) score was determined at the interface of the 12th and 13th ribs and was evaluated by subjective comparison of the amount of fat within the longissimus muscle with photographic standards (National Livestock and Meat Board 1981). Subcutaneous fat depth was recorded at the 12th rib at a point three-fourths the width of the longissimus muscle from its chine bone end. The amount of KPH was estimated and recorded as a percentage of carcass weight. Development and management of the Wagyu–Limousin reference population and measurement and definition of these phenotypes were described previously [15,16].
Gene, mutation, genotyping and association
Both cDNA and genomic DNA sequences of the bovine UTS2 and UTS2R were retrieved from the GenBank databases using a comparative approach, as previously described [17]. Alignment between cDNA and genomic DNA sequences was used to determine genomic organization for each of these two genes. A total of 11 pairs of primers (Table 1) were designed based on the genomic DNA sequences and used to screen genetic polymorphisms in the promoter, coding and untranslated regions of both bovine UTS2 and UTS2R genes. Approximately 50 ng of genomic DNA each from six Wagyu x Limousin F1 bulls was amplified in a final volume of 10 μl that contained 12.5 ng of each primer, 150 μM dNTPs, 1.5 mM MgCl2, 50 mM KCl, 20 mM Tris-HCl and 0.25U of Platinum Taq polymerase (Invitrogen, Carlsbad, CA). The following PCR conditions were used: 94oC for 2 min, 35 cycles of 94oC for 30 sec, 60oC for 30 sec and 72oC for 30 sec, followed by a final 5 min extension at 72oC. PCR products were then sequenced on ABI 3730 sequencer in the Laboratory for Biotechnology and Bioanalysis (Washington State University) using a standard protocol and polymorphisms were detected.
A total of 5 mutations were identified in UTS2 and 14 in UTS2R gene, respectively. Two SNPs (single nucleotide polymorphisms), one in the promoter and one in intron 2 of UTS2, and two mutations, one INDEL (insertion/deletion) in the promoter and one coding SNP of UTS2R, were selected for genotyping on a Sequenom iPLEX assay using services provided by the Children's Hospital Oakland Research Institute, Oakland, California. The phenotypic data for all measurements described above were analyzed in a fixed effects model that included the effects of year, gender, age at harvest (linear) and the genotype for each marker using the GLM (general linear model) procedure of SAS v9.1 (SAS Institute Inc., Gary, NC). Pair-wise comparisons of least squares means were performed using a protected t-test.
Promoter activity assay
Among the genetic polymorphisms discovered above, three SNPs were located in the promoter region of UTS2 and one insertion/deletion in the promoter of UTS2R gene, respectively. The effects of these mutations on promoter activities were then examined using a Dual-Luciferase Report Assay System (Promega, Madison, WI). The forward and reverse gene-specific primers that were used to amplify the promoter regions (Table 1) were then engineered with a 5' BglII and 3' HindIII site plus a 5' tail of CTTC, respectively, for directional cloning into the BglII/HindIII site of pGL3-basic (Premega, Madison, WI). Two types of haplotypes at three polymorphic sites in the UTS2 promoter: A-C-A and C-A-G; and two types of INDELs in the UTS2R promoter: TA insertion and TA deletion were prepared for the promoter constructs. The human lung carcinoma H1299 cells were transfected with each of the recombinant pGL3 plasmids containing the constructs described above. pRL-CMV plasmid was also co-transfected into H1299 cells as a transfection control. The human cells were collected 28 hours post-transfection and firefly luciferase and Renilla luciferase activities were measured with the Dual Luciferase Reporter Assay system according to the manufacturer's protocol. Light emission was quantified with a Multilabel Counter (Wallace 1420 Victor 2, Turku, Finland). Triplicate data were collected from three independent experiments. The ratios of firefly luciferase activity to Renilla luciferase activity were calculated and used to compare the differences in activity between two constructs for each gene.
Primers designed for mutation detection in the bovine UTS2 and UTS2R genes.
