10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(5):500-516. doi:10.7150/ijbs.5.500 This issue Cite
Review
The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass
1. Biorefining Research Initiative, Lakehead University, 955 Oliver Rd, Thunder Bay, Ontario P7B5E1, Canada.
2. Department of Biology, Lakehead University, 955 Oliver Rd, Thunder Bay, Ontario P7B5E1, Canada.
Received 2009-6-5; Accepted 2009-7-21; Published 2009-7-29
Abstract
Lignocellulosic biomass is a renewable and abundant resource with great potential for bioconversion to value-added bioproducts. However, the biorefining process remains economically unfeasible due to a lack of biocatalysts that can overcome costly hurdles such as cooling from high temperature, pumping of oxygen/stirring, and, neutralization from acidic or basic pH. The extreme environmental resistance of bacteria permits screening and isolation of novel cellulases to help overcome these challenges. Rapid, efficient cellulase screening techniques, using cellulase assays and metagenomic libraries, are a must. Rare cellulases with activities on soluble and crystalline cellulose have been isolated from strains of Paenibacillus and Bacillus and shown to have high thermostability and/or activity over a wide pH spectrum. While novel cellulases from strains like Cellulomonas flavigena and Terendinibacter turnerae, produce multifunctional cellulases with broader substrate utilization. These enzymes offer a framework for enhancement of cellulases including: specific activity, thermalstability, or end-product inhibition. In addition, anaerobic bacteria like the clostridia offer potential due to species capable of producing compound multienzyme complexes called cellulosomes. Cellulosomes provide synergy and close proximity of enzymes to substrate, increasing activity towards crystalline cellulose. This has lead to the construction of designer cellulosomes enhanced for specific substrate activity. Furthermore, cellulosome-producing Clostridium thermocellum and its ability to ferment sugars to ethanol; its amenability to co-culture and, recent advances in genetic engineering, offer a promising future in biofuels. The exploitation of bacteria in the search for improved enzymes or strategies provides a means to upgrade feasibility for lignocellulosic biomass conversion, ultimately providing means to a 'greener' technology.
Keywords: bacteria, bioconversion, cellulases, cellulosome
Introduction
The combustion of petroleum-based fossil fuels has become a concern with respect to global climate change due to accelerated carbon emissions [1]. Burning of fossil fuels has also created a concern for unstable and uncertain petroleum sources, as well as, the rising cost of fuels [2]. These concerns have shifted global efforts to utilize renewable resources for the production of a 'greener' energy replacement which can also meet the high energy demand of the world. The Canadian renewable fuel standard has been raised so that fuel will contain 5% ethanol by 2010 [3]; the US Environmental Protection Agency raised their renewable fuel standard to 10.21% ethanol mixed fuels by 2009 [4]; while, the current mandate for mixing ethanol in fuel for Brazil is 25% (set in 2007) [5].
Currently, the US and Brazil are leaders in the production of starch/sugar-based fuels from corn and sugarcane crops, respectively. This is the production of first generation fuel from food-crop sugars using conventional technologies; however, starch raw materials will not be sufficient enough to meet increasing demand and are a controversial resource for bioconversion [6]. Also the reduction in greenhouse gases is low for starch-based ethanol and thus, second generation fuels based on non-edible crops (lignocellulosic biomass), is gaining immense global and scientific attention.
Lignocellulosic biomass, ('plant biomass'), is a great potential resource for the production of biofuels because it is largely abundant, inexpensive and production of such resources is environmentally sound. Agricultural residues are a great source of lignocellulosic biomass which is renewable, chiefly unexploited, and inexpensive. Such resources include: leaves, stems, and stalks from sources such as corn fibre, corn stover, sugarcane bagasse, rice hulls, woody crops, and forest residues. Also, there are multiple sources of lignocellulosic waste from industrial and agricultural processes, e.g., citrus peel waste, sawdust, paper pulp, industrial waste, municipal solid waste, and paper mill sludge. In addition, dedicated energy crops for biofuels could include perennial grasses such as Switchgrass and other forage feedstocks such as Miscanthus, Bermuda grass, Elephant grass, etc [6]. Approximately 70% of plant biomass is locked up in 5- and 6-carbon sugars. These sugars are found in lignocellulosic biomass, which is comprised of mainly cellulose (a homologous polymer comprised of long chains of glucose); less so, hemicelluloses (heterologous polymer of 5- and 6-carbon sugars); and least of all lignin (a complex aromatic polymer). The major component cellulose, is a homopolysaccharide comprised of glucose units, linked by β-(1→4) glycosidic bonds. Cellobiose is the smallest repeating unit of cellulose and can ultimately be converted into glucose. Hemicellulose is a heterogeneous polymer, which varies in composition from plant to plant and within different parts of the same plant. It is made up of mainly pentoses (D-xylose, D-arabinose), hexoses (D-mannose, D-glucose, D-galactose) and sugar acids. In hardwoods hemicellulose contains mainly xylans, while in softwood mainly glucomannans are present. Hydrolysis of hemicelluloses requires various types of enzymes. Briefly, xylan degradation requires endo-1-4,-β-xylanase, β-xylosidase, α-glucuronidase, α-L-arabinofuranosidase, as well as acetylxylan esterases. In glucomannan degradation β-mannanase and β-mannosidase are required to cleave the polymer backbone.
There are several advantages for the production of biosolvent fuels such as bioethanol: 1) produced from a variety of raw materials; 2) it is non-toxic; and 3) easily introduced into the existing infrastructure [7]. However, the path to sustainable and economically feasible biofuels is hampered. There are a few major bottlenecks with the current production of biofuels; one being, there is a lack of biocatalysts that can work efficiently and inexpensively at high temperatures and/or low pH conditions used in the bioconversion of lignocellulosic material to bioethanol. Moreover, there is a great need for cost-effective fermentation of derived sugars from cellulose and hemicellulose. Currently, industrial bioconversions of lignocellulose requires the application of high temperature and acidic or sometimes basic conditions to break down lignin, decrease crystallinity, increase pore volume and solubilise cellulose and hemicellulose to allow enzymatic hydrolysis of target polysaccharides [8]. This process is both expensive and inefficient. It is therefore important that enzymes be stable and active at high temperatures and/or low or high pH conditions.
Additionally, industrial bioconversion of lignocelluloses to ethanol occurs in multiple steps, where hydrolyzing enzymes are added after pre-treatment of the lignocelluloses (saccharification) and then in an additional step, microorganisms capable of fermentation are added to the resulting monosaccharides generated during hydrolysis to ferment sugars to bioethanol. The multiplicity of the current biorefining process makes it both time-consuming and costly. By combining saccharification with fermentation in a process referred to as consolidated bioprocessing (CBP) or saccharification-co-fermentation (SCF), using a whole-cell(s) based approach, costs of fermentation and hydrolysis could be reduced [9,10]. Some additional rate-limiting steps in the bioconversion of lignocelluloses are the crystalline recalcitrance of cellulose and the limited number of cellulases. That is, all cellulolytic strains identified are low in one or more type of glycoside hydrolases (GH) required for efficient cellulose hydrolysis (endo-/exo-glucanases, β-glucanases). In attempts to improve the feasibility of the bioconversion of lignocellulose to biofuel, enzymes must have high adsorption capabilities, high catalytic efficiencies, high thermal stability and low end-product inhibition.
Both fungi and bacteria have been heavily exploited for their abilities to produce a wide variety of cellulases and hemicellulases. Most emphasis has been placed on the use of fungi because of their capability to produce copious amounts of cellulases and hemicellulases which are secreted to the medium for easy extraction and purification. In addition, the enzymes are often less complex than bacterial glycoside hydrolases and can therefore be more readily cloned and produced via recombination in a rapidly growing bacterial host such as E. coli. However, the isolation and characterization of novel glycoside hydrolases from Eubacteria are now becoming widely exploited. There are several reasons for these shifts, for one, bacteria often have a higher growth rate than fungi allowing for higher recombinant production of enzymes. Secondly, bacterial glycoside hydrolases are often more complex and are often expressed in multi-enzyme complexes providing increased function and synergy. Most importantly, bacteria inhabit a wide variety of environmental and industrial niches, which produce cellulolytic strains that are extremely resistant to environmental stresses. These include strains that are thermophilic or psychrophilic, alkaliphilic or acidiophilic, and, strains that are halophilic. Not only can these strains survive the harsh conditions found in the bioconversion process, but they often produce enzymes that are stable under extreme conditions which may be present in the bioconversion process and this may increase rates of enzymatic hydrolysis, fermentation, and, product recovery. Researchers are now focusing on utilizing, and improving these enzymes for use in the biofuel and bioproduct industries.
This review will focus on aspects rarely covered by other reviews, such as bacterial screening techniques, and new bacterial cellulases by comparing different cellulase-producing bacteria. Moreover, it will examine how these rare cellulases can help overcome some of the major bottlenecks in the biofuel industry. In addition, this review will address how some novel bacterial strategies in biotechnology can advance the growing field of biorefining.
