Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(1):34-43. doi:10.7150/ijbs.5.34 This issue Cite
Research Paper
Molecular cloning and characterization of pigeon (Columba liva) ubiquitin and ubiquitin-conjugating enzyme genes from pituitary gland library
The key laboratory of animal breeding and genetics & reproduction, Shanxi Agriculture University, Taigu, 030801, Shanxi, China
Received 2008-10-22; Accepted 2008-12-14; Published 2008-12-18
Abstract
In the study of the regulation of incubation, broodiness and laying performance in pigeons (Columba liva), a cDNA library, which was enriched with full-length brooding-related genes, was constructed by SMART LD-PCR techniques using the pituitary glands of incubating White King pigeons. The titers of optimal primary libraries were 1.54×106 pfu/mL and 1.80×106 pfu/mL and the titers of amplified libraries were 1.89×108 pfu/mL and 2.32×109 pfu/mL. The percentages of recombinant clones of primary libraries and amplified libraries were all over 90%. A positive clone was sequenced and named ubiquitin based on the highly similar from other species. The fragment has the four initial codons of ATG, a termination codon of TAA and a signal sequence of AATAAA for adding the poly-A tail. The open reading frame of 918bp encodes 305 amino acids (NCBI accession number is EU981283). Recombinant pigeon ubiquitin protein was efficiently expressed with the form of insoluble inclusion bodies in E. coli BL21 transformed with a pET28a+ expression vector containing the DNA sequence encoding mature pigeon ubiquitin. The molecular weight of expressed protein is the same as predicted size of approximately 35kD. To improve the efficiency of cloning full-length cDNA, strategies of RACE combined with cDNA library were used. The length of pigeons ubiquitin-conjugating enzyme gene obtained was 1263 bp containing a complete open reading frame of 435 bp that encodes 144 aa (NCBI accession number is EU914824). The results of this study not only provide a starting point for further study of ubiquitin function in pigeon species, but also provide a starting point for investigating the brooding mechanisms of pigeons.
Keywords: cDNA cloning, ubiquitin, ubiquitin-conjugating enzyme, fusion expression, pigeon (Columba liva)
1. Introduction
Broodiness, which means a hen sits on her eggs for the purpose of hatching the embryos, is observed in most breeds of domestic fowl with the exception of the White Leghorn, which has undergone long-term artificial selection to minimize phenotypic expression of this behavior. Due to the adoption of artificial incubation technology, the broodiness is no longer necessary in modern poultry production. Extensive studies on the mechanisms of broodiness have been carried out to inhibit incubation behavior and improve the egg production of domestic fowl, such as chicken [1], turkey [2], quail [3], goose [4], and so on.
The pituitary gland secretes several proteins involved in the function of egg laying in birds. For example, prolactin is secreted from the anterior pituitary at a high level at the onset of egg laying in chickens and quail [5-7]. Estradiol, progesterone, latinizing hormone (LH) and follicle-stimulating hormone (FSH) are the most important hormones involved in regulating ovulation [8,9]. Growth hormone and thyroid-stimulating hormone-subunit mRNA increase as embryonic development proceeds [10].
Eukaryocytic gene expression was controlled in several steps. The degradation of protein is very important in the expression regulation on the level of post-translation. Ubiquitin-mediated proteolysis plays a universal role in the irreversible negative regulation of various signaling pathways [11], including those induced by pituitary hormones [12]. The polyubiquitination of proteins leads to their degradation by 26S proteasome complexes; the oligoubiquitination of plasma membrane proteins targets them for endocytosis and degradation in lysosomes [13]. It has been reported that most cytokine receptors, for instance, prolactin receptor [14], progesterone receptor [15] and growth hormone receptor [16,17], and their downstream signaling molecules, are regulated by the ubiquitin-proteasome system. The important roles of ubiquitination in controlling the activities and turnover of key signaling proteins suggest potential roles in controlling brooding mechanisms of avians.
To date, very limited full-length cDNAs of pigeon were deposited to the public database. To identify more genes of pigeon, including the characterization of specific expressed, new or unknown genes and further study their functions, construction of cDNA libraries of pigeons is an efficient method.
To look into this aspect, a full-length cDNA library was constructed using the pituitary gland of white king pigeon with SMART LD-PCR techniques [18,19]. Several clones of encoding sequences were obtained and we studied one of these clones in detail. The amino acid sequence derived from the cDNA sequence revealed that the protein is ubiquitin, a highly conserved protein that appears to play a role in the chromatin structure and in an ATP-dependent proteolytic degradation of unstable cellular proteins [20]. We also over expressed it in E. coli using Pet28a+ plasmids. By combining the methods of RACE [21] with cDNA library, a full-length ubiquitin-conjugating enzyme gene of pigeons was cloned. The constructed cDNA libraries and the improvement of gene cloning methods will fascinate the identification and cloning of the brooding related genes to further elucidate the molecular mechanism of the incubation of pigeon.