3. Results and Discussion
Comparative annotation of UTS2 and UTS2R genes
The cDNA sequences of human UTS2 (NM_021995.1, transcript variant 1) and UTS2R (NM_018949.1) genes were used as references for BLAST searches against GenBank “nr” database, thus retrieving orthologous cDNA sequences for both bovine genes (CO872879 for UTS2 and NM_001040484.1 for UTS2R). BLAST searches using the bovine cDNA sequences as queries then obtained their genomic DNA sequences (AAFC03010889 for UTS2 and AAFC03013715 for UTS2R) from the bovine genome sequencing database (Build 3.1).
For the bovine UTS2 gene, alignment between the cDNA and the genomic DNA sequences revealed that it consists of four exons only, which correspond to the transcript variant 2 of the human ortholog (NM_006786) (Figure 1A). However, alignment between the human transcript variant 1 (NM_021995) and the same bovine genomic DNA contig detected a relic for exon 1 of the variant, but it seems that its expression in cattle is totally destroyed by an unusual intron splicing site of CT instead of GT (see AAFC03010889.1:g.9800C). In fact, GenBank databases show that the UTS2 variant 1 does not exist in either mouse (NM_011910) or rat (NM_019160).
Genomic organizations of UTS2 (A) and UTS2R (B) between human and cattle.
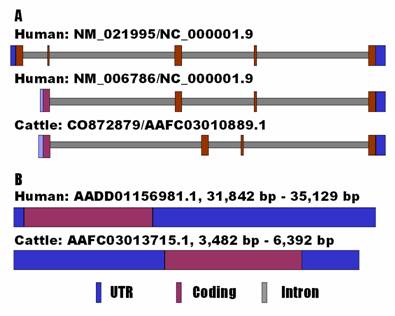
In mammals, the UTS2R is an intronless gene. However, the current GenBank information shows that the bovine UTS2R gene contains a 5'UTR of 89 bp and a 3'UTR of 384 bp (NM_001040484.1), while the human ortholog has none of them (NM_018949.1). Therefore, we decided to re-annotate the gene in both human and cow using an electronic rapid amplification of cDNA ends (e-RACE) approach. Basically, we used the genomic DNA sequences flanking the coding sequences as queries for BLAST searches to determine both 5'UTR and 3'UTR ends of the genes against the EST (expressed sequence tag) databases. The e-RACE approach retrieved an EST BF513269 for the 5'UTR end and AA621666 for the 3'UTR end of the human UTS2R gene. The same approach also obtained an EST EE388038 for the 5'UTR end and CK775836 for the 3'UTR end of the bovine ortholog. As a result, we complied a tentative full-length cDNA sequence of 3,288 bp for the human gene (see AADD01156981.1 from position 31,842 bp to 35,129 bp) and 2,911 bp for the bovine gene (see AAFC02086367.1 from position 3,482 bp to 6,392 bp). The former cDNA sequence has a 5'UTR of 95 bp, a coding sequence of 1170 bp and a 3'UTR of 2,023 bp, while the latter cDNA sequence contains a 5'UTR of 1,270 bp, a coding sequence of 1,155 bp and a 3'UTR of 486 bp (Figure 1B). In addition, a polyadenylation site (AAUAAA) was also identified in the newly annotated cDNA sequences of both human and bovine UTS2R genes.
Genetic polymorphisms
Direct sequencing of PCR products on 6 F1 bulls identified three SNPs in the promoter and two SNPs in intron 2 of the UTS2 gene: AAFC03010889.1:g.9408A>C, g.9552C>A, g.9628G>A, g.13294G>A and g.13900A>C, respectively. Interestingly, we observed in these 6 F1 bulls that three promoter SNPs and two intronic SNPs have no historical recombination by forming two haplotypes (ACA and CAG) in the former region and two haplotypes (GA and AC) in the latter region. Therefore, only one SNP from the promoter and one SNP from the intron 2 were chosen for genotyping.
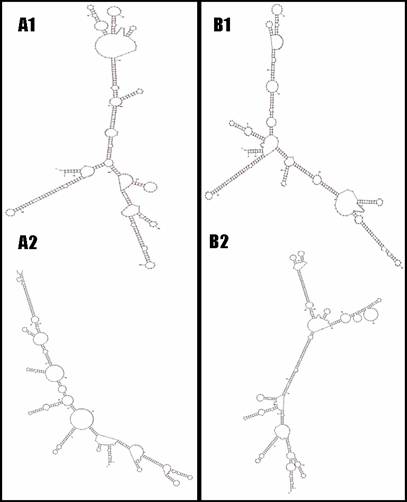
For the bovine UTS2R, one insertion/deletion (INDEL) with two nucleotides of TA (AAFC03013715.1:g.2935-36TA>--) was detected in the promoter region, while 13 SNPs were identified in the coding and 3'UTR regions of the gene, including AAFC03013715.1:c.6446T>C, c.6506C>T, c.6593T>C, c.6749G>A, c.6830T>C, c.6842A>G, c.7232G>A, c.7359C>T, g.7466G>C, g.7632A>G, g.7692C>T, g.7714G>A and g.7720G>A, respectively. Although the first eight SNPs in the bovine UTS2R gene are coding SNPs, none are missense mutations. However, the Mfold web server [18] predicted that these 13 SNPs do cause mRNA secondary structure changes in the bovine gene (Figure 2). In addition to the promoter INDEL, only one of these 13 SNPs was chosen for genotyping, because they form just two haplotypes as described in Figure 2.
Associations of UTS2 and UTS2R with body fat deposition
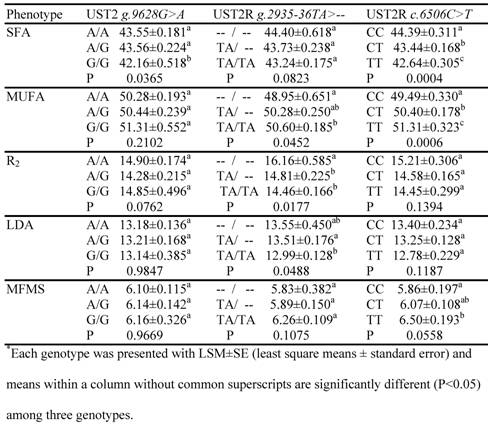
The association analysis involved four mutations, i.e., AAFC03010889.1: g.9628G>A and g.13900A>C in the bovine UTS2 gene and AAFC03013715.1:g.2935-36TA>-- and c.6506C>T in the bovine UTS2R gene, which were genotyped on a Sequenom iPLEX assay. As two SNPs in the former genes had no combination in these F2 progeny by forming two haplotypes AA and CG, only three markers were used in the data analysis. The phenotypes that had at least one suggestive (P=0.05 – 0.06) or significant (P<0.05) association are listed in Table 2. The UTS2 gene had a significant effect on one trait only, i.e., the relative amount of saturated fatty acids. In particular, animals with GG genotypes had 1.39 units (P=0.0365) less SFA than animals with AA genotypes. For the bovine UTS2R gene, the promoter INDEL polymorphism was significantly associated with MUFA (P=0.0452), R2 (P=0.0177) and LDA (P=0.0488) (Table 2). The homozygous animals with the insertion allele (TA/TA) had 1.65 units higher MUFA, but 1.70% lower R2 activity and 0.56 in2 smaller LD muscle area than the homozygous animals with TA deleted (Table 2). The coding SNP in the bovine UTS2R gene had significant impacts on both SFA and MUFA, plus a suggestive effect on MFMS (Table 2). The TT animals had an additional 0.64 units of muscle fat deposition and 1.82 units of MUFA, but 1.75 less units of SFA than the CC animals.
The UTS2R mRNA local secondary structures between haplotype TCTGTAGCGACGG and haplotype CTCACGATCGTAA based on two-segment analysis (A1 vs. B1 and A2 vs. B2).