Bacterial cellulases
Cellulases are comprised of independently folding, structurally and functionally discrete units called domains or modules, making cellulases modular [11]. A typical free cellulase is composed of a carbohydrate binding domain (CBD) at the C-terminal joined by a short poly-linker region to the catalytic domain at the N-terminal. There are only two modes of action for the hydrolysis of cellulose by cellulases, either inversion or retention of the configuration of the anomeric carbon. At least two amino acids with carboxyl groups located within the active site catalyze the reaction by acid-base catalysis. The commonly described mode of action for cellulases on polymers is either exo- or endo-cleavage, and all cellulases target the specific cleavage of β-1,4-glycosidic bonds [12]. Using this classification system, cellobiohydrolases (exoglucanases) were classified as exo-acting based on the assumption that they all cleave β-1,4-glycosidic bonds from chain ends. As well, those enzymes truly exo-acting often have a tunnel-shaped closed active site which retains a single glucan chain and prevents it from re-adhering to the cellulose crystal [13-15]. While endoglucanases on the other hand, are often classified as endo-acting cellulases because they are thought to cleave β-1,4-glycosidic bonds internally only and appear to have cleft-shaped open active sites. Endoglucanase are active on amorphous regions of cellulose and thus their activity can be assayed using soluble cellulose substrates; i.e., the carboxymethylcellulase assay (CMCase). However, there is now supporting evidence that some cellulases display both modes of action, endo- and exo- [16]. Thus classification has changed; cellobiohydrolases (exoglucanases) are described as active on the crystalline regions of cellulose; whereas, endoglucanases are typically active on the more soluble amorphous region of the cellulose crystal. There is a high degree of synergy seen between cellobiohydrolases (exoglucanases) and endoglucanases, and it is this synergy that is required for the efficient hydrolysis of cellulose crystals.
CBD is the most common accessory module of cellulases and there are 54 distinct families [17]. The major function of CBDs is to deliver its resident catalytic domain to crystalline cellulose. Binding brings the catalytic domain into close contact with the crystalline cellulose for efficient hydrolysis. Binding of the cellulase via CBD is extremely stable, yet still allows the enzymes to diffuse laterally across the surface of the substrate and in some cases CBD has also been shown to catalyze the disruption of noncovalent interactions between cellulose chains of crystalline cellulose. Some other CBDs are preferential to binding noncrystalline cellulose [18-20].
Interestingly, the family 9 cellulase of aerobic Thermomonospora fusca has a family IIIc CBD with a different function that gives family 9 cellulases their distinctive theme [21]. This unique CBD does not bind crystalline cellulose but instead directly assists the catalytic function of the cellulase by binding a single cellulose chain and ultimately feeding this chain into the active site of the enzyme [22,23]. This contributes to the overall processivity of the family 9 cellulase, that is, the sequential cleavage of the cellulose chain. Additionally, a second type of CBD must be associated with this cellulase to bind it to the crystalline cellulose. Moreover, the family 9 cellulase of T. fusca provides strong evidence for enzymes that can exhibit both endoglucanase and exoglucanase activities accentuating the equivocacy of these terms.
The products of exoglucanases and cellobiohydrolases, that are cellobiose and cellodextrans, respectively, are inhibitory to their activity. Thus, efficient cellulose hydrolysis requires the presence of β-glucosidases to cleave the final glycosidic bonds producing glucose. Typically cellobiose and cellodextrins are taken up by the bacteria and internally cleaved via cellodextrin phosphorylases or cellobiose phosphorylases to create glucose monophosphate, which is energetically favoured. Some bacteria also produce inter- or extra-cellular β-glucosidases to cleave cellobiose and cellodextrins and produce glucose to be taken up by or assimilated by the cell.
Screening and isolation of cellulase-producing bacteria
Over the years, culturable, cellulase-producing bacteria have been isolated from a wide variety of sources such as composting heaps, decaying plant material from forestry or agricultural waste, the feces of ruminants such as cows, soil and organic matter, and extreme environments like hot-springs, to name a few [24]. Screening for cellulase production can be done by enrichment growth on microcrystalline cellulose as a sole source of carbon, followed by the extraction of 16S rDNA/RNA to determine the molecular community structure of the environment and analyze whether families containing cellulase-producing species are present. Strains with cellulase potential can be isolated by subculturing from the enrichment culture on cellulose as a sole carbon source. This method was used to identify cellulase-producing bacteria in the deep subsurface of the Homstake gold mine, Lead, South Dakota, USA [25].
Moreover, efficient plate-screening methods are a prerequisite. Screening for bacterial cellulase activity in microbial isolates is typically performed on carboxymethylcellulose (CMC) containing plates [26]. This method can be timely and zones of hydrolysis are not easily discernable. Recently, Kasana and colleagues found that Gram's iodine for plate flooding in place of hexadecyltrimethyl ammonium bromide or Congo red, gave a more rapid and highly discernable result [27]. However, plate-screening methods using dyes are not quantitative or sensitive enough due to poor correlation between enzyme activity and halo size. This has sparked the development of short cellooligosaccharides possessing modified reducing terminal with chromogenic/fluorogenic groups due to achievement of higher sensitivity and quantification. Several examples such as fluorescein, resorufin and 4-methylumbelliferone are well-established [28-33]. A major limitation of the incorporation of fluorescent substrates into agar plates is the tendency for hydrolysis products to diffuse widely and therefore these kinds of compounds are not as readily used. Today, new substrates, 2-(2'-benzothiazolyl)-phenyl (BTP) cellooligosaccharides with degree of polymerization (d.p.) 2-4 (BTPG2-4) were synthesized for the screening of microbial cellulolytic activity in plate assays. The usefulness of the 2-(2'-benzothiazolyl)-phenyl substrates was shown during purification of the Bacillus polymyxa cellulolytic complex, which consists of at least three types of the enzymes: cellobiohydrolase, endo-β-D-glucanase and β-glucosidase [34].
Nonetheless, these methods are mainly limited to culturable cellulase-producing bacteria and the full cellulase-potential of the site (culturable and nonculturable microorganisms) is not being fully examined. Researchers have now focused on the identification and exploitation of cellulase genes from unculturable microorganisms found in more extreme environments in hopes that the enzymes isolated will be novel and have specific applications in the biorefining industry due to a higher resistance to harsh environmental conditions. These enzymes may contribute to a decrease in the current cost of bioconversion of lignocellulose to ethanol by being more resistant to acids or bases used and by retaining activity at higher temperatures. To identify novel cellulases from all species present, culturable and nonculturable in a swift manner, a metagenomic clone library should be created and then functionally screened; the key feature of this technique is the functional screening. Screening requires knowledge or rather an objective for the isolation of a specific enzyme with specific activity whether it be exoglucanases with activity on microcrystalline cellulose or endoglucanases with activity on soluble cellulose such as carboxymethyl cellulose (CMC). Depending on the objective different assays can be used to screen the recombinant proteins produced in E. coli. This is a quick and efficient method to screen a wide population which has been used recently to identify novel cellulase-producing bacteria from the rumen of buffalo and from pulp and paper mill effluent sediments by screening for crystalline and soluble cellulase activity [35,36]. Using different screening methods, a variety of cellulases with novel characteristics have been identified and are still being identified to date.
The isolation and identification of cellulases has been limited in the past to culturable microorganisms. However, recent advances in molecular techniques, such as the creation of metagenomic libraries will widen the pool of cellulolytic enzymes available for biofuel research. This approach will allow exploition of cellulases and related enzymes from otherwise unculturable microorganisms which may produce enzymes with novel characteristics.
Novel cellulase producing bacteria
Isolation, screening and selection have favoured the discovery of several novel cellulase-producing bacteria from a wide variety of environments as previously discussed. Due to the vast diversity among bacteria the identification of novel cellulases remains a currently explored route to the improvement of biorefining industries. Here will be discussed briefly some of the new bacterial isolates and/or newly discovered and characterized cellulases, with potential use in the biorefining industry.
Recently, the bacterial strain B39, previously isolated from poultry manure compost in Taichung, Taiwan, was identified through 16S rRNA gene sequencing and phylogentic analysis to be a novel cellulose-degrading Paenibacillus sp. strain. A high-molecular weight (148 kDa) cellulase, possessing both CMCase and Avicelase activities, was found to be secreted by this isolate into the media. The CMCase activity of the newly isolated cellulase was much higher than the activity on Avicel or filter paper and this cellulase was found to have maximum CMCase activity at 60°C, pH 6.5. Due to the promising thermostability and slight acidic tolerance of this enzyme, it has good potential for industrial use in the hydrolysis of soluble cellulose as well as activity on microcrystalline sources of cellulose [37]. Furthermore a novel cellulase-producing Paenibacillus campinasensis BL11 was isolated in 2006, from black liquor of brownstock at washing stage of the Kraft pulping process. This black liquor environment is strongly alkaline and therefore highly unfavourable to bacterial growth, isolation of a cellulase-degrading species from this environment provides plausibility that the enzymes produced by such a species could be tolerant to some of the harsh conditions used in the different pretreatments of lignocellulosic biomass. P. campinasensis BL11 is a thermophilic, spore-forming bacterium which was found to grow between 25 and 60°C over a wide range of pH. Optimal growth is around neutral pH, at 55°C. This isolate used a variety of saccharides and polysaccharides and produced multiple extracellular saccharide-degrading enzymes including: a xylanase, two cellulases, a pectinase and a cyclodextrin glucanotransferase. The physiological properties of this strain and the vast number of free glycosyl hydrolases produced give this strain potential for use in the biorefining industry [38].
More recently, a thermostable cellulase was found in newly isolated Bacillus subtilis DR, extracted from a hot spring. The high temperature environment allowed for the production of a thermostable endocellulase CelDR with an optimum temperature at 50°C. It was found to retain 70% of its maximum activity (CMCase) at 75°C after incubation for 30 minutes. This strain offers a potentially more valuable thermostable enzyme for the biorefining industry due to extreme heat tolerance [39]. Cultivation of thermophiles offers several advantages, it reduces the risk of contamination, reduces viscosity thus making mixing easier, and leads to a high degree of substrate solubility while reducing the cost of cooling. This is a greatly sot after property for cellulases in industrial applications like the bioconversion of lignocellulose. Also recently, a novel thermophilic, cellulolytic bacterium was isolated from swine waste and identified as Brevibacillus sp. strain JXL. It was found to use a broad spectrum of substrates such as crystalline cellulose, CMC, xylan, cellobiose, glucose and xylose. The enzymes appeared to retain 50% of their activity after 1h at 100°C, making them highly thermostable [40]. Furthermore, a salt-activated endoglucanase was recently isolated from another Bacillus strain, alkaliphilic Bacillus agaradhaerens JAM-KU023 which was shown to have increased optimal thermostability from 50°C to 60°C with the addition of 0.2M NaCl and optimal pH range from 7-9.4 [41].