2. Materials and Methods
Animal
All the birds used in the present study were white-king pigeon, which were purchased from a commercial pigeon farm and were raised according to the standard program, which was approved by Department of the Animal Science and Veterinary Medicine of Shanxi Agriculture University. A photoperiod of 14 L:10 D was maintained. 13 pairs of laying pigeons were killed at the day 2 or 3 of incubation (body weights on average were 0.664±0.208 kg and 0.568±0.446 kg for male and female, respectively). Pituitary glands were rapidly removed, wrapped in foil, frozen in liquid nitrogen and then stored at -70℃ until the extraction of the total RNA.
Total RNA extraction and cDNA library construction
The total RNA was extracted from pituitary glands using TRIzolR reagent (Invitrogen) according to the manufacturer's protocol. RNA concentrations were determined from the OD260 and OD280 readings on a ND-1000 spectrophotometer (Nano drop). To determine the integrity and stability of total RNA, before electrophoresis a 5 μl fresh sample and another 5 μl sample which was incubated at 70℃ for 30 min, were run on a 1.2% denaturing formaldehyde TBE agarose gel, stained with ethidium bromide, and photographed under UV light using an Gel Doc 2000 Imaging System (BIO-RAD). Poly A+ mRNA was isolated from the total RNA using Oligotex mRNA midi Kits (QIAGEN) according to the manufacturer's protocol. The titer and purity of poly A+ mRNA were checked by measuring the absorbance of ultraviolet light at A260 and A280 on ND-1000 spectrophotometer (Nanodrop). Approximately 3 μg of poly A+ mRNA was reverse-transcripted into first-strand cDNA by PowerScriptTM reverse transcriptase. Subsequently approximately 2 μl of the first-strand cDNA sample was amplified using long-distance PCR (LD-PCR). The product was analyzed by running a 1.2% agarose gel alongside DNA size markers. The double-stranded cDNA (ds-cDNA) was treated with protein K, and followed by Sfi I digestion and size fractionation. The first three peak fractions containing cDNA (>500 bp) were pooled together for packaging. Three parallel ligation reactions were performed using three different ratios of cDNA to the λTripIEx2 vector (1:0.5, 1:1, 1:1.5) to ensure the optimal possible library was obtained. Each of the three ligations was packaged with Gigapack III Cold Packaging Extract (Stratagene).
Titration of the primary library and determining the percentage of recombinant clones
The primary library was diluted by 1:5, 1:20, 1:50 and 1:100. The number of clones was counted to calculate the library titer as follows:
Insert-containing phages were identified by transforming E. coli XL1-Blue and screening for white plaques on medium containing IPTG and X-gal. The recombination efficiency of primary and amplified libraries were calculated the ratio of white (recombinant) to blue (no recombinant) plaques.
Library amplification and conversion of cDNA library to plasmid
The primary libraries were amplified on 20 plates, each 12 cm in diameter. The titer and recombinant efficiency were calculated using the same method as the above.
One hundred clones were randomly picked to check the insert fragments by PCR using 5′ sequencing primer and 3′sequencing primer of vector. Sequencing analysis was completed by an ABI 3730 automatic DNA sequencer.
cDNA cloning of pigeon ubiquitin gene
One full-length pigeon cDNA was obtained using random sequencing analysis and was identified using the NCBI Basic Local Alignment Search Tool (BLAST) in order to find homologues from other species.
Phylogenetic analysis of ubiquitin
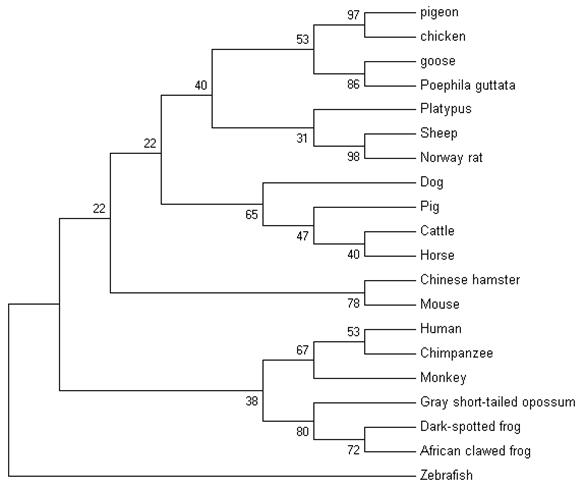
The nucleotide sequence of pigeon ubiquitin obtained in the current work was aligned with the nucleotide sequences of the ubiquitin (UB) of other avians and mammals including chicken (XM_00123337), goose (DQ420616), poephila guttata (DQ216253), mouse (NM_019639), rat (NM_138895), Chinese hamster (AB003732), cattle (XM_874550), sheep (NM_00100920), human (NM_021009), rhesus monkey (XM_001102090), chimpanzee (XM_001136911), pig (XM_001925239), horse (XM_001499082), dog (XM_847967), platypus (XM_001508854), zebrafish (XM_001923619), African clawed frog (NM_001086120), gray short-tailed opossum (XM_001366716) and dark-spotted frog (DQ520795) using Clustal X [22]. The alignment was manually refined using the BioEdit Sequence Alignment Editor v7.0.9.0 [23]. A neighbour-joining tree was produced in MEGA v4.0 [24]. The optimal tree with the sum of branch length (0.02799395) is shown.
Cloning of pigeon ubiquitin-conjugating enzyme cDNA using RACE method
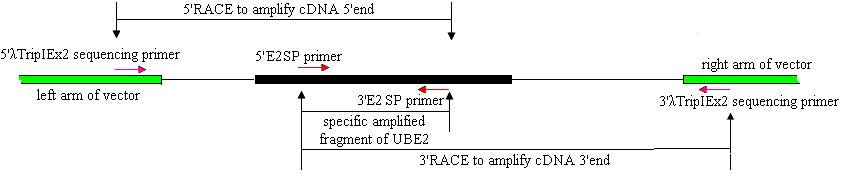
Cloning of full-length ubiquitin-conjugating enzyme (UBE2) cDNAs was performed using 3′- and 5′-RACE and Reverse Transcription (RT)-PCR. The strategies are shown in Fig.1.
Based on the UBE2 cDNA sequence of chicken (NM_001031018), two UBE2 specific primers (E2SP), 5′E2SP primer (5′-TCGTAATTTTCGCTTGTTG-3′) and 3′E2SP primer (5′- CTAACACTGGTATGCTCCGT-3′) were designed. The reaction mixture (50 μl) contained 1.5 μl pituitary SMARTTM cDNA (titer:0.875 μg/μl), 1μl each of the primers (10 μmol/L), 2.5 U high LA Taq DNA polymerase ( TaKaRa, Japan), 5 μl 10 × LA Taq DNA polymerase buffer, 5 μl dNTPs (2.5 mmol/L), added ddH2O to the final volume. The reaction was initiated by a denaturation step at 94℃ for 2 min, followed by 30 cycles of 94℃ for 30 s, 57.8℃ for 30 s and 72℃ for 30 s, and a final 5 min extension at 72℃.
A strategy of RACE combined with cDNA library was used to clone the full-length cDNA. λTripIEx2 5′ LD-Insert screening primer (5′-CTCGGGAAGCGCGCCATTGT-3′) and the 3′E2SP primer were used to amplify the cDNA 5′end. 5′E2SP primer and the 3′λTripIEx2 sequencing primer (5′-TAATACGACTCACTATAGGG-3′) were used to amplify the cDNA 3′end (Fig.1). The reaction was initiated by a denaturation step at 94℃ for 2 min, followed by 34 cycles of 94℃ for 30 s, 50-60℃ for 30 s and 72℃ for 50 s, and a final 7 min extension at 72℃. All PCR reactions were undertaken on a GeneAmp PCR System 9700.
Strategies for cloning full-length E2 cDNA of pigeon.
The 5′and 3′RACE products were separated by the agarose gel (1.2 %) electrophoresis and purified with the columns (BioDev, China). 1μl purified products were used as template in a nest amplification using 5′and 3′E2SP primers. The amplification conditions were the same as above. After verifying, the nest PCR products were about 300 bp. The 5′and 3′ RACE products were cloned into pUCM-T vector (Sangon, China), and transformed into E. coli strain DH5α using hot-shock method (Sambrook, 1989). The positive clones, characterized by blue/white screening, were sequenced with an ABI 3730 automatic DNA sequencer.
Generation of Expression Vector
DNA encoding mature pigeon ubiquitin protein was amplified with the ORF primers designed according to the ubiquitin sequence isolated in this study. The forward primer (5′-GGAATTCCAGCAAAATGCAGATCTT-3′) contained the EcoR I site and reverse primer (5′-GAGAGCTCCTGGAAGAATTTAATACC-3′) contained the Xho I site. The amplified fragment was digested by EcoR I and Xho I and dubcloned into the multiple cloning sites of pET-28a+ (Novagen, Germany) expression vector. The generated expressed plasmid was designated as pET28a-UB and transformed into the competent cells of E.coli BL21 for fusion expression.
Fusion expression of pigeon ubiquitin
A single colony of BL21 containing pET28a-UB was culture overnight at 37℃ in an LB medium containing 100 mg/mL Kanamycin. Cultures were then diluted (1:20) in a fresh 50 mL LB medium and grown at 37℃ for approximately 2 h until the OD reached approximately 0.6. Expression was initiated by the addition of isopropyl-b-D-thiogalactoside (IPTG) (Sigma) to a final concentration of 2.0 mM. The recombinant protein samples were induced with time gradient of 0, 0.5, 1, 2 and 4 hours, a 1mL culture was collected and centrifuged (12,000g, 1 min). The pellet was dissolved in 100 mL 1×SDS sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 0.1 M DTT, 10% glycerol, 0.1% bromophenol blue), heated at 100℃ for 5 min, and resolved on 12% SDS polyacrylamide gel electrophoresis (SDS-PAGE). PET28a-UB without IPTG induction and BL21 served as controls. Gel was analyzed by Commassie blue staining.
Western blot analysis
The samples were electrophoresed on a 12% SDS polyacrylamide gel, and electro-blotted over to a PVDF membrane. The membrane was incubated with mouse anti-His antibody (TIANGEN, China) at 1:1000 dilutions. The secondary antibody was horseradish peroxidase-conjugated goat anti-mouse IgG (TIANGEN, China) at 1:1000 dilution. The membrane was incubated with chemiluminescent substrate for 5 min before exposing to photographic film. Exposure times generally varied from 1 sec to 30 sec.
3. Results
Total RNA extraction
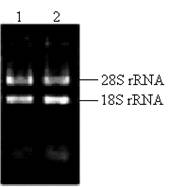
The key to construct an excellent quality cDNA library is to prepare high quality RNA. In this study, the ratio of the A260/A280 for total RNA was approximately 1.98. The concentration of total RNA was approximately 0.875 μg/μl according to the absorbance of ultraviolet light at 260nm. Two bright bands of 18S rRNA and 28S rRNA can be seen clearly (Fig.2), and a little difference between the sample incubated at 70℃ for half an hour and the fresh sample existed (Fig.2), which indicated that the total RNA isolated from the pituitary glands of pigeon was pure, integrated and stable for cDNA library construction.
Total RNA isolated from pituitary of pigeon. Lane 1: fresh sample of 5 μl total RNA; Lane 2: sample incubated at 70℃ for half an hour.
Synthesis of cDNA
Using 2 μg high quality total RNAs, first-strand cDNAs were synthesized according to the protocol of SMARTTM cDNA Library Construction Kit (Clontech, USA). Then, 1/5 of the ss cDNA were used to synthesize the ds cDNAs. After 22 cycles, 5μL of 100μL was analyzed by agarose gel electrophoresis (Fig. 3). The bands of ds cDNAs were dispersed and the length of ds cDNA was mainly bounded between 500 bp- 3000 bp. Fragments smaller than 500 bp and longer than 3000 bp were eliminated by cDNA fractionation to avoid the library having a preponderance of very small inserts and/or apparently non-recombinant clones.
The products of LD-PCR. Lane 1: the product of LD-PCR with 22 cycles; Marker: 1 kb DNA ladder (Gibco-BRL).

Characterization of cDNA library
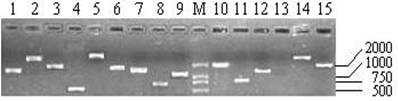
As shown in Table 1, the titers of three primary libraries constructed with three different ratios of cDNA to vector (1:0.5, 1:1, and 1:1.5) were 0.39×106 pfu/mL, 1.54×106 pfu/mL, and 1.80×106 pfu/mL, respectively, which showed that the optimal ratio was 1:1.5. The primary libraries with titers of over 1.0×106 pfu/mL were both amplified. The titers of amplified libraries were 1.89×108 pfu/mL and 2.32×109 pfu/mL, respectively. The recombination efficiencies of the primary and amplified libraries were all over 90%. The insert ratio and the average length of inserted fragments were measured by PCR (Fig.4). The average size was approximately 1.3 kb and the majority inserts were all over 0.5 kb.
Characterization of the primary and amplified cDNA libraries of pigeon.
| Number of primary libraries | Ligation ratio of cDNA to vector | Titer of primary libraries (pfu/mL) | Recombination efficiency of primary libraries (%) | Titer of amplified libraries (pfu/mL) | Recombination efficiency of amplified libraries (%) |
|---|---|---|---|---|---|
| (1) | 1:0.5 | 0.39×106 | 92.31 | --- | --- |
| (2) | 1:1 | 1.54×106 | 95.47 | 1.89×108 | 92.11 |
| (3) | 1:1.5 | 1.80×106 | 97.82 | 2.32×109 | 96.46 |
Part of the inserted cDNA fragments selected randomly from the amplified cDNA library. Lane 1-15: 15 inserted cDNA fragments; M: DL2000 (TakaRa, Japan).
Sequence analysis of the ubiquitin gene
A random positive clone was sequenced and a fragment of 1037 bp was obtained. BLASTn search showed that the cDNA sequence was highly similar to a goose ubiquitin C (accession number: DQ420616) and chicken ubiquitin C (accession number: NM_001079474). The sequence informations are shown in Fig. 5. The fragment has four initial codons of ATG, a termination codon TAA and a signal sequence of AATAAA for adding the poly-A tail. The open reading frame (ORF) of 918 bp (from the 68th bp to the 986th bp) encodes 305 amino acids (aa). Four repeated units (U1 from the 68th bp to the 295th bp; U2 from the 296th bp to the 523th bp; U3 from the 524th bp to the 751th bp and U4 from the 752th bp to the 986th bp) were shown in the sequence, of which the frist three units are all 228 bp encoding 76 aa and the last unit consists of 234 bp encoding 77 aa. The nucleotide sequence data has been submitted to GenBank under the accession number EU981283.
The nucleotide sequence of pigeon ubiquitin was compared with 19 vertebrates' ubiquitins. Ubiquitin is well conserved among vertebrates. Following the multiple sequence alignment of ubiquitin nucleotide from pigeons, chickens, goose and several other vertebrates, a phylogenetic tree of ubiquitin was constructed using nerghbor-joining methods (NJ). The neighbor-joining gene tree for the remaining 20 sequences is shown in Figure 6. Phylogenetic analysis of the ubiquitin family indicated that pigeon, chicken, goose and poephila guttata ubiquitin sequences were clustered with strong bootstrap values (bootstrap value was shown in Fig.6).
Cloning of the ubiquitin-conjugating enzyme E2
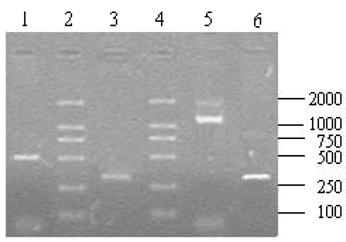
A total of 10 μl aliquots of the amplification products were separated on 1.2% agarose gel after the RACE amplification. The 5′ RACE product was about 500 bp (Fig.7, lane 1) and 3′ end fragment was about 1200 bp (Fig.7, Line 5) in length. Using the 5′ and 3′ UBSP primers, both recovered 5′ and 3′ products could produce an expected single band of about 300 bp (Fig.7, line 3,6) after the nest PCR that indicated that the products of 5′ and 3′ RACE were the target fragments.
Full-length cDNA sequence and deduced amino acid sequence of ubiquitin gene of pigeon. The underlined site indicates the start codon (AUG); the double underlined site indicate the stop codon (UAA); bold underline indicates the multiple AATAAA sequence described in the text.

The phylogenetic tree describing evolutionary distances between the ubiquitin in various species with nerghbor-joining (NJ) methods.
The results of 5′, 3′ RACE and nest PCR. Lane 1: 5′ RACE product; Lane 2, 4: DL2 000 DNA Marker (TaKaRa, Japan); Lane 5: 3′ RACE product; Lane3, 6: Products derived by the nest PCR with recovery of 5′ and 3′ RACE products as temperate respectively.
Sequencing and alignment search results
Sequencing results showed that 5′ and 3′ RACE products were 404 bp and 1149 bp excluding the vector sequence, respectively. Conjugation of the 5′ and 3′ fragments produced a cDNA with full-length of 1263 bp containing a 435 bp ORF, an 88bp noncoding region at 5′ end, an untranslated flank at 3′ end that includes two polyadenylation signal AATAAA which locate 94 bp and 12 bp to 5′ side of the poly A tail and three ATTTA sequences, which are, in specific cases, involved in the targeting of mRNAs for rapid turnover and present in the 3′ noncoding regions of transcripts with short half-lives (Fig. 8). The ORF (from the 89th bp to the 523th bp) encodes 144 aa. The nucleotide sequence data has been submitted to GenBank (Accession number: EU914824).
Expression of fusion protein
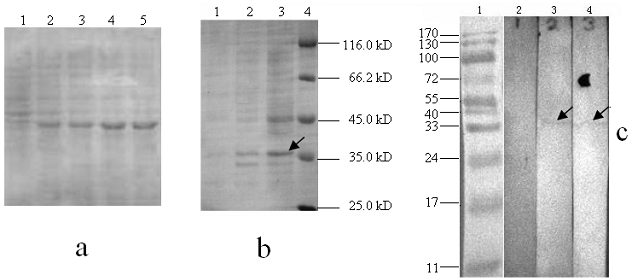
To express the recombinant ubiquitin in E.coli, the DNA encoding pigeon mature ubiquitin was amplified and inserted downstream of the His·taq coding region of pET-28a+. Cells with 2.0 mM IPTG induction were analyzed on SDS-PAGE. Apparently, the recombinant protein was expressed after half an hour of induction and then after 2 hours reached the highest level (Fig. 9a). As shown in SDS-PAGE of Fig. 9b, distinct protein bands could be visualized in samples induced by IPTG, but not in the samples without IPTG induction or the host strain BL 21. The molecular weight of expressed protein is the same as the predicted, which is about 35 kD. To further confirm His·tag fusion expression, anti-His antibody was used to probe expressed protein by western blot. As expected, a distinct signal in the position of 35 kD was observed in the extracts with IPTG induction, but not in the host BL21 strain (Fig. 9c).
Nucleotide and deduced amino acid sequence of UBE2. The double underlined sites indicate the two polyadenylation signal sequences. Bold underlines indicate the multiple ATTTA sequence described in the text.

Electrophoresis analysis of ubiquitin expressed products. (a): Lane 1-5 indicates the recombinant protein bands induced by IPTG with 0, 0.5, 1, 2 and 4 hours, respectively. (b): Lane 1, total protein extracts from the host BL21 strain; Lane 2, total protein extracts without IPTG containing pET-28a-UB; Lane 3, total protein extracts with IPTG containing pET-28a-UB; M, protein molecular weight marker. (c): Lane 1, prestained protein molecular weight marker; Lane 2, total protein extracts of host BL21 strain; Lane 3 and 4: total protein extracts with IPTG induction.
4. Discussion
To date, the sequencing and characterization of complementary DNA (cDNA), which represents a direct link to functional genomics, is a powerful means of identifying genetic polymorphisms and is essential for the determination of different gene expressions [25,26], not only in humans but also in other species. Although a variety of cloning methods without cDNA libraries have been established, a cDNA library enriched with broodiness-related genes of pigeon is essential for approaching the molecular interaction among hormones.
The quantity and quality of cDNA are keys to construct a high quality cDNA library [27]. The quantity and integrity of total RNA detected by spectrophotometer and by electrophoresis on a denaturing formaldehyde agarose gel was good. In this paper, the recombination efficiency of primary and amplified libraries was all over 90%. The primary library had no less than 1.5×106 clones, which in principle was sufficient for including the most mRNA of rarely expressed. The full-length cDNA library constructed from the pituitary glands of pigeons conformed to the requirements of a standard library. This library provided a useful resource for the functional genomic research of the laying pigeon.
There were 100 pigeon gene fragments obtained in our laboratory through screening the library. A scheduled cloning of the full-length genes and function research are undergoing. The ubiquitin of pigeon obtained in the library as a full-length cDNA showed similar characteristics with ubiquitin of other species. Ubiquitin is one of the most conserved proteins known in eukaryotic cells and plays a pivotal role in many aspects of eukaryota [28, 29]. It can be expressed as polyubiquitin, or as a fusion with other proterins that are usually 52 aa or 76-81 aa ribosomal proteins [30]. In the sequence of pigeon ubiquitin (Fig. 5), the four nucleotide similarities repeat units: U1 and U2 show 94.22 % nucleotide identity; U3 has 17 different nucleotides from U1 and they have 92.54% identity; U4 and U1 are 92.11 % homologous. The nucleotide mutations among the four repeats are at the second and/or third nucleotide positions that are silent variations. Interestingly, there were 7 c/t transitions at the first nucleotide position but there were silent variations as well. It is unclear if this is due to experimental error or if it is truly a specialized change in this species.
In the text, to express the pigeon ubiquitin protein, DNA encoding mature protein was inserted between EcoR I and Xho I sites of pET28a+ vector. Under the control of T7 inducible promoter, the vector used is a fusion expression vector designed for maximizing the yield of proteins fused with 6-His·tag, under the control of T7 inducible promoter. Due to a thrombin cleavage site designed downstream of His·tag, the expressed protein was then easy to be separated from the His·tag fusion protein. Because of a high level of fusion expression, recombinant Ubiquitin protein was aggregated into a form of insoluble inclusion bodies losing their biological activity. Therefore, solubilizing, purifying, and refolding expressed protein would be needed to obtain a bioactive ubiquitin protein for further activity studies. Solubilizing Ubiquitin-His fusion protein with a proper method and obtaining purified ubiquitin separated from fused protein are being conducted in our laboratory.
Screening libraries by the probe methods is time-consuming and tedious, and cDNA with uncompleted 5′translate region (5′UTR) is frequently obtained [31]. The PCR-based method seems to be the most simple and efficient way to clone genes. To improve the efficiency, SMART libraries combined with RACE methods were used in our experiments. Using the λ phage DNA as template, two gene specific primers designed according to the sequences of the gene fragment and two primers of the λ phage vector arm were used to amplify the cDNA 5′and 3′end separately. We have cloned full-length cDNA of pigeon UBE2 using the above strategies. The result suggested that a combination of the library and RACE is a rapid and effective method for cloning full-length cDNA. 3 copies of the addl motif (ATTTA) were found in the 3′-UTR of the pigeon UBE2 cDNA. The addl motif of mRNA is implicated in the post-transcriptional control of mRNA stability and is found in a number of transiently expressed mRNAs [32,33]. Among the 13 UBE2 mRNA sequences analyzed, pigeon, pig and cattle have the same number of ATTTA repeats at the 3′ end, which suggests they have similar stabilities in vivo.
In some cases, due to undesired gene specific primers, it might difficult to clone cDNA using the above strategies. In this condition, screening libraries by probe would be a better method. To get rid of this problem, it is wise to use fewer PCR cycles with high fidelity and efficient Taq DNA polymerase and repeat sequencing for cloning high fidelity cDNA.
In conclusion, we are the first to construct a high quality pigeon cDNA library using the SMART method. A number of genes were expressed in the pituitary gland of the laying pigeon. Pigeon ubiquitin was successfully expressed in E.coli. Further demonstration of the functions of genes discovered in the current study will add great value to the understanding of the reproductive biology of the pigeon.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Liu HK, Bacon WL. Changes in egg production rate induced by progesterone injection in broiler breeder hens. Poult Sci. 2005;84(2):321-7
2. Liu HK, Long DW, Bacon WL. Concentration change patterns of luteinizing hormone and progesterone and distribution of hierarchical follicles in normal and arrested laying turkey hens. Poult Sci. 2001;80(10):1509-18
3. Liu HK, BaconW L. Effect of chronic progesterone injection on egg production in Japanese quail. Poult Sci. 2004;83(12):2051-8
4. Chen WH, Li DJ, Chen J. Role of serotonin in the broodiness in SI-JI goose. Journal of Nanjing Agriculture University. 1994;17(2):75-8
5. Sharp PJ, Scanes CG, Williams JB, Harvey S, Chadwick A. Variations in concentrations of prolactin, luteinizing hormone, growth hormone and progesterone in the plasma of broody bantams (Gallus domesticus). J. Endocrinol. 1979;80:51-7
6. Yen CF, Lin HW, Hsu JC, Lin C, Shen TF, Ding ST. The expression of pituitary gland genes in laying geese. Poult Sci. 2006;85:2265-9
7. Leclerc B, Zadworny D, Bedecarrats G, Kuhnlein U. Development of a real-time (Q) PCR assay to measure variation in expression of prolactin receptor mRNA in the hypothalamus and pituitary gland during late embryogenesis in turkeys and chickens. Gen Comp Endocrin. 2007;15(2):319-25
8. Wong EA, Silsby JL, Ishii S, Halawani MEEl. Pituitary luteinzing hormone and prolactin messenger ribonucleic acid levels are inversely related in laying and incubating turkey hens. Biol Reprod. 1992;47:598-602