Associations of UTS2 and UTS2R genes with muscle fat phenotypes*
Mutations and promoter activities
As indicated above, two types of haplotypes at three polymorphic sites in the UTS2 promoter: A-C-A and C-A-G; and two types of INDELs in the UTS2R promoter: TA insertion and TA deletion, were prepared for the promoter constructs to investigate how these mutations affect promoter activities of these two genes. In the UTS2 promoter, the C-A-G construct produced less promoter activities than the A-C-A construct by 27% in the H1299 cells (P=0.0234) (Figure 3). In the UTS2R promoter, the deletion allele yielded lower promoter activities than the insertion constructs by 16% in the same cell lines (P=0.0436) (Figure 3). A search for transcription binding sites using MatInspector [19] indicated AAFC03010889.1:g.9408A in the UTS2 promoter gained a site for FAST-1 SMAD interacting proteins, while AAFC03010889.1:g.9408C gained a site for neuron-specific-olfactory factor. At the g.9552C>A polymorphic site, the former allele had a transcription binding site for AP4 and related proteins only, while the latter allele possessed one site for MAF/AP1 related factors and one site for octamer binding protein. There were a total of four binding sites for MAF and AP1 related factors, TCF11 transcription factor, TALE homeodomain class recognizing TG motifs and MYT1 C2HC zinc finger protein when an A allele is located at position AAFC03010889.1:g.9628. When the g.9628A is substituted by g.9628G, only one site was created for activator/repressor binding to transcription initiation site. Overall, the A-C-A haplotype in the UTS2 promoter had two additional binding sites than the C-A-G haplotype. However, the MatInspector program failed to detect any gain/loss of transcription binding sites for the insertion or deletion allele in the promoter of UTS2R gene although they showed a difference in promoter activities as described above.
Affects of mutations on promoter activity in the H1299 cells.
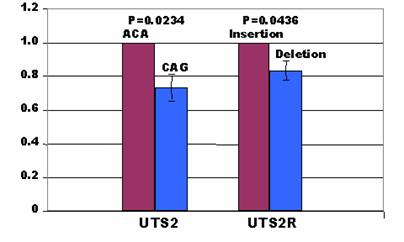
In summary, our present study provided evidence that both UTS2 and UTS2R genes are involved in skeletal muscle fat deposition and fatty acid metabolism. Such evidence is supported by the fact that none of the mutations genotyped in the population were associated with either subcutaneous fat depth (P=0.2074 – 0.9847) or the fat percentage surrounding the kidney, pelvic and heart fat (P=0.3278 – 0.7059) (data not shown). Instead, the UTS2 gene was significantly associated with relative amount of saturated fatty acids, while its receptor had significant effects on relative amounts of saturated and monounsaturated fatty acids, Δ9 desaturase activity for converting 16:0 into 16:1, overall fat deposition in muscle and LD muscle area (Table 2). Although mutations in the promoter regions altered the promoter activities in both genes (Figure 3) and coding SNPs might affect the mRNA stability in UTS2R gene (Figure 2), how these genes regulate fat deposition and fatty acid composition in skeletal muscle remains unknown.
Acknowledgements
This work was supported by NIH grant RO1 CA104470 to N.S.M. and Merial Ltd. Animal Genomics Research Fund to Z.J. This activity was also funded, in part, with an Emerging Research Issues Internal Competitive Grant from the Washington State University, College of Agricultural, Human, and Natural Resource Sciences, Agricultural Research Center to Z.J.
Conflict of interests
The authors have declared that no conflict of interest exists.
References
1. Bern HA, Pearson D, Larson BA. et al. Neurohormones from fish tails: the caudal neurosecretory system. I. "Urophysiology" and the caudal neurosecretory system of fishes. Recent Prog Horm Res. 1985;41:533-52
2. Ong KL, Wong LY, Cheung BM. The role of urotensin II in the metabolic syndrome. Peptides. 2007 [Epub ahead of print]
3. Marchese A, Heiber M, Nguyen T. et al. Cloning and chromosomal mapping of three novel genes, GPR9, GPR10, and GPR14, encoding receptors related to interleukin 8, neuropeptide Y, and somatostatin receptors. Genomics. 1995;29(2):335-44
4. Ames RS, Sarau HM, Chambers JK. et al. Human urotensin-II is a potent vasoconstrictor and agonist for the orphan receptor GPR14. Nature. 1999;401(6750):282-6
5. Sugo T, Murakami Y, Shimomura Y. et al. Identification of urotensin II-related peptide as the urotensin II-immunoreactive molecule in the rat brain. Biochem Biophys Res Commun. 2003;310(3):860-8
6. Ong KL, Wong LY, Man YB. et al. Haplotypes in the urotensin II gene and urotensin II receptor gene are associated with insulin resistance and impaired glucose tolerance. Peptides. 2006;27(7):1659-67
7. Langham RG, Kelly DJ, Gow RM. et al. Increased expression of urotensin II and urotensin II receptor in human diabetic nephropathy. Am J Kidney Dis. 2004;44(5):826-31
8. Silvestre RA, Egido EM, Hernandez R. et al. Urotensin-II is present in pancreatic extracts and inhibits insulin release in the perfused rat pancreas. Eur J Endocrinol. 2004;151(6):803-9
9. Clozel M, Hess P, Qiu C. et al. The urotensin-II receptor antagonist palosuran improves pancreatic and renal function in diabetic rats. J Pharmacol Exp Ther. 2006;316(3):1115-21
10. Goodpaster BH, Wolf D. Skeletal muscle lipid accumulation in obesity, insulin resistance, and type 2 diabetes. Pediatr Diabetes. 2004;5(4):219-26
11. Pan DA, Lillioja S, Kriketos AD. et al. Skeletal muscle triglyceride levels are inversely related to insulin action. Diabetes. 1997;46:983-8
12. Forouhi NG, Jenkinson G, Thomas EL. et al. Relation of triglyceride stores in skeletal muscle cells to central obesity and insulin sensitivity in European and South Asian men. Diabetologia. 1999;42(8):932-5
13. Suzuki ST, Zhao B, Yang J. Enhanced muscle by myostatin propeptide increases adipose tissue adiponectin, PPAR-alpha, and PPAR-gamma expressions. Biochem Biophys Res Commun. 2008;369(2):767-73
14. Elshourbagy NA, Douglas SA, Shabon U. et al. Molecular and pharmacological characterization of genes encoding urotensin-II peptides and their cognate G-protein-coupled receptors from the mouse and monkey. Br J Pharmacol. 2002;136(1):9-22
15. Alexander LJ, Macneil MD, Geary TW. et al. Quantitative trait loci with additive effects on palatability and fatty acid composition of meat in a Wagyu-Limousin F2 population. Anim Genet. 2007;38(5):506-13
16. Jiang Z, Kunej T, Michal JJ. et al. Significant associations of the mitochondrial transcription factor A promoter polymorphisms with marbling and subcutaneous fat depth in Wagyu x Limousin F2 crosses. Biochem Biophys Res Commun. 2005;334(2):516-23
17. Jiang Z, Michal JJ, Williams GA. et al. Cross species association examination of UCN3 and CRHR2 as potential pharmacological targets for antiobesity drugs. PLoS ONE. 2006;1:e80
18. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:406-15
19. Quandt K, Frech K, Karas H. et al. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878-48
Author contact
![]() Correspondence to: Zhihua Jiang, Department of Animal Sciences, Washington State University, Pullman, WA 99164–6351, USA. Tel: +509 335 8761; Fax: +509 335 4246; E-mail: jiangzedu
Correspondence to: Zhihua Jiang, Department of Animal Sciences, Washington State University, Pullman, WA 99164–6351, USA. Tel: +509 335 8761; Fax: +509 335 4246; E-mail: jiangzedu

 Global reach, higher impact
Global reach, higher impact