In addition, bacteria are capable of producing more complex protein structures supporting enzymes for the hydrolysis of cellulose, such as the cellulosome, xylosome and bifunctional or multifunctional enzymes which are currently gaining a lot of attention. If these enzymes can be recombinantly produced on mass or produced in situ by the bacterial strains naturally encoding them, then they may have great potential in improving the cost of hydrolysis for the production of biofuels by reducing the need for production of multiple enzymes for efficient hydrolysis. For example, a bifunctional endoglucanase/endoxylanase was isolated from Cellulomonas flavigena providing potential for use in different industrial processes such as biofuel production. This bifunctional enzyme was found to have optimum cellulase and xylanase activity at pH 6 and 9, respectively, with a general optimum temperature at 50°C [42]. Similarly, in 2007, a multifunctional enzyme was found to be produced by Terendinibacter turnerae T7902, which is a bacterial symbiont isolated from the wood-boring marine bivalve Lydrodus pedicellatus. This CelAB was found to have two catalytic and two carbohydrate-binding domains. It binds both cellulose and chitin and possesses cellobiohydrolase and beta-1,4(3) endoglucanase activity allowing it to degrade multiple complex polysaccharides. This enzyme is marginally acid-tolerant at an optimum pH of 6 and mesophilic with a temperature optimum of 42°C. Additionally, this enzyme was able to reduce viscosity of CMC approximately 40% after 25 minutes, displaying promising characteristics for the biofuel industry [43]. All of these recently isolated enzymes and many more provide the framework needed to characterize and build highly efficient hydrolysis systems to be used in the biorefining industry. Isolation and characterisation of cellulase-producing bacteria will continue to be an important aspect of biofuel research.
Improvement of bacterial cellulases
Despite the broad spectrum of cellulases being isolated, no single enzyme is completely suitable as it is, for the hydrolysis of cellulose in the biorefining industry. However, these enzymes offer a good starting point for the improvement of cellulases in steps towards enhancing the overall economics of biofuel production. Typically, the use of protein engineering technology has been directed towards the study of cellulase catalytic function. Mutagenesis has provided a means for studying the role of different amino acids within the catalytic domain. More recently, modifications to bacterial cellulases through the use of protein engineering is taking a stage in the production of efficient hydrolytic enzymes used in a broad scope of industries and includes targeting structural amino acids, beyond amino acids in the catalytic site. There are two major strategies for the improvement of a cellulase or cellulase component: 1) rational design and 2) directed evolution.
Rational design
Rational design involves 1) choice of a suitable enzyme, 2) identification of the amino acid sites to be changed, based usually on a high resolution crystallographic structure, and 3) characterization of the mutants [44]. The use of rational design requires detailed knowledge of the protein structure: what makes the catalytic site active, a theoretical molecular structure-based model of the protein, and most ideally structure-function relationship. With at least part of this knowledge, modification of amino acid sequence can be achieved using site-directed mutagenesis, in some cases elements of secondary structure can be altered and even exchange of whole domains and/or generation of fusion proteins [44]. However, the vast majority of enzymes do not have structural information available. Despite the fact that some target cellulases are well characterized, the molecular mutation required for the desired function cannot always be achieved [45]. To date, there are no general rules for site-directed mutagenesis strategies for the enhancement of cellulase activity and it therefore remains at present in a trial-like state. There is still limited knowledge about the properties of insoluble cellulosic substrates which differ based on pretreatment technologies; the interactions between cellulases and cellulose; and, the synergistic relationship among cellulase components. These factors hamper the ability of using rational design for improving bacterial cellulases for the biofuel industry.
To date, there are a few reports where site-directed mutagenesis was used to increase the catalytic activity of a bacterial cellulase. Mahadevan et al. [46] subjected the amino acids around the active site of endoglucanase Cel5A from Thermotoga maritima creating the N147E mutant which displayed 10% higher activity than the wild-type Cel5A. This group also showed a correlation between binding ability and the activity of the enzyme. By binding two CBDs, one from Trichoderma reesei and the other from Clostridium stercorarium, to Cel5A this CBD-engineered Cel5A displayed 14 to18-fold higher hydrolytic activity towards Avicel [46]. In addition, the mutation of the conserved residue F476 to Y476 from Cel9A of Thermobifida fusca displayed 40% improved activity in assays with soluble and amorphous cellulose such as CMC and swollen cellulose. This was achieved through the integration of computer modeling with site-directed mutagenesis [47]. Furthermore, enzymatic activity was increased by 80% for a mutant Cel5Z endoglucanase of Pectobacterium chrysanthemi compared to the wild-type. However, this mutant enzyme was created by the use of a nonsense mutation which removed the C-terminal region creating a truncated Cel5Z containing 280 amino acids compared to the native Cel5Z which has 426 amino acids. Without the CBD this enzyme would not be efficient for hydrolysis of crystalline cellulose but could offer potential for solubilised cellulose [48]. Likewise, the Cel5Z::Omega mutant Cel5Z of P. chrysanthemi hydrolyzed CMC with 1.7-fold higher activity than the intact Cel5Z cellulase [49]. Similarly, a complex multifunctional enzyme Cel44C-Man26A secreted by Paenibacillus polymyxa GS01, was truncated from 1352 amino acids down to 549 amino acids. The truncated enzyme maintained cellulase, xylanase, mannanase and lichenase activities but on the contrary activity was not enhanced, however truncation allows the recombinant production of this multifunctional enzyme with more ease [50].
Furthermore, Baker and colleagues (2005) were able to design and mutate Tyr 245 of Cel5A of Acidothermus cellulolyticus to Gly and this was found to decrease the inhibition of the endoglucanase by cellobiose. Solubilised sugars were hydrolyzed 40% greater by the mutant Cel5A (with T. reesei cellobiohydrolase-I) compared to the wild-type. Structural and kinetic studies correlated increased enzymatic activity to reduced product inhibition [51]. In addition, mutation of a single active-site cleft tyrosyl residue to a glycyl residue significantly changed the mixture of products released from phosphoric acid-swollen cellulose (PSC) from the catalytic domain of the endoglucanase-I from A. cellulolyticus. The percentage of glucose found in the product stream was approximately 40% greater for the Y245G mutant they created compared to the wild-type enzyme [52]. Bacterial cellulases improved by rational design are summarized in Table 1.
Classical chemical mutagenesis does not require knowledge of the protein structure and selection of desired traits becomes a guiding force for the development of improved enzymes. The maximum product yield of an endoglucanase from Cellulomonas biazotea mutant 51 was 1.5- to 2.5-fold more than was produced by the wild-type cells and was twice that reported by previous researchers on CMC [53]. Similarly, the highest productivity of β-glucosidase by a mutant of C. biazotea was 2.5-fold more than that of the parent organism and the mutation stabilized the enzyme thermophilically [54]. This type of random mutation, although more crude, is indicative of the ideas directed evolution was based upon.
Directed Evolution
Contrary to rational design, irrational design or directed evolution is an approach to non-informational protein engineering which utilizes the power of natural selection to evolve proteins and select for those with desired traits. Specifically, directed evolution requires the use of DNA techniques such as error-prone PCR (epPCR) and DNA shuffling to randomly generate a large library of gene variants. It has a great advantage over rational design because it is independent of enzyme structure and of the interactions between enzyme and substrate. Nonetheless, a major challenge of this method is developing a means to accurately evaluate the performance of mutants generated by recombinant DNA techniques and the selection of high-performance mutants. Screening methods typically include such tests as CMC agar with Congo red staining or the use of chromogenic or fluorogenic substrates, as previously mentioned. The more quantitative these methods are the greater chance of improving the directed evolution for improving bacterial cellulases. The success of directed evolution relies on a large library of gene variants, the larger, the greater the chance of mutants with desired properties.
The method of directed evolution was used to improve the thermal stability of Clostridium cellulovorans cellulosomal endoglucanase (EngB) in vitro by DNA recombination with non-cellulosomal endoglucanase EngD. The screening was done using CMC agar and staining with Congo red [55]. Further, DNA shuffling was used to create a library of mutated endoglucanases from B. subtilis. Interestingly, a bacterial surface display method was used to selectively screen for variants with improved activity on CMC agar with Congo red staining. By fusing the genes with the ice nucleation protein (Inp) the resulting fusion proteins would be displayed on the bacterial cell surface for easy screening [56].
Furthermore, directed evolution using epPCR and family shuffling was used to successfully increase thermal stability of β-D-glucosidases from Paenibacillus polymyxa, desired mutants were screened using a chromogenic substrate [57,58]. Likewise, the catalytic activity of 1,4-β-D-glucan glucohydrolase A from Thermotoga neapolitana was improved using epPCR to generate the gene variant library [59]. While additionally, catalytic activity of a hyperthermostable β-glucosidase CelB from Pyrococcus furiosus was improved by family shuffling. Catalytic activity was increased by 3- and 5-fold compared to the wild-type; screening for successful mutants was accomplished by a chromogenic substrate [60]. Finally, directed evolution of a glycosynthase from Agrobacterium sp. increased its catalytic activity dramatically and expanded its substrate usage, the successful mutants were screened by fluorogenic substrate [61]. Continued advancements in technology may increase the ease of using rational design in attempts to improve cellulolytic enzymes. However, irrational design or random mutagenesis will continue to be a dominant technique to alter cellulases because there is still much to be learned about predicting protein structure and function. Some bacterial cellulases improved by rational design and directed evolution are summarized in Table 1.
A list of bacterial strains and cellulases or related enzymes from these microorganisms which have been improved using rational design or directed evolution (Modified from Percival Zhang et al., 2006).
| Bacterial Strain | Enzyme | Property Altered | Method | Reference |
|---|---|---|---|---|
| Rational Design | ||||
| Acidothermus cellulolyticus | Endoglucanase | Type of products released | Site-directed mutation | [52] |
| Acidothermus cellulolyticus | Endoglucanase | Product inhibition | Site-directed mutation | [51] |
| Pectobacterium chrysanthami | Endoglucanase | Activity | Nonsense mutation | [48] |
| Pectobacterium chrysanthami | Endoglucanase | Activity | Insertional truncation | [49] |
| Thermobifida fusca | Processive Endoglucanase | Activity | Site-directed mutation | [47] |
| Thermotoga maritime | Endoglucanase | Activity | Site-directed mutation, CBD engineering | [46] |
| Directed Evolution | ||||
| Agrobacterium sp. | Mutated α-glucosidase | Activity | epPCR | [61] |
| Bacillus subtilis | Endoglucanase | Activity | DNA shuffling | [56] |
| Clostridium cellulovorans | Endoglucanase | Thermal stability | Family shuffling | [55] |
| Paenibacillus polymyxa | α-D-glucosidase | Thermal stability | epPCR | [58] |
| Paenibacillus polymxa | α-D-glucosidase | Thermal stability | epPCR + family shuffling | [57] |
| Pyrococcus furiousus | α-glycosidase | Activity | Family shuffling | [60] |
| Thermotoga neapolitana | α-D-glucosidase | Activity | epPCR | [59] |
Hemicellulase-producing bacteria and engineering hemicellulases
Hemicellulose is the second most abundant renewable biomass, accounting for approximately 25-35% of lignocellulosic biomass and therefore bacterial enzymes involved in its degradation have also been the focus of several hemicellulase engineering studies using either rational design or directed evolution.
Rational design has been used to improve thermostability and functionality of several hemicellulases, however to date; rational design has not been successful in directly improving enzymatic activity. Disulphide bridges were constructed, using computer modeling, in the xylanase of Bacillus circulans and in GH-AA xylanase of Thermobacillus xylanolyticus. The half life of each mutant was 69°C for 120 mins and 70°C for 80 mins, respectively. Disulphide bonds increased thermostability but did not improve activity at elevated temperatures. The number of disulphide bonds was also shown to play a key role in thermostability [62,63]. Increasing enzyme thermostability is one step towards lowering biofuel production costs.
More so, increasing the versatility of a single hemicellulolytic enzyme will have a great reduction on enzyme production cost. Lu and Feng [64], created a bifunctional xylanase by creating an optimized flexible peptide linker between β-glucanase (Glu) of Bacillus amyloliquefaciens and the xylanase (Xyl) of B. subtilis. The catalytic efficiencies of Glu and Xyl moieties increased 304-426% and 82-143%, respectively, compared to an end-end fusion of Glu and Xyl that they have previously created [65]. Similarly, Fan and colleagues [66], also using a flexible peptide linker, created a multifunctional xylan-degrading enzyme. The xylanase domain of the xylanase XynZ from Clostridium thermocellum, was fused to a dual functional arabinofuranosidase/xylosidase (DeAFc; isolated from a compost starter mixture). The resulting trifunctional enzyme was more active in the hydrolysis of natural xylans and corn stover and retained pH, temperature optima, and, kinetics of the parental enzymes [66]. Increasing the versatility of enzymes may increase activity through the synergistic action of fused enzymes and offer a greater production-cost savings.
Directed evolution, without the knowledge of enzyme structures, has been used to enhance thermostability, pH optima and specific activity of hemicellulases. A family shuffling technique referred to as degenerated oligonucleotide gene shuffling was created by Gibbs and colleagues [67], to reduce regeneration of unshuffled parental genes. One round of this technique was used after epPCR to generate a gene variant library and ultimately improve thermostability and pH optima (alkaline > 8.5 pH) of a family-11 xylanase (XynB) from Dictyoglomus thermophilum [67]. More recently, epPCR followed by 1 round of DNA shuffling was used to increase the melting temperature by 20°C for the xylanase XylA of B. subtilis. Screening of efficient variants was done using 1% oat spelt xylan and Congo red staining [68]. Consequently, epPCR has also been used to increase specific activity of xylanase Xys1 from Streptomyces halstedii JM8. By the random mutagenesis, two structural mutations were created (G133D and N148D) outside the catalytic centre. This slight structural change resulted in a 22-25% increase in specific activity of Xys1 towards xylan compared to the wild-type [69]. This study not only displays results towards creating more efficient enzymes for use in lignocellulosic biomass conversion; it also lends insight to key residues that are not directly involved in the catalytic site but play a indirect role in the active site function. Some bacterial hemicellulases improved by rational design and directed evolution are summarized in Table 2.
Rational design and directed evolution are helping to improve not only cellulases, but also hemicellulases by providing important insights about enzyme structure and function. Each contribution, no matter how large, is a step closer to improving lignocellulose biomass conversion.
A list of bacterial strains and hemicellulases from these microorganisms which have been improved using rational design or directed evolution.
| Bacterial strain | Enzyme | Property altered | Method | Reference |
|---|---|---|---|---|
| Rational design | ||||
| Bacillus circulans | Xylanase | Thermostability | Site-directed mutagenesis | [62] |
| Bacillus amyloliquefaciens Bacillus subtilis | Bifunctional: xylanase-β-glucosidase | Substrate usage | Peptide linker fusion | [64] |
| Clostridium thermocellum Anaerobic digester unknown | Trifunctional: xylanase, arabinofuranosidase/ xylosidase | Substrate Usage | Peptide linker fusion | [66] |
| Thermobacillus xylanolyticus | GH-AA xylanase | Thermostability | Site-directed mutagenesis | [63] |
| Directed evolution | ||||
| Bacillus subtilis | Xylanase XylA | Thermostability | epPCR, DNA shuffling | [68] |
| Dictyoglomus thermophilum | Xylanase XynB | Thermostability Alkalinity | DOGS, epPCR | [67] |
| Streptomyces halstedii | Xylanase Xys1 | Activity | epPCR | [69] |
Cellulosomes
Cellulosomes are multienzyme complexes produced mainly by anaerobic bacteria, many from the class clostridia. However, evidence suggests the presence of cellulosomes in at least one aerobic bacterium and a few anaerobic fungi from species such as Neocallimastix, Piromyces, and Orpinomyces [70,71]. It is speculated that several other cellulolytic bacteria may also produce cellulosomes which have yet to be described [40]. Production of cellulosomes by mainly anaerobic microorganisms is thought to be an evolutionary advantage which may counteract the low energy production by fermentation. Therefore, anaerobes produce this highly efficient multienzyme complex which allows for fine control over metabolic activities.
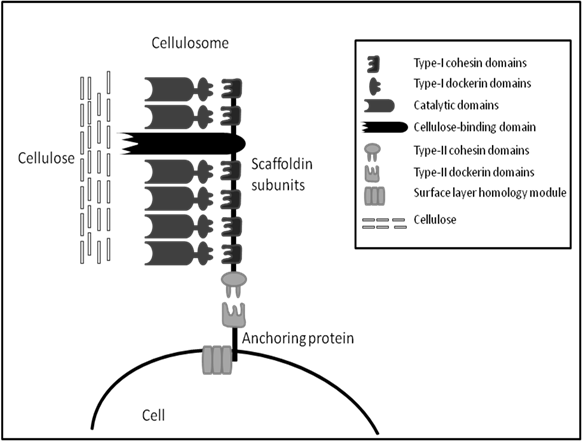
The cellulosome was first identified in 1983 from the anaerobic, thermophilic, spore-forming Clostridium thermocellum [72]. Unlike fungal cellulases, the C. thermocellum cellulase complex has very high activity on crystalline cellulose; this activity is termed “true cellulase activity” or Avicelase, characterized by its ability to completely solubilise crystalline forms of cellulose such as cotton and Avicel [73]. The cellulosome of C. thermocellum is commonly studied along with cellulosomes from the anaerobic mesophiles, C. cellulolyticum and C. cellulovorans. All cellulosomes share similar characteristics, they all contain a large distinct protein referred to as the scaffoldin which allows binding of the whole complex to microcrystalline cellulose via a nonspecific carbohydrate binding module (CBM). Also, the cellulosome scaffolding expresses type I cohesins which allow binding of a wide variety of cellulolytic and hemicellulolytic enzymes within the complex via the expression of complementary type I dockerins on enzymes. Similarly, at the C-terminal the scaffolding expresses type II cohesins which allow the binding of the cellulosomes to the cell through type II dockerins on surface layer-homology proteins (SLH) (Figure 1). The structure and function of bacterial cellulosomes have been reviewed several times elsewhere and will not be discussed in greater detail here [74-77].
The cellulosome eliminates the wasteful expenditure of energy of microorganisms continuously producing copious amounts of free enzymes along with which, the products get diluted in the bulk solution. There are several other advantages for microorganisms to naturally produce cellulosomes; specific characteristics of cellulosomes give rise to efficient cellulose hydrolysis. Firstly, synergism is optimized by the correct ratio between components, which is determined by the composition of the complex. Secondly, non-productive adsorption is avoided by the optimal spacing of the components working together in the synergistic fashion. Thirdly, competitiveness in binding to a limited number of binding sites in the biomass surface is avoided by binding the whole complex to a single site through a strong binding domain with low specificity. Moreover, the close proximity of the cell to the substrate during enzymatic hydrolysis allows the close monitoring of inhibitory products and mediates the passage of cellobiose and cellodextrins into the cell for metabolism. Finally, a halt in hydrolysis on depletion of one structural type of cellulose at the site of adsorption is avoided by the presence of other enzymes with different specificity [74]. With all these advantages in mind and the available knowledge of these structures, cellulosomes may provide great potential for use in the biofuel industry.
A simplified schematic of general cellulosome composition, connection with cell surface and interaction with substrate based on knowledge of Clostridium sp. cellulosomes. (Modified from Shoham et al. [74]).
Mini-cellulosome chimeras
The complete genome sequence is available for C. thermocellum and the well known solvent-producing C. acetobutylicum, in the NCBI Genbank (NC_009012 and NC_003030, respectively). Additionally, several sequences for different cellulosomal scaffoldin are also now available. The sequencing of these genomes and various cellulosomal scaffoldin genes has opened the door for future enhancement of clostridia for cellulose hydrolysis and in some strains additionally fermentation. Researchers recognize the value of cellulosomes for the efficient hydrolysis of microcrystalline cellulose and have begun to focus research on creating designer cellulosomes for recombinant expression for industry and to advance our knowledge of true cellulolytic activity. Murashima and colleagues [55] created the first in vitro recombinant minicellulosomes with a specific function, using the scaffoldin structure of C. cellulovoran and the knowledge of cohesion-dockerin self assembly. The mini cellulosomes contained the enzymatic subunit EngB and the scaffolding unit, mini-CbpA, cellulose binding domain, a putative cell wall binding domain, and two cohesin units [55]. The full-length EngB containing the dockerin domain was expressed by B. subtilis WB800, which is deficient in eight extracellular proteases, to prevent the proteolytic cleavage of the enzymatic subunit between the catalytic and dockerin domains that was observed in previous attempts to express EngB with Escherichia coli. The mini-CbpA and cohesins were expressed by E. coli. This paved the way for in vivo synthesis of the EngB enzymatic subunit and mini-CbpA scaffolding unit by co-expression in B. subtilis [78]. Moreover, Perret and colleagues [79] created an enriched, highly specific cellulosome by cloning and overexpression of the Man5K gene in C. cellulovorans. Due to the high expression levels, Man5K was heavily incorporated into the cellulosome resulting in an increased activity towards galactomannans and ultimately reducing specific activity on crystalline cellulose [79]. This is further evidence that the enzymatic composition of cellulosomes can be altered towards a specific activity.
Creating designer cellulosomes also allows us to examine properties of the cellulosome that may contribute to its efficiency. In 'bifunctional' designer cellulosomes, two divergent cohesion-dockerin devices from C. thermocellum and C. cellulolyticum of a CBD containing scaffoldin were compared to the function of similar minicellulosomes lacking CBD and free enzymes with and without CBDs. The result was higher cellulase activity on crystalline cellulose for minicellulosomes with CBD, however no apparent advantage over free enzymes on soluble substrate. The proximity of enzymes and the presence of CBD on the scaffoldin appear to contribute significantly and almost equally to the efficiency of the cellulosome on recalcitrant substrate [80]. Additionally, Fierobe and colleagues [81] used the same principle to create 'trifunctional' designer cellulosomes with the addition of a third divergent cohesin-dockerin device from Ruminococcus flavefaciens. The trifunctional cellulosome chimera was found to be considerably more active than their previous bifunctional cellulosome, in addition to free enzymes. Their work also suggests that the cellulases from family-48 and -9 glycoside hydrolases are prominent and crucial for crystalline cellulose degradation. Also, co-operation and synergistic action between cellulases and hemicellulases of different organisms within designer cellulosomes, does exist and contribute to overall efficiency [81]. The largest designer cellulosome created using CbpA of C. cellulovorans contained four cohesins and was compared with activities of designer cellulosomes containing one and two cohesins. The incorporation of endoglucanase EngB and endoxylanase XynA enzymes in CbpA1234 again exemplified the importance of clustering for efficiency of cellulose degradation [82]. This research also provides evidence for the construction of more specific and larger designer cellulosomes with high activity.
Moreover, fungal cellulases were recently fused with dockerin sequences matching bacterial cohesins, and shown to be incorporated in vivo into mini bacterial cellulosomes alongside bacterial cellulases. These enzymes, despite species difference still showed increased synergy when bound in minicellulosomes further demonstrating the importance of synergy and enzyme proximity [83]. Similarly, the activities of free exoglucanases from T. fusca were compared to the activity of these enzymes incorporated into minicellulosomes. Incorporation showed a marked increase in cellulase activity due to increased synergy of the enzymes and close contact compared to free enzymes diluted in the bulk solution [84]. These cellulosome chimeras offer an opportunity to take efficient cellulases or hemicellulases to further increase lignocellulosic hydrolysis.
In contrast to these studies, Mingardon and colleagues [83] deviated from designing cellulosome chimeras based on the general native structure of cellulosomes. Instead, they designed novel cellulosome chimeras which exhibited atypical geometries. Family-48 and -9 enzymes were modified to contain cohesins/dockerins and CBD's, additional to the CBD and cohesins of the scaffoldin. This resulted in novel, oddly shaped cellulosomes. The number of protein-protein interactions within these complexes diminished the hydrolytic activity, due to the reduced mobility of the catalytic domains. Similarly, the presence of numerous CBD's also restricted the activity and it appears that the native structure of the cellulosome is critical because it maximizes enzyme mobility [83].
The recent development of designer cellulosomes has unlocked key knowledge for the exploitation of cellulosomes in the bioconversion of lignocellulose. Designer cellulosomes offer a means to create specificity towards substrates and enhance enzyme activity with incorporation of efficient enzymes from a broad range of hosts. The next step is to find a means to develop cellulosome chimeras in a biologically and economically feasible manner.
The potential for cellulosome-producing C. thermocellum
Due to the production of highly versatile cellulosomes and the anaerobic, thermophilic, ethanologenic nature, of C. thermocellum, it is an excellent candidate for consolidated bioprocessing (CBP). CBP features the production of cellulases and hemicellulases, hydrolysis of cellulose and hemicellulose, and, fermentation of hydrolysis products, all in one step. Using a strain such as C. thermocellum means less time for cooling and easy removal of ethanol at higher temperatures. It also means no addition of oxygen during the biorefining process and fermentation of glucose to produce ethanol and organic acids [85]. The compromise to using such a strain is the slow growth rate of anaerobic thermophiles; the possibility of spore-formation during biorefining; and, the fact that C. thermocellum does not metabolize the 5-carbon sugars it produces during hydrolysis. Albeit, this strain is highly amenable to co-culture and co-culturing would allow growth with second or third party strains to enhance fermentation by the utilization of 5-carbon sugars as has been suggested [86,87].
Further, it has been observed that cellulase production in C. thermocellum is rapidly depressed by increasing concentrations of cellobiose [88]. However, the addition of exogenous β-glucosidase such as that purified from Aspergillus niger, can increase cellulosome activity up to 10-fold and offers a potential solution towards reducing cellulase inhibition [89]. In addition, the lower growth produced by anaerobic thermophiles can also be exploited as an advantage because it allows for higher ethanol production and less end-product inhibition to the hydrolysis enzymes because of a lack of overgrowth.
An additional limiting factor to the exploitation of C. thermocellum for CBP for biomass conversion has been its recalcitrance to genetic modification. C. thermocellum has a strict restriction endonuclease system and is described as having a Dam+ phenotype [90,91]. However, several breakthrough DNA recombinant technologies are being developed for genetic engineering of the anaerobic clostridia. There are also DNA transformation protocols optimized specifically for C. thermocellum [92,93]. It has been shown that if DNA is Dam methylated it can provide protection to DNA from the restriction endonuclease system of C. thermocellum; therefore, if DNA is Dam methylated prior to electrotransformation a higher number of successful transconjugates should be seen [90]. A large number of plasmids have been developed for engineering thermophilic anaerobic bacteria [94-96]. The pIMK1 plasmid developed from the replicons of C. acetobutylicum and Escherichia coli, was used to successfully express kanamycin in Thermoanaerobacterium saccharolyticum, an anaerobic, thermophilic strain and close relative of C. thermocellum. Therefore this plasmid offers great potential framework for recombinant gene expression in C. thermocellum [97].
One final limiting factor for use of C. thermocellum in CBP is the inhibition of cell growth and metabolism by toxic by-products such as the production of acetic and lactic acids during fermentation to produce ethanol. Blocking or knocking out genes involved in acetic and lactic acid production (e.g. acetate kinase and phosphotransacetylase), could help solve this problem. However, again the recalcitrance of clostridia to genetic modification has impeded this development. Nonetheless, a new very recent technology has been developed for efficient gene knockout in clostridia: The ClosTron. The ClosTron utilizes Targetron technology which is a mobile group II intron originating from Lactococcus lactis L1 (LtrB intron). The LtrB intron allows a double-cross over event which is highly stable compared to previously used single-cross over events [98,99]. Successful transformants are selected based on erythromycin resistance and can be made in as short as 10 to 14 days for a variety of clostridia tested. Six knockout mutants of C. acetobutylicum were created and five knockout mutants of C. difficile were created, exceeding the number of mutants ever published for these species. Genes were also inactivated for the first time in C. botulinum and C. sporogenes [98]. These results make the ClosTron universally applicable to the clostridium genus and should therefore be of use in creating knockout mutants of C. thermocellum.
With recent great advancements in genetic technologies, overcoming the stumbling blocks of using C. thermocellum for a CBP process in the bioconversion of lignocellulosic biomass is a good concept with great potential. It may one day offer the most economically feasible means to create lignocellulosic derived ethanol.
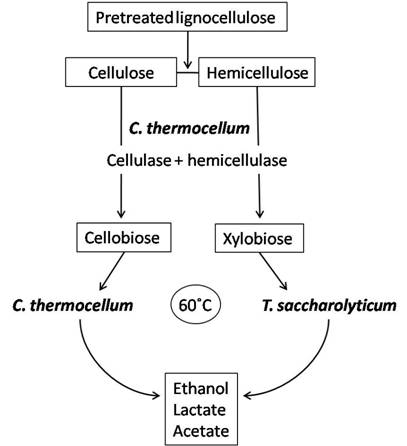
Simplified process using C. thermocellum and T. saccharolyticum in co-culture for ethanol production. C. thermocellum produces the cellulases and hemicellulases for hydrolysis of lignocelluloses to sugars such as cellobiose and xylobiose. In addition, C. thermocellum can utilize hexose sugars derived from celluloses to produce ethanol. While, the hemicelluloses derived pentoses can be utilized by T. saccharolyticum. T. saccharolyticum also contributes to cellobiose reduction and is a good ethanol producer (modified from Demain et al. [85]).
Co-culture
Bacterial co-cultures can offer a means to improve hydrolysis of cellulose as well as enhance product utilization and thus increase desirable fermentation products. Clostridium thermocellum has gained special interest for co-culture with organisms capable of fermenting pentose sugars to ethanol because C. thermocellum can only ferment hexose sugars. Hence, C. thermocellum has been co-cultivated with other anaerobic thermophilic clostridia or close relatives such as Clostridium thermosaccharolyticum (now classified as Thermoanaerobacterium saccharolyticum) [100-102], Clostridium thermohydrosulfuricum [86,102,103], Thermoanaerobacter ethanolicus [104] and Thermoanaerobium brockii [105]. These organisms can share a syntrophic relationship with C. thermocellum which exploits its cellulases and hemicellulases to hydrolyze cellulose to cellobiose and cellodextrans, and hemicelluloses to mainly xylobiose, arabinoxylans and xylooligosaccharides. C. thermocellum will then convert cellulose breakdown products to ethanol while the latter strains will utilize hemicellulose hydrolysis products to produce ethanol; this avoids the competition for substrates between species and maximizes product formation (Figure 2). The current challenge with this type of co-culture application is the increased production of by-products such as acetate and lactate which decrease ethanol production by slowing the growth rate of cells [106].
Developing bacterial co-cultures can be a tedious task. To establish a stable co-culture, media and growth requirements, such as temperature, atmosphere and carbon source, must be fine-tuned to permit equal growth of each strain. Stable co-cultures may not only depend on the media and growth requirements of each strain, but may also be controlled more specifically by metabolic interactions (i.e. syntrophic relationships or alternatively competition for substrates) and other interactions (i.e. growth promoting or growth inhibiting such as antibiotics). Criteria for structurally stable bacterial communities have been established, where 1) all the members must persist over more than 20 times subculturing and 2) the abundance ratio of members does not change even after subculturing. This is represented by reproducible growth, that is, subsequent subcultures without overgrowth or growth failure [107,108].
The alternative of bacterial co-culture would be to engineer one microorganism to complete an entire task from start to finish itself. In the case of C. thermocellum, this would mean metabolically engineering this strain to ferment pentose sugars in addition to hexose sugars. This is a difficult task as far as molecular engineering goes in clostridia due the recalcitrance of clostridia to genetic manipulation. Also to consider, if one could successfully engineer C. thermocellum to utilize pentose sugars, would this have an alternative effect on the ethanol yield produced from hexose sugars? Co-cultivation has advantage because it reduces the number of exogenous elements produced by a single bacterial population and therefore reduces the chance of metabolic imbalance for host cells. Additionally, division of labor will simplify the optimization of each reaction pathway [109]. Although bacterial co-culture is not an uncommon concept, its use in the bioconversion of lignocellulosic biomass is still premature and offers great potential.
Closing comments
Bacteria present an attractive potential for the exploitation of cellulases and hemicellulases due to their rapid growth rate, enzyme complexity and extreme habitat variability. The development of rapid and reliable methods for the screening of cellulases from microorganisms within inhospitable environments will allow a greater number of novel bacterial cellulases to be isolated with purpose for industrial use. None of the enzymes isolated to date, are fully resistant to the harsh environmental conditions used in the bioconversion process such as high temperature, acidic and or alkali pretreatments. However, these novel enzymes can be further engineered using available knowledge of enzyme structure and function through rational design. Or, they can be improved using random mutagenesis techniques with focus on selection of ideally augmented traits through directed evolution. Furthermore, novel or improved enzymes can be incorporated into designer minicellulosomes, which can further enhance the hydrolytic activity of individually efficient enzymes through synergy. Beyond free bacterial cellulases is the opportunity for whole cells in bacterial co-culture and the use of strains with multiple exploitable characteristics to reduce time and cost of current bioconversion processes. The future may hold great prospects for lignocellulosic biofuel; by combining our knowledge of excellent cellulolytic and hemicellulolytic systems such as the cellulosome of C. thermocellum with technologies such as directed evolution and co-culture, the future of lignocellulolytic biofuel looks potentially feasible.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Schneider SH. The Greenhouse Effect: Science and Policy. Science. 1989;243:771-781
2. US Department of Energy. Annual Energy Outlook 1996 with Projections to 2015; US Department of Energy, Energy Information Administration: DOE/EIA-0383(96). Washington, DC: US Department of Energy. 1996
3. House of commons passes milestone biofuels bill, 2009. Canadian Renewable Fuels Association. http://www.greenfuels.org/
4. EPA Proposes New Regulations for the National Renewable Fuel Standard Program for 2010 and Beyond. Revised 26 May 2009 US Environmental Protection Agency. http://www.epa.gov/OMS/renewablefuels/
5. October 2008. AgMRC Renewable Energy Newsletter. http://www.agmrc.org/renewable_energy/agmrc_renewable_energy_newsletter.cfm/brazils_ethanol_
6. Greene et al. Growing energy. How biofuels can help end America's oil dependence. Nat Res Def Council Rep. 2004:1-86
7. Hill J, Nelson E, Tilman D, Polasky S, Tiffany D. Environmental, economic, and energetic costs and benefits of biodiesel and ethanol biofuels. Proc Natl Acad Sci USA. 2006(314):1598-1600
8. Wyman CE, Dale BE, Elander RT, Hotzapple M, Ladisch MR, Lee YY. Coordinated development of leading biomass pretreatment technologies. Bioresour Technol. 2005(96):1959-1966
9. Philippidis GP, Smith TK, Wyman CE. Study of the enzymatic hydrolysis of cellulose for production of fuel ethanol by the simultaneous saccharification and fermentation process. Biotechnol Bioeng. 1993;41(9):846-853
10. Lynd LR, van Zyl WH, McBride JE, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16(5):577-583
11. Henrissat B, Teeri TT, Warren RAJ. A scheme for designating enzymes that hydrolyse the polysaccharides in the cell walls of plants. FEBS Lett. 1998(425):352-354
12. Wood TM, McCrae SI. Synergism between enzymes involved in the solubilization of native cellulose. Adv Chem Ser. 1979(181):181-209
13. Rouvinen J, Bergfors T, Teeri TT, Knowles JKC, Jones TA. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990(249):380-386
14. Divne C, Stahlberg J, Reinikainen T, Ruohonen L, Petterson G, Knowles JKC, Teeri T, Jones TA. The three dimensional structure of cellobiohydrolase I from Trichoderma reesei. Science. 1994(265):524-528
15. Divne C, Stahlberg J, Teeri T, Jones TA. High-resolution crystal structures reveal how a cellulose chain is bound in the 50Å long tunnel of cellobiohydrolase I from Trichoderma reesei. J Mol Biol. 1998(275):309-325
16. Davies GJ, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995(3):853-859
17. Revised 22 June 2009. CAZy database - Carbohydrate-Active Enzymes database. http://www.cazy.org/
18. Din N, Gilkes NR, Tekant B, Miller RCJr, Warren RAJ, Kilburn DG. Non-hydrolytic disruption of cellulose fibres by the binding domain of a bacterial cellulase. Bio/Technology. 1991(9):1096-1099
19. Johnson PE, Tomme P, Joshi MD, McIntosh LP. Interaction of soluble cellooligosaccharides with the N-terminal cellulose-binding domain of Cellulomonas fimi CenC. 2. NMR and ultraviolet absorption spectroscopy. Biochemistry. 1996(35):13895-13906
20. Jervis EJ, Haynes CA, Kilburn DG. Surface diffusion of cellulases and their isolated binding domains on cellulose. J Biol Chem. 1997;272(38):24016-24023
21. Sakon J, Irwin D, Wilson DB, Karplus PA. Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca. Nat Struct Biol. 1997;4(10):810-818
22. Gal L, Gaudin C, Belaich A, Pages S, Tardif C, Belaich JP. CelG from Clostridium cellulolyticum: a multidomain endoglucanase acting efficiently on crystalline cellulose. J Bacteriol. 1997;179(21):6595-601
23. Irwin D, Shin DH, Zhang S, Barr BK, Sakon J, Karplus PA, Wilson DB. Roles of the catalytic domain and two cellulose binding domains of Thermomonospora fusca E4 in cellulose hydrolysis. J Bacteriol. 1998;180(7):1709-1714
24. Doi RH. Cellulases of mesophilic microorganisms: cellulosome and nocellulosome producers. Ann NY Acad Sci. 2008(1125):267-279
25. Rastogi G, Muppidi GL, Gurram RN, Adhikari A, Bischoff KM, Hughes SR, Apel WA, Bang SS, Dixon DJ, Sani RK. Isolation and characterization of cellulose-degrading bacteria from the deep subsurface of the Homestake gold mine, Lead, South Dakota, USA. J Ind Microbiol Biotechnol. 2009;36(4):585-598
26. Hankin L, Anagnostakis S. Solid media containing carboxymethyl cellulose to detect CM celllulase activity of microorganisms. J Gen Microbiol. 1977(98):109-115
27. Kasana RC, Salwan R, Dhar H, Dutt S, Gulati A. A rapid and easy method for the detection of microbial cellulases on agar plates using gram's iodine. Curr Microbiol. 2008;57(5):503-507
28. Wittrup KD, Bailey JE. A single-cell assay of β-galactosidase activity in Saccharomyces cerevisiae. Cytometry. 1988(9):394-404
29. Huang ZJ. Kinetic assay of fluorescein mono-β-D-galactoside hydrolysis by β-galactosidase: a front-face measurement for strongly absorbing fluorogenic substrates. Biochemistry. 1991(930):8530-8534
30. Eggertson MJ, Craig DB. β-Galactosidase assay using capillary electrophoresis laser-induced fluorescence detection and resorufin-β-D-galactopyranoside as substrate. Biomed Chromatogr. 1999(13):516-519
31. Irwin JA, Morrissey PE, Ryan JP, Walshe A, O'Neill SM, Carrington SD, Matthews E, Fitzpatrick E, Mulcahy G, Corfield AP, Dalton JP. Glycosidase activity in the excretory-secretory products of the liver fluke Fasciola hepatica. Parasitology. 2004(129):465-472
32. Xu H, Ewing AG. A rapid enzyme assay for β-galactosidase using optically gated sample introduction on a microfabricated chip. Anal Bioanal Chem. 2004(378):1710-1715
33. Fia G, Giovani G, Rosi I. Study of β-glucosidase production by wine-related yeasts during alcoholic fermentation. A new rapid fluorimetric method to determine enzymatic activity. J Appl Microbiol. 2005(99):509-517
34. Ivanen DR, Rongjina NL, Shishlyannikov SM, Litviakova GI, Isaeva-Ivanova LS, Shabalin KA, Kulminskaya AA. Novel precipitated fluorescent substrates for the screening of cellulolytic microorganisms. J Microbiol Methods. 2009;76(3):295-300
35. Xu YQ, Duan CJ, Zhou QN, Tang JL, Feng JX. Cloning and identification of cellulase genes from uncultured microorganisms in pulp sediments from paper mill effluent. Wei Sheng Wu Xue Bao. 2006;46(5):783-788
36. Duan CJ, Xian L, Zhao GC, Feng Y, Pang H, Bai XL, Tang JL, Ma QS, Feng JX. Isolation and partial characterization of novel genes encoding acidic cellulases from metagenomes of buffalo rumens. J Appl Microbiol. 2009 [Epub ahead of print]
37. Wang CM, Shyu CL, Ho SP, Chiou SH. Characterization of a novel thermophilic, cellulose-degrading bacterium Paenibacillus sp. strain B39. Lett Appl Microbiol. 2008;47(1):46-53
38. Ko CH, Chen WL, Tsai CH, Jane WN, Liu CC, Tu J. Paenibacillus campinasensis BL11: a wood material-utilizing bacterial strain isolated from black liquor. Bioresour Technol. 2007(14):2727-2733
39. Li W, Zhang WW, Yang MM, Chen YL. Cloning of the thermostable cellulase gene from newly isolated Bacillus subtilis and its expression in Escherichia coli. Mol Biotechnol. 2008(2):195-201
40. Liang Y, Yesuf J, Schmitt S, Bender K, Bozzola J. Study of cellulases from a newly isolated thermophilic and cellulolytic Brevibacillus sp. strain JXL. J Ind Microbiol Biotechnol. 2009 [Epub ahead of print]
41. Hirasawa K, Uchimura K, Kashiwa M, Grant WD, Ito S, Kobayashi T, Horikoshi K. Salt-activated endoglucanase of a strain of alkaliphilic Bacillus agaradhaerens. Antonie Van Leeuwenhoek. 2006;89(2):211-219
42. Pérez-Avalos O, Sánchez-Herrera LM, Salgado LM, Ponce-Noyola T. A bifunctional endoglucanase/endoxylanase from Cellulomonas flavigena with potential use in industrial processes at different pH. Curr Microbiol. 2008;57(1):39-44
43. Ekborg NA, Morrill W, Burgoyne AM, Li L, Distel DL. CelAB, a multifunctional cellulase encoded by Teredinibacter turnerae T7902T, a culturable symbiont isolated from the wood-boring marine bivalve Lyrodus pedicellatus. Appl Environ Microbiol. 2007;73(23):7785-7788
44. Percival Zhang YH, Himmel ME, Mielenz JR. Outlook for cellulase improvement: screening and selection strategies. Biotechnol Adv. 2006;24(5):452-81
45. Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001(409):253-257
46. Mahadevan SA, Wi SG, Lee DS, Bae HJ. Site-directed mutagenesis and CBM engineering of Cel5A (Thermotoga maritima). FEMS Microbiol Lett. 2008;287(2):205-211
47. Escovar-Kousen JM, Wilson D, Irwin D. Integration of computer modeling and initial studies of site-directed mutagenesis to improve cellulase activity on Cel9A from Thermobifida fusca. Appl Biochem Biotechnol. 2004(113-116):287-297
48. Lim WJ, Hong SY, An CL, Cho KM, Choi BR, Kim YK, An JM, Kang JM, Lee SM, Cho SJ, Kim H, Yun HD. Construction of minimum size cellulase (Cel5Z) from Pectobacterium chrysanthemi PY35 by removal of the C-terminal region. Appl Microbiol Biotechnol. 2005;68(1):46-52
49. Park SR, Cho SJ, Kim MK, Ryu SK, Lim WJ, An CL, Hong SY, Kim JH, Kim H, Yun HD. Activity enhancement of Cel5Z from Pectobacterium chrysanthemi PY35 by removing C-terminal region. Biochem Biophys Res Commun. 2002;291(2):425-430
50. Cho KM, Math RK, Hong SY, Asraful Islam SM, Kim JO, Hong SJ, Kim H, Yun HD. Changes in the activity of the multifunctional beta-glycosyl hydrolase (Cel44C-Man26A) from Paenibacillus polymyxa by removal of the C-terminal region to minimum size. Biotechnol Lett. 2008;30(6):1061-1068
51. Baker JO, McCarley JR, Lovett R, Yu CH, Adney WS, Rignall TR, Vinzant TB, Decker SR, Sakon J, Himmel ME. Catalytically enhanced endocellulase Cel5A from Acidothermus cellulolyticus. Appl Biochem Biotechnol. 2005(121-124):129-148
52. Rignall TR, Baker JO, McCarter SL, Adney WS, Vinzant TB, Decker SR, Himmel ME. Effect of single active-site cleft mutation on product specificity in a thermostable bacterial cellulase. Appl Biochem Biotechnol. 2002(98-100):383-94
53. Rajoka MI, Ashraf Y, Khalid AM. Kinetic of improved production and carboxymethyl cellulose hydrolysis by an endo-glucanase from a derepressed mutant of Cellulomonas biazotea. Biotechnol Lett. 2004;26(17):1329-1333
54. Rajoka MI, Durrani IS, Khalid AM. Kinetics of improved production and thermostability of an intracellular beta-glucosidase from a mutant-derivative of Cellulomonas biazotea. Biotechnol Lett. 2004;26(4):281-285
55. Murashima K, Kosugi A, Doi RH. Thermostabilization of cellulosomal endoglucanase EngB from Clostridium cellulovorans by in vitro DNA recombination with non-cellulosomal endoglucanase EngD. Mol Microbiol. 2002;45(3):617-626
56. Kim YS, Jung HC, Pan JG. Bacterial cell surface display of an enzyme library for selective screening of improved cellulase variants. Appl Environ Microbiol. 2000;66(2):788-793
57. Arrizubieta MJ, Polaina J. Increased thermal resistance and modification of the catalytic properties of a beta-glucosidase by random mutagenesis and in vitro recombination. J Biol Chem. 2000;275(37):28843-28848
58. Gonzalez-Blasco G, Sanz-Aparicio J, Gonzalez B, Hermoso JA, Polaina J. Directed evolution of beta -glucosidase A from Paenibacillus polymyxa to thermal resistance. J Biol Chem. 2000;275(18):13708-13712
59. McCarthy JK, Uzelac A, Davis DF, Eveleigh DE. Improved catalytic efficiency and active site modification of 1,4-beta-D-glucan glucohydrolase A from Thermotoga neapolitana by directed evolution. J Biol Chem. 2004;279(12):11495-502
60. Kaper T, Lebbink JH, Pouwels J, Kopp J, Schulz GE, van der Oost J, de Vos WM. Comparative structural analysis and substrate specificity engineering of the hyperthermostable beta-glucosidase CelB from Pyrococcus furiosus. Biochemistry. 2000;39(17):4963-70
61. Kim YW, Lee SS, Warren RA, Withers SG. Directed evolution of a glycosynthase from Agrobacterium sp. increases its catalytic activity dramatically and expands its substrate repertoire. J Biol Chem. 2004;279(41):42787-42793
62. Wakarchuk WW, Sung WL, Campbell RL, Cunningham A, Watson DC, Yaguchi M. Thermostabilization of the Bacillus circulans xylanase by the introduction of disulfide bonds. Protein Eng. 1994;7(11):1379-1386
63. Paës G, O'Donohue MJ. Engineering increased thermostability in the thermostable GH-11 xylanase from Thermobacillus xylanilyticus. J Biotechnol. 2006;125(3):338-350
64. Lu P, Feng MG. Bifunctional enhancement of a beta-glucanase-xylanase fusion enzyme by optimization of peptide linkers. Appl Microbiol Biotechnol. 2008;79(4):579-87
65. Lu P, Feng MG, Li WF, Hu CX. Construction and characterization of a bifunctional fusion enzyme of Bacillus-sourced beta-glucanase and xylanase expressed in Escherichia coli. FEMS Microbiol Lett. 2006;261(2):224-230
66. Fan Z, Werkman JR, Yuan L. Engineering of a multifunctional hemicellulase. Biotechnol Lett. 2009;31(5):751-757
67. Gibbs MD, Nevalainen KM, Bergquist PL. Degenerate oligonucleotide gene shuffling (DOGS): a method for enhancing the frequency of recombination with family shuffling. Gene. 2001;271(1):13-20
68. Ruller R, Deliberto L, Ferreira TL, Ward RJ. Thermostable variants of the recombinant xylanase A from Bacillus subtilis produced by directed evolution show reduced heat capacity changes. Proteins. 2008;70(4):1280-1293
69. Díaz M, Rodriguez S, Fernández-Abalos JM, De Las Rivas J, Ruiz-Arribas A, Shnyrov VL, Santamaría RI. Single mutations of residues outside the active center of the xylanase Xys1 Delta from Streptomyces halstedii JM8 affect its activity. FEMS Microbiol Lett. 2004;240(2):237-243
70. Fanutti C, Ponyi T, Black GW, Hazlewood GP, Gilbert HJ. The conserved noncatalytic 40-residue sequence in cellulases and hemicellulases from anaerobic fungi functions as a protein docking domain. J Biol Chem. 1995;270(49):29314-29322
71. Li XL, Chen H, Ljungdahl LG. Two cellulases, CelA and CelC, from the polycentric anaerobic fungus Orpinomyces strain PC-2 contain N-terminal docking domains for a cellulase-hemicellulase complex. Appl Environ Microbiol. 1997;63(12):4721-4728
72. Lamed R, Setter E, Kenig R, Bayer EA. The cellulosome: a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol Bioeng. 1983(13):163-181
73. Johnson E.A, Madia A, Demain A.L. Chemically defined minimal medium for the growth of the anaerobic cellulolytic thermophile Clostridium thermocellum. Appl Environ Microbiol. 1981(41):1060-1062
74. Shoham Y, Lamed R, Bayer EA. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 1999;7(7):275-281
75. Schwarz WH. The cellulosome and cellulose degradation by anaerobic bacteria. Appl Microbiol Biotechnol. 2001;56(5-6):634-649
76. Bayer EA, Belaich JP, Shoham Y, Lamed R. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol. 2004;58:521-554
77. Bayer EA, Lamed R, White BA, Flint HJ. From cellulosomes to cellulosomics. Chem Rec. 2008;8(6):364-377
78. Cho HY, Yukawa H, Inui M, Doi RH, Wong SL. Production of minicellulosomes from Clostridium cellulovorans in Bacillus subtilis WB800. Appl Environ Microbiol. 2004;70(9):5704-7
79. Perret S, Bélaich A, Fierobe HP, Bélaich JP, Tardif C. Towards designer cellulosomes in Clostridia: mannanase enrichment of the cellulosomes produced by Clostridium cellulolyticum. J Bacteriol. 2004;186(19):6544-6552
80. Fierobe HP, Bayer EA, Tardif C, Czjzek M, Mechaly A, Bélaïch A, Lamed R, Shoham Y, Bélaïch JP. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277(51):49621-49630
81. Fierobe HP, Mingardon F, Mechaly A, Bélaïch A, Rincon MT, Pagès S, Lamed R, Tardif C, Bélaïch JP, Bayer EA. Action of designer cellulosomes on homogeneous versus complex substrates: controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J Biol Chem. 2005;280(16):16325-16334
82. Cha J, Matsuoka S, Chan H, Yukawa H, Inui M, Doi RH. Effect of multiple copies of cohesins on cellulase and hemicellulase activities of Clostridium cellulovorans mini-cellulosomes. J Microbiol Biotechnol. 2007;17(11):1782-1788
83. Mingardon F, Chanal A, López-Contreras AM, Dray C, Bayer EA, Fierobe HP. Incorporation of fungal cellulases in bacterial minicellulosomes yields viable, synergistically acting cellulolytic complexes. Appl Environ Microbiol. 2007;73(12):3822-3832
84. Caspi J, Irwin D, Lamed R, Li Y, Fierobe HP, Wilson DB, Bayer EA. Conversion of Thermobifida fusca free exoglucanases into cellulosomal components: comparative impact on cellulose-degrading activity. J Biotechnol. 2008;135(4):351-357
85. Demain AL, Newcomb M, Wu JHD. Cellulase, Clostridia, and Ethanol. Microbiol Mole Biol Rev. 2005;69(1):124-154
86. Ng TK, Ben-Bassat A, Zeikus JG. Ethanol Production by Thermophilic Bacteria: Fermentation of Cellulosic Substrates by Cocultures of Clostridium thermocellum and Clostridium thermohydrosulfuricum. Appl Environ Microbiol. 1981;41(6):1337-1343
87. Mori Y. Characterization of a Symbiotic Coculture of Clostridium thermohydrosulfuricum YM3 and Clostridium thermocellum YM4. Appl Environ Microbiol. 1990;56(1):37-42
88. Brener D, Johnson BF. Relationship Between Substrate Concentration and Fermentation Product Ratios in Clostridium thermocellum Cultures. Appl Environ Microbiol. 1984;47(5):1126-1129
89. Lamed R, Kenig R, Morgenstern E, Calzada JF, De Micheo F, Bayer EA. Efficient cellulose solubilisation by a combined cellulosome-β-glucosidase system. Appl Biochem Biotechnol. 1990(27):173-183
90. Klapatch TR, Demain AL, Lynd LR. Restriction endonuclease activity in Clostridium thermocellum and Clostridium thermosaccharolyticum. Appl Microbiol Biotechnol. 1996;45(1-2):127-131
91. Ozkan M, Desai SG, Zhang Y, Stevenson DM, Beane J, White EA, Guerinot ML, Lynd LR. Characterization of 13 newly isolated strains of anaerobic, cellulolytic, thermophilic bacteria. J Ind Microbiol Biotechnol. 2001;27(5):275-280
92. Tyurin MV, Sullivan CR, Lynd LR. Role of spontaneous current oscillations during high-efficiency electrotransformation of thermophilic anaerobes. Appl Environ Microbiol. 2005;71(12):8069-8076
93. Tyurin MV, Desai SG, Lynd LR. Electrotransformation of Clostridium thermocellum. Appl Environ Microbiol. 2004;70(2):883-90
94. Mather MW, Fee JA. Development of plasmid cloning vectors for Thermus thermophilus HB8: Expression of a heterologous, plasmid-borne kanamycin nucleotidyltransferase gene. Appl Environ Microbiol. 1992(58):421-425
95. Mai V, Wiegel J. Advances in development of a genetic system for Thermoanaerobacterium spp.: Expression of genes encoding hydrolytic enzymes, development of a second shuttle vector, and integration of genes into the chromosome. Appl Environ Microbiol. 2000(660):4817-4821
96. Moreno R, Zafra O, Cava F, Berenguer J. Development of a gene expression vector for Thermus thermophilus based on the promoter of the respiratory nitrate reductase. Plasmid. 2003(490):2-8
97. Mai V, Lorenz WW, Wiegel J. Transformation of Thermoanaerobacterium sp. strain JW/SL-YS485 with plasmid pIKM1 conferring kanamycin resistance. FEMS Microbiol Lett. 1997(148):163-167
98. Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. The ClosTron: a universal gene knock-out system for the genus Clostridium. J Microbiol Methods. 2007;70(3):452-464
99. Shao L, Hu S, Yang Y, Gu Y, Chen J, Yang Y, Jiang W, Yang S. Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum. Cell Res. 2007;17(11):963-965
100. Wang DIC, Avgerinos GC, Biocic I, Wang S-D, Fang H-Y. Ethanol from cellulosic biomass. Phil Trans R Soc London. 1983(300):323-333
101. Venkateswaren S, Demain AL. The Clostridium thermocellum-Clostridium thermosaccharolyticum ethanol production process: nutritional studies and scale-down. Chem Eng Commun. 1986(45):53-60
102. Saddler JN, Chan MKH. Conversion of pretreated lignocellulosic substrates to ethanol by Clostridium thermocellum in mono- and coculture with Clostridium thermosaccharolyticum and Clostridium thermohydrosulfuricum. Can J Microbiol. 1985(30):212-220
103. Germain P, Toukourou F, Donaduzzi L. Ethanol production by anaerobic thermophilic bacteria: regulation of lactate dehydrogenase activity in Clostridium thermohydrosulfuricum. Appl Microbiol Biotechnol. 1986(24):300-305
104. Weigel J, Ljungdahl LG. Ethanol as fermentation product of extreme thermophilic, anaerobic bacteria. In: (ed.) H. Dellweg H. In Klassische Prozesse mit Neun Aussichten. Berlin, germany: Verlag Versuchs und Lehranstalt fuer Spiritusfabrikation and Fermentation Technologie im insituut fuer Gaerungsgewerbe und Biotechnologie. 1979:117-127
105. Lamed R, Zeikus JG. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol. 1980(144):569-578
106. Herrero AA, Gomez RF, Roberts MF. 31P NMR studies of Clostridium thermocellum. Mechanism of end product inhibition by ethanol. J Biol Chem. 1985;260(12):7442-7451
107. Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y. Network relationships of bacteria in a stable mixed culture. Microb Ecol. 2008;56(3):403-11
108. Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl Environ Microbiol. 2005;71(11):7099-106
109. Brenner K, Lingchong Y, Arnold FH. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;66(9):483-489
Author contact
![]() Correspondence to: Wensheng Qin, Email: wqinca. Phone: (807) 343-8840 Fax: (807) 346-7796
Correspondence to: Wensheng Qin, Email: wqinca. Phone: (807) 343-8840 Fax: (807) 346-7796

 Global reach, higher impact
Global reach, higher impact