Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(6):570-577. doi:10.7150/ijbs.5.570 This issue Cite
Research Paper
Disruption of the BMEI0066 gene attenuates the virulence of Brucella melitensis and decreases its stress tolerance
College of Veterinary Medicine, China Agricultural University, Beijing 100193, P.R. China.
Received 2008-4-18; Accepted 2009-8-25; Published 2009-9-1
Abstract
Brucella melitensis is a facultative intracellular pathogen. An operon composed of BMEI0066, which encodes a two-component response regulator CenR, and BMEI0067, which encodes a cAMP-dependent protein kinase regulatory subunit, has been predicted to exist in many bacterial species. However, little is known about the function of this operon. In order to characterize this operon and assess its role in virulence, we constructed a marked deletion mutant of BMEI0066. The mutant was less able to withstand hyperosmