10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2008; 4(6):415-421. doi:10.7150/ijbs.4.415 This issue Cite
Research Paper
cDNA, genomic sequence and overexpression of crystallin alpha-B Gene (CRYAB) of the Giant Panda
1. College of Agriculture, Sichuan Agricultural University, 46# Xinkang Road, 625014, Ya-an, China.
2. College of Life Science, China West Normal University, 44# Yuying Road, 637002, Nanchong, China.
Received 2008-8-31; Accepted 2008-11-11; Published 2008-11-14
Abstract
αB-crystallin, a small heat-shock protein, has been shown to prevent the aggregation of other proteins under various stress conditions. Here we have cloned the cDNA and the genomic sequence of CRYAB gene from the Giant Panda (Ailuropoda melanoleuca) using RT-PCR technology and Touchdown-PCR, respectively. The length of cDNA fragment cloned contains an open reading frame of 528bp encoding 175 amino acids and the length of the genomic sequence is 3189bp, containing three exons and two introns. Alignment analysis indicated that the nucleotide sequence and the deduced amino acid sequence are highly conserved to other four species studied, including Homo sapiens, Mus musculus, Rattus norvegicus and Bos taurus. The homologies for nucleotide sequences of Giant Panda CRYAB to that of these species are 93.9%, 91.5%, 91.5% and 95.3%, respectively, and the homologies for amino acid sequences are 98.3%, 97.1%,97.7% and 99.4%, respectively. Topology prediction shows that there are only four Casein kinase II phosphorylation sites in the CRYAB protein of the Giant Panda. The cDNA of CRYAB was transfected into E. coli, and the CRYAB fused with the N-terminally His-tagged protein gave rise to the accumulation of an expected 24KDa polypeptide, which accorded with the predicted protein. The expression product obtained could be used for purification and study of its function further.
Keywords: cDNA cloning, CRYAB, the Giant Panda (Ailuropoda melanoleuca), genomic cloning, overexpression.
1. Introduction
The lens proteins consist of about 95% crystallins. The lens continues to grow throughout life, forming new fiber cells at the periphery of lens without ever replacing the older cells. As a consequence, the cells in the nucleus of the lens contain proteins that are as old as the animal itself. To warrant transparency, the light-scattering organelles, including the nuclei, are broken down in the later stages of differentiation [1-4]. The transparency and high refractive index of the normal lens is achieved by a regular arrangement of the lens fiber cells during growth of the lenticular body and by the high concentration and the supramolecular organization of the lens-specific proteins, the crystallins, within each fiber cell [5,6]. In the mammalian lens, 3 major classes of crystallins are distinguished: alpha, beta, and gamma. The largest, alpha-crystallins, which composed of 2 primary gene products--alpha-A and alpha-B, are some of the most abundant soluble proteins in the lens and, along with the other lens crystallins, play an important role in establishing and maintaining the optical properties of the lens [7]. αB-crystallin, an abundant eye lens protein, is also present in other tissues and is heat- and stress-inducible, where as αA-crystallin, the other eye lens protein, is not heat-inducible [8-11]. Both αA and αB-crystallin (subunitMolecular mass≈20 kDa) form homo- as well as hetero-multimers of various sizes [12] and exhibit molecular-chaperone-like activity in preventing aggregation of other proteins [13-16], with αB-crystallin being more efficient than αA-crystallin [15,16]. α-crystallins are associated with some type of cancers such as basal-like breast carcinomas [17] , and belong to the sHsp (small heat-shock protein) family [18].
The Giant Panda (Ailuropoda melanoleuca) is a rare species currently found only in China. They are known as a “living fossil”. For many years, studies on the Giant Panda have been mainly concentrated on fields such as breeding and propagation, ecology, morphology, taxology, physiology and pathological biochemistry. Recently, researches on genetic diversity, parentage and phylogenesis et al. have been developed, while reports on functional gene are handful [19-27].
In the present study, we have amplified the cDNA sequence of CRYAB gene from the total RNAs extracted from the skeleton muscle of the Giant Panda, and then analyzed the sequence characteristics of the protein encoded by the cDNA and compared it with those of human and other mammalian species reported. The overexpression was also done in E. coli using pET28a plasmids successfully. The study provided scientific data for inquiring into the hereditary traits of the gene from Giant Panda and formulating the protective strategy for the Giant Panda.
2. Materials and methods
Materials and RNA isolation
Skeletal muscle was collected from a dead giant panda at the Wolong Conservation Center of the Giant Panda, Sichuan, China. The collected skeletal muscle was frozen in liquid nitrogen and then used for RNA isolation.
Total RNAs were isolated from about 400mg of muscle tissue using the Total Tissue/Cell RNA Extraction Kits (Waton Inc., Shanghai, China) according to the manufacturer's instructions. The total RNAs extracted were dissolved in DEPC (diethypyrocarbonate) water, and kept at-70℃.
Primers Design, RT-PCR, Cloning of RT-PCR Products and Sequencing
The PCR primers were designed by Primer Premier 5.0, basing on the mRNA sequence of alpha-B from Homo sapiens (NM_001885), Mus musculus (NM_009964), Rattus norvegicus (NM_012935). The specific primers of alpha-B are as follows:
alpha-B-F: 5′-GCCGCCATGGACATCGCCA -3′;
alpha-B-R: 5′-CTACTTCTTGGGGGCTGCAG-3′
Total RNAs were synthesized into the first-stranded cDNAs using a reverse transcription kit with Oligo dT as the primers according to the manufacturer's instructions (Promega). The 20μL of first-strand cDNA synthesis reaction system included 1μg of total RNAs, 5mM of MgCl2, 1mM of dNTPs, 0.5μg of Oligo dT15, 10U/μL of RNase inhibitor, and 15U of AMV reverse transcriptase, and was incubated at 42℃ for 60 minutes.
The first-strand cDNA synthesized was used as a template. The total reaction volume for DNA amplification was 25μL. Reaction mixtures contained 1.5mM of MgCl2, 200μM of each of dATP, dGTP, dCTP and dTTP (Omega), 0.3μM of each primer, 5.0 units of Taq plus DNA polymerase (Sangon Co., Shanghai, China). DNA amplification was performed using a MJ Research thermocycler, Model PTC-200 (Watertown, MA) with a program of 4 minutes at 94.0℃, followed by 30 cycles of 1 minute at 94.0℃, 0.5 minute at 45℃ and 1.5 minutes at 72.0℃, and then ended with the final extension for 10 minutes at 72.0℃. After amplification, PCR products were separated by electrophoresis in 1.5% agarose gel with 1× TAE (Tris-acetate-EDTA) buffer, stained with ethidium bromide and visualized under UV light. The expected fragments of PCR products were harvested and purified from gel using a DNA harvesting kit (Omega, China), and then ligated into a pET28a vector at 22℃ for 12 hours. The recombinant molecules were transformed into E. coli complete cells (JM109), and then spread on the LB-plate containing 50μg/mL ampicillin, 200mg/mL IPTG (isopropyl-beta-D-thiogalactopyranoside), and 20mg/mL X-gal. Plasmid DNA was isolated and digested by PstI and ScaII to verify the insert size. Plasmid DNA was sequenced by Huada Zhongsheng Scientific Corporation (Beijing, China).
Cloning the Genomic sequence of CRYAB
The PCR primers were the same as the alpha-B -F and alpha-B -R presented above. The genomic sequence of the CRYAB gene was amplified using Touchdown-PCR with the following conditions: 94℃ for 30s, 62℃ for 45s, 72℃ for 4 minutes in the first cycle and the anneal temperature deceased 0.5℃ per cycle; after 20 cycles conditions changed to 94℃ for 30s, 52℃ for 45s, 72℃ for 4 minutes for another 20 cycles. The fragment amplified was also purified, ligated into the clone vector and tansformed into the E. coli competent cells. Finally, the recombinant fragment was sequenced by Sangon (Shanghai, China).
Construction of the Expression Vector and Overexpression of Recombinant CRYAB
PCR fragment corresponding to the CRYAB polypeptide was amplified from the CRYAB cDNA clone with the forward primer, 5'- TCG AAT TCA TGG ACA TCG CCA -3' (EcoRI) and reverse primer, 5'- CTA AGC TTG CGT CTA CTT CTT -3' (HindIII), respectively. The PCR was performed at 94℃ for 3 minutes; 35 cycles of 30s at 94℃, 45s at 53℃ and 1 minutes at 72℃;10 minutes at 72℃. The amplified PCR product was cut and ligated into corresponding site of pET28a vector (Stratagen). The resulting construct was transformed into E.coli BL21(DE3) strain (Novagen) and used for the induction by adding IPTG (isopropyl-b-D-thiogalactopyranoside) at an OD600 of 0.6 and culturing further for 4 hours at 37℃, using the empty vector transformed BL21(DE3) as a control. The recombinant protein samples were induced after 1, 2 and 3 hours, and then separated by SDS-PAGE and stained with Commassie blue R 250.
SDS-PAGE analysis
The culture was centrifuged at 10000 g for 5 min at room temperature after it was induced for 0.5,1,2,3,4 hours, respectively. The culture supernatant was concentrated with methanol and chloroform (3:1,v/v),and SDS-PAGE(SDS polyacrylamide gel electrophoresis)was performed to investigate protein production and purity using slab gels containing 12%(w/v) polyacrylamide on a mini-protean II slab cell apparatus (Bio-Rad,Hercules,CA). Protein samples were visualized by Coomassie brilliant blue R-250 staining. The samples from the SDS-PAGE gels were transferred to a membrane of polyvinylidene difluoride (PVDF, Millipore, Billerica, MA ), as previously described [28].
Data analysis
The sequence data were analyzed by GenScan software (http://genes.mit.edu/GENSCAN.html). Homology research of the Giant Panda CRYAB compared with the gene sequences of other species ware performed using Blast 2.1 (http://www.ncbi.nlm.nih.gov/blast/). ORF of the DNA sequence was searched using ORF finder software (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Protein structure of the CRYAB sequence cloned was deduced using PredictProtein software (http://cubic.bioc.columbia.edu/predictprotein/).
3. Results
Analysis of the cDNA of CRYAB from the Giant Panda
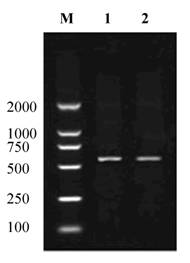
About 530bp of cDNA fragment was amplified from the Giant Panda with the primers CRYAB -F and CRYAB -R (Fig. 1). The length of the cDNA cloned is 534bp. Blast research showed that the cDNA sequence cloned is highly homologous with the CRYAB from Homo sapiens and some other mammals reported. On the basis of the high identity, we concluded that the cDNA isolated is the cDNA encoding the Giant Panda CRYAB protein. The CRYAB sequence has been submitted to Genbank (accession number: FJ169483). An ORF of 528bp encoding 175 amino acids was found in the cDNA sequence (Fig. 2).
Reverse Transcription Polymerase Chain Reaction Products of the Giant Panda CRYAB. M: Molecular ladder DL2000; 1, 2: the amplified CRYAB.
Nucleotide and deduced amino acid sequences of cDNA enoding the Giant Panda CRYAB.

Analysis of the genomic sequence of CRYAB from the Giant Panda
A fragment of 3189 bp was amplified from genonic DNA of the Giant Panda using primers alpha-B -F and alpha-B -R. Comparison between the cDNA sequence and this DNA fragment indicated that the cDNA sequence is a full cDNA corresponding to three exons in the the CRYAB genomic sequence of the Giant Panda. The genomic sequence of the CRYAB gene has been submitted to Genbank (accession number: FJ439506).
Prediction and analysis of protein functional sites in CRYAB protein of the Giant Panda
Primary structure analysis revealed that the molecular weight of the putative CRYAB protein of the Giant Panda is 20.0668kDa with a theoretical pI 6.76 (Table 2). Topology prediction shows there are only four Casein kinase II phosphorylation sites in the CRYAB protein of the Giant Panda (Fig. 3).
Functional Sites in CRYAB Among the Giant Panda, Homo sapiens, Mus musculus, Rattus norvegicus and Bos Taurus. rat: Rattus norvegicus; bos: Bos Taurus; homo: Homo sapiens; mus: Mus musculus; pd: the Giant panda; ▬: Casein kinase II phosphorylation site, ▒: polymorphic sites.

Overexpression of the CRYAB gene in E. coli
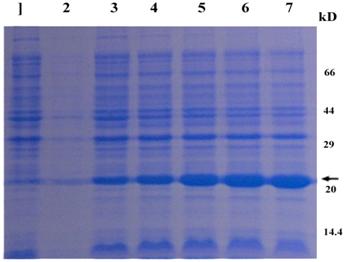
The CRYAB gene was overexpressed in E. coli, using pET28a plasmid, which carries a strong promoter and terminator sequences derived from phage T7. For this purpose, the CRYAB gene was amplified individually by PCR and cloned into the pET28a plasmid, resulting in a gene fusion coding for a protein bearing a GST-tag extension at the N terminus. Expression was tested by SDS-PAGE analysis of protein extracts from recombinant in E. coli BL21 strains (Fig. 4). The results indicated that the protein CRYAB fusion with the N-terminally His-tagged form gave rise to the accumulation of an expected 24kDa polypeptide that formed inclusion bodies. Apparently, the recombinant protein was expressed after half an hour of induction and the after 3 hours reached the highest level. These results suggested that the protein is active and it is just the protein encoded by the CRYAB from the Giant Panda. The expression product obtained could be used to purify the protein and study its function further.
Protein extracted from recombinant E. coli strains were analyzed by SDS-PAGE gel stained with Commassie blue R 250. Numbers on right shows the molecular weight, and the arrow indicates the recombinant protein bands induced by IPTG with 0, 0.5, 1, 2 ,3 and 4 hours (lane 2-7), respectively. The lane 1 represents the products of the E. coli strains with the empty vectors.
4. Discussions
Alignment analysis of CRYAB among the Giant Panda and those of Homo sapiens, Mus musculus, Rattus norvegicus and Bos taurus, indicated that both the nucleotide sequence and the deduced amino acid sequence are highly conserved. There is not any deletion and insertion of nucleotide and amino acid residue. Among them, the Giant Panda shares the highest homology for nucleotide sequence from Bos taurus; and the same highest homology for amino acid sequences (Table 1). Physical and chemical analysis showed that the molecular weight of the putative protein among the five mammalians is very close and that the theoretical pI is exactly identical (Table 2).
Comparison of Nucleotide and Amino Acid Sequences among 5 Mammal Species.
| A. melanoleuca | H.sapiens | M. musculus | R. norvegicus | B. taurus | |
|---|---|---|---|---|---|
| A. melanoleuca | 93.9% | 91.5% | 91.5% | 95.3% | |
| H. sapiens | 98.3% | 88.5 | 88.4% | 91.1% | |
| M. musculus | 97.1% | 97.7% | 93.1% | 86.5% | |
| R. norvegicus | 97.7% | 97.1% | 98.3% | 87.1% | |
| B. taurus | 99.4% | 97.7% | 96.6% | 97.1% |
Note: The homology matrix of CRYAB encoding sequence is above the diagonal, the homology matrix of protein sequence is below the diagonal.
Molecular Weight and pI of CRYAB of the Giant Panda and Other Four Mammals.
| A. melanoleuca | H. sapiens | M. musculus | R. norvegicus | B. taurus | |
|---|---|---|---|---|---|
| Molecular weight(kDa) | 20.0668 | 20.1589 | 20.0688 | 20.0888 | 20.0368 |
| pI | 6.76 | 6.76 | 6.76 | 6.76 | 6.76 |
The genomic sequence of CRYAB is 3189 bp in size, containing three exons and two introns. Compared with some mammals incluing Homo sapiens (NC_000011), Mus musculus (NC_000075), Rattus norvegicus (NC_005107) and Bos taurus (NC_007313 ), the length of the genomic, the first intron, the second intron, the 5′-untranslated sequence and the 3′-untranslated sequence are different (see Table 3 ).
Comparison of CRYAB genomic among 5 Mammal Species.
| Length of genomic | Number of exons | Number of introns | Length of 5′-untranslated sequence | Length of 3′-untranslated sequence | Length of first intron | Length of second intron | |
|---|---|---|---|---|---|---|---|
| Ailuropoda melanoleuca | 3189 | 3 | 2 | 6 | 0 | 1113 | 1542 |
| Homo sapiens | 3124 | 3 | 2 | 25 | 138 | 1074 | 1359 |
| Mus musculus | 3876 | 3 | 2 | 519 | 148 | 1034 | 1667 |
| Rattus norvegicus | 4253 | 3 | 2 | 579 | 137 | 1029 | 1970 |
| Bos taurus | 3226 | 3 | 2 | 0 | 81 | 1127 | 1480 |
It is known that αB-crystallins undergo posttranslational modifications, including truncation of both the N terminus and the C terminus, deamidation, racemization, phosphorylation, methionine oxidation, glycation, disulfide formation, addition of O-GlcNAc, and the addition of 72 mass units to the C-terminal lysine of B-crystallin [29-34]. Some of these activities, such as phosphorylation and specific cleavage, may be functionally important; others are likely the result of aging and detrimental stresses. aB-crystallin has polar, flexible C-terminal extensions that is thought to contribute to the solubility of this crystallin and that has been implicated in the chaperone-like activity of this crystallin [35]. Topology prediction revealed there are four Casein kinase II phosphorylation sites in the CRYAB proteins of the five mammalian species. Our analysis indicate that the same number and patter functional sites are located in the same locations in CRYAB protein of the Giant Panda, Homo sapiens, Rattus norvegicus, Mus musculus and Bos Taurus (Fig. 3) and the amino acid sequence deduced from Giant Panda shares the highest homology with that of Bos Taurus. Further analysis detected eight polymorphic sites in the amino acid sequences of the five species compared. These polymorphic sites are located irregularly in the amino acid sequences all of which result from the transversion or transition of the corresponding codons without any deletion and insertion of base. Among these polymorphic sites, only site 61 is located in Casein kinase II phosphorylation site, but it does not result in any differences from human and other four mammalian species in the functional site. The fact shows that the variation of sites has no affect on the structure and function of CRYAB protein. However, what changes caused by other mutations outside the functional sites in the structure and the function of CRYAB need further studies.
In addition, it was reported that in the interactive β3 sequence, 73DRFSVNLDVKHFS85 of human αB crystallin, Ser-76, Asn-78, Lys-82, and His-83 were identified as nonconserved residues on the exposed surface of the a crystallin core domain. Chaperone activity was influenced by the amount of unfolding of the target proteins and independent of complex size. It was reported that the importance of the exposed side chains of Glu-78, Lys-82, and His-83 in the interactive β3 sequence of the α crystallin core domain in aB crystallin for chaperone function. The β3 strand was conserved at 5 of 8 positions, and 4 of the 5 conserved residues were buried inside the β sandwich structure of the α crystallin core domain where the side chains were positioned to form intramolecular bonds. In contrast, the side chains of the nonconserved residues were surface exposed, suggesting their involvement in protein-protein interactions with homologous sHSP subunits and/or unfolded chaperone target proteins at the surface of the a crystallin core domain [36,37]. It was reported that that amino acid differences at positions 76, 78, 82, and 83 in the β3 sequence could account for the difference in chaperone activities. On the basis of numbers of research findings above, we speculate that the most important sequence of our predicted CRYAB protein of the Giant Panda for its structure and function is the sequence: 73DRFSVNLDVKHFS85. A comparison of the β3 sequence of Giant Panda aB crystalline with those of Homo sapiens, Mus musculus, Rattus norvegicus and Bos Taurus shows that amino acid share the same homology and the position Ser-85 is located in the first site of a Casein kinase II phosphorylation site (Fig. 3). It indicates that the β3 sequence in αB crystallin may be an interactive site that selects target proteins for chaperone activity on the basis of the degree of unfolding and may be a chaperone site that is selective for partially and completely unfolded proteins.
In Summary, the cDNA and the complete coding sequence of CRYAB gene has been cloned and the CRYAB cDNA is expressed efficiently in prokaryotic organism using pET28a plasmids. The gained fusion protein is in accordance with the expected 24kDa polypeptide. These results suggest that the protein is active and it is just the protein encoded by the CRYAB from the Giant Panda. Further study on CRYAB protein will contribute to the genetic polymorphism and the protection for gene resources of Giant Panda.
Acknowledgements
This work is supported by the Key Chinese National Natural Science Foundation (30470261), Application Technology Project in Sichuan Prov ince (2006J13-057), Key Scientific Research Foundation of Educational Committee of Sichuan Province (07ZA120) and Key Discipline Construction Project in Sichuan Province (SZD0420).
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. de Jong WW, Hendriks W, Mulders JW, Bloemendal H. Evolution of eye lens crystallins: the stress connection. Trends Biochem Sci. 1989;14:365-368
2. Piatigorsky J. Lens crystallins and their genes: diversity and tissue-specific expression. FASEB J. 1989;3:1933-1940
3. Reddy VN, Katsura H, Arita T, Lin LR, Eguchi G, Agata K, Sawada K. Study of crystallin expression in human lens epithelial cells during differentiation in culture and in non-lenticular tissues. Exp Eye Res. 1991;53:367-374
4. Iwaki T, Kume-Iwaki A, Goldman JE. Cellular distribution of alpha B-crystallin in non-lenticular tissues. J Histochem Cytochem. 1990;38:31-39
5. Ghosh JG, Clark JI. Insights into the domains required for dimerization and assembly of human alphaB crystallin. Protein Sci. 2005;14:684-695
6. Kim KK, Kim R, Kim SH. Crystal structure of a small heatshock protein. Nature. 1998;394:595-599
7. Delaye M, Tardieu A. Short-range order of crystallin proteins accounts for eye lens transparency. Nature. 1983;302:415-417
8. Dasgupta S, Hohman TC, Carper D. Hypertonic stress induces alpha B-crystallin expression. Exp. Eye Res. 1992;54:461-470
9. Bhat SP, Nagineni CN. Alpha B subunit of lens-specific protein alpha-crystallin is present in other ocular and non-ocular tissues. Biochem. Biophys. Res. Commun. 1989;158:319-325
10. Clark JI, Muchowski PJ. Small heat-shock proteins and their potential role in human disease. Curr. Opin. Struct. Biol. 2000;10:52-59
11. Sax CM, Piatigorsky J. Expression of the alpha-crystallin/small heat-shock protein/molecular chaperone genes in the lens and other tissues. Adv. Enzymol. Relat.Areas Mol. Biol. 1994;69:155-201
12. van den Oetelaar PJ, van Someren PF, Thomson JA, Siezen RJ, Hoenders HJ. A dynamic quaternary structure of bovine alpha-crystallin as indicated from intermolecular exchange of subunits. Biochemistry. 1990;29:3488-3493
13. Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. U.S.A. 1992;89:10449-10453
14. Sun TX, Das BK, Liang JJ. Conformational and functional differences between recombinant human lens alphaA- and alphaB-crystallin. J. Biol. Chem. 1997;272:6220-6225
15. Datta SA, Rao CM. Differential temperature-dependent chaperone-like activity of alphaA- and alphaB-crystallin homoaggregates. J. Biol. Chem. 1999;274:34773-34778
16. Raman B, Rao CM. Chaperone-like activity and quaternary structure of alpha-crystallin. J. Biol. Chem. 1994;269:27264-27268
17. Moyano JV, Evans JR, Chen F, Lu ML, Werner ME, Yehiely F, Diaz LK, Turbin D, Karaca G, Wiley E, Nielsen TO, Perou CM, Cryns VL. αB-Crystallin is a novel oncoprotein that predicts poor clinical outcome in breast cancer. The Journal of Clinical Investigation. 2006;116(1):261-270
18. Clark JI, Muchowski PJ. Small heat-shock proteins and their potential role in human disease. Curr Opin Struct Biol. 2000;10:52-59
19. Hou WR, Chen Y, Peng ZS, Wu X, Tang ZX. cDNA cloning and sequences analysis of ubiquinol-cytochrome c reductase complex ubiquinone-binding protein (QP-C) from giant panda. Acta Theriologica Sinica. 2007;27(2):190-194
20. Hou WR, Du YJ, Chen Y, Wu X, Peng ZS, Yang J, Zhou CQ. Nucleotide Sequence of cDNA Encoding the Mitochondrial Precursor Protein of the ATPase Inhibitor from the Giant Panda (Ailuropoda melanoleuca). DNA AND CELL BIOLOGY. 2007;26(11):799- 802
21. Hou WR, Luo XY, Du YJ, Chen Y, Wu X, Peng ZS, Yang J, Zhou CQ. cDNA Cloning and Sequences analysis of RPS15 from the Giant Panda. Recent Patent on DNA Sequence. 2007;2(2):16- 19
22. Du YJ, Luo XY, Hao YZ, Zhang T, Hou WR. Cloning and Overexpression of Acidic Ribosomal Phosphoprotein P1 Gene (RPLP1) from the Giant Panda. International Journal of Biological Sciences. 2007;3(7):428- 433
23. Du YJ, Hou WR, Peng ZS, Zhou CQ. cDNA Cloning and Sequences Analysis of Acidic Ribosomal Phosphoprotein P1 (RPLP1) from Giant Panda. Acta Theriologica Sinica. 2008;28(1):75- 80
24. Liao MJ, Zhu MY, Zhang ZH, Zhang AJ. Cloning and sequence analysis of FSH and LH in the giant panda (Ailuropoda melanoleuca). Anim Reprod Sci. 2003;77:107- 116
25. Liao MJ, Zhu MY, Zhang ZH, Zhang AJ. cDNA cloning of growth hormone from giant panda (Ailuropoda melanoleuca) and its expression in Escherichia coli. Comp Biochem Phys B. 2003;135:109- 116
26. Montali RJ. Causes of neonatal mortality in giant panda. Tokyo Zoological Park Society. 1990:83- 94
27. Wu ZA, Liu WX, Murphy C, Gall J. Satellite DNA sequence from genomic DNA of the giant panda. Nucleic Acids Res. 1990;18(4):1054
28. Towbin H, Staehelin T, Gordon J. Eletrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76(9):4350-4354
29. Chiesa R, Gawinowicz-Kolks MA, Kleiman NJ, Spector A. The phosphorylation sites of the B2 chain of bovine alpha-crystallin. Biochem Biophys Res Commun. 1987;144:1340-1347
30. de Jong WW, Mulders JW, Voorter CE, Berbers GA, Hoekman WA, Bloemendal H. Post-translational modifications of eye lens crystallins: crosslinking, phosphorylation and deamidation. Adv Exp Med Biol. 1988;231:95-108
31. Roquemore EP, Dell A, Morris HR, Panico M, Reason AJ, Savoy LA, Wistow GJ, Zigler JS Jr, Earles BJ, Hart GW. Vertebrate lens alpha-crystallins are modified by Olinked N-acetylglucosamine. J Biol Chem. 1992;267:555-563
32. Smith JB, Sun Y, Smith DL, Green B. Identification of the posttranslational modifications of bovine lens alpha Bcrystallins by mass spectrometry. Protein Sci. 1992;1:601-608
33. Groenen PJ, Merck KB, de Jong WW, Bloemendal H. Structure and modifications of the junior chaperone alphacrystallin: from lens transparency to molecular pathology. Eur J Biochem. 1994;225:1-19
34. Lin P, Smith DL, Smith JB. In vivo modification of the C-terminal lysine of human lens alphaB-crystallin. Exp Eye Res. 1997;65:673-680
35. Carver JA, Lindner RA. NMR spectroscopy of alpha crystallin: insights into the structure, interactions and chaperone action of small heat-shock proteins. Int J Biol Macromol. 1998;22:197-209
36. Ghosh JG, Clark JI. Insights into the domains required for dimerization and assembly of human alphaB crystallin. Protein Sci. 2005;14:684-695
37. Ghosh JG, Estrada MR, Clark JI. Interactive domains for chaperone activity in the small heat shock protein, human alphaB crystallin. Biochemistry. 2005;44:14854-14869
Author contact
![]() Correspondence to: Wan-ru Hou, Ph. D, College of Life Science, China West Normal University, 44# Yuying Road, 637002, Nanchong, China. Phone/Fax: +86-0817-2568653. E-mail: hwr168com.cn. Or to: Zheng-long Ren, Ph. D, College of Agriculture, Sichuan Agricultural University, 46# Xinkang Road, 625014, Ya-an, China. Phone/Fax: +86-0835-2882320. E-mail: cnzsjyedu.cn.
Correspondence to: Wan-ru Hou, Ph. D, College of Life Science, China West Normal University, 44# Yuying Road, 637002, Nanchong, China. Phone/Fax: +86-0817-2568653. E-mail: hwr168com.cn. Or to: Zheng-long Ren, Ph. D, College of Agriculture, Sichuan Agricultural University, 46# Xinkang Road, 625014, Ya-an, China. Phone/Fax: +86-0835-2882320. E-mail: cnzsjyedu.cn.

 Global reach, higher impact
Global reach, higher impact