9. Bacon WL, Liu HK. Progesterone Injection and Egg Production in Turkey Hens. Biol Reprod. 2004;71(3):878-86
10. Laura EE, Wilfrid C, Michael M, Sultan AJ, Xiaofei W, Larry AC, Tom EP. Gene expression profiling during cellular differentiation in the embryonic pituitary gland using cDNA microarrays. Physiol. Genomics. 2006;25:414-25
11. Weissman AM. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001;2:169-78
12. Strous GJ, Gent J. Dimerization, ubiquitylation and endocytosis go together in growth hormone receptor function. FEBS Lett. 2002;529:102-9
13. Hicke L. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2001;2:195-201
14. Gayathri S, Bentley V, Chellappagounder T, Christopher JC, Alexander P, Suresh KKG, Elizabeth MJ, Charles VC, Vincent G, Luqin D, Stuart JF, Serge YF. Prolactin stimulates ubiquitination, initial internalization, and degradation of its receptor via catalytic activation of Janus kinase 2. Endocrinology. 2008;196:R1-R7
15. Bebington C, Doherty FJ, Ndukwe G, Fleming SD. The progesterone receptor and ubiquitin are differentially regulated within the endometrial glands of the natural and stimulated cycle. Mol. Hum. Reprod. 2000;6(3):264-8
16. Leung DW, Spencer SA, Cachianes G, Hammonds RG, Collins C, Henzel WJ, Barnard L, Waters MJ, Wood WI. Growth hormone receptor and serum binding protein purification, cloning and expression. Nature. 1987;330:537-44
17. Strous GJ, van Kerkhof P, Govers R, Ciechanover A, Schwartz AL. The ubiquitin conjugation system is required for ligandinduced endocytosis and degradation of the growth hormone receptor. EMBO J. 1996;15:3806-12
18. Barnes WM. PCR amplification of up to 35kb DNA with high fidelity and high yield fromă bacteriophage template. Proc Natl Acad Sci USA. 1994;91(6):2216-20
19. Chenchik A, Zhu YY, Diatchenko L, Li R, Hill J, Siebert PD. Generation and use of high-quality cDNA from small amounts of total RNA by SMART PCR. Gene cloning and analysis by RT-PCR. MA: BioTechniques Books. 1998:305-19
20. Hershko A. Ubiquitin: roles in protein modification and breakdown. Cell. 1983;34:11-2
21. Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNA from rare transcripts: Amplification using a single gene-specific Oligonucletide primer. Proc Natl Acad Sci USA. 1988;85(23):8998-9002
22. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876-82
23. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95-8
24. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596-9
25. Draper MP, August PR, Connolly T, Packard B, Call KM. Efficient cloning of full-length cDNAs based on cDNA size fractionation. Genomics. 2002;79(4):603-7
26. Wiemann S, Mehrle A, Bechtel S, Wellenreuther R, Pepperkok R, Poustka A. CDNAs for functional genomics and proteomics: The German consortium. CR Biol. 2003;326(10-11):1003-9
27. Urmenyi TP, Bonaldo MF, Soares MB, Rondinelli E. Construction of a normalized cDNA library for the Trypanosoma cruzi genome project. J. Eukaryot Microbiol. 1999;46(5):542-4
28. Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695(1-3):189-207
29. Staub O, Rotin D. Role of ubiquitylation in cellular membrane transport. Physiol Rev. 2006;86(2):669-707
30. Jentsch S, Seufert W, Hauser HP. Genetic analysis of the ubiquitin system. Biochim Biophys Acta. 1991;1089(2):127-39
31. D'Alessio JM, Gerard GF. Second-strand cDNA synthesis with E.coli DNA polymerase I and RNase H: the fate of information at the mRNA 5'terminus and the effect of E. coli DNA ligase. Nucleic Acids Res. 1988;16(5):1999-2014
32. Nakagawa O, Ogawa Y, Itoh H, Suga S, Komatsu Y, Kishimoto I, Nishino K, Yoshimasa T, Nakao K. Rapid transcriptional activation and early mRNA turnover of brain natruretic peptide in cardiocyte hypertrophy. Clin Invest. 1995;96:1280-7
33. Loannidis P, Havredaki M, Courtis N, Trangas T. In vivo generation of 3' and 5' truncated species in the process of c-mys mRNA decay. Nucleic Acids Res. 1996;24:4969-77
Author contact
![]() Correspondence to: Qin-de Wang, the key laboratory of animal breeding and genetics & reproduction, Shanxi Agriculture University, Taigu, 030801, Shanxi, China. Phone/Fax: +86-0354-6288153. E-mail: wqd1952edu.cn
Correspondence to: Qin-de Wang, the key laboratory of animal breeding and genetics & reproduction, Shanxi Agriculture University, Taigu, 030801, Shanxi, China. Phone/Fax: +86-0354-6288153. E-mail: wqd1952edu.cn

 Global reach, higher impact
Global reach, higher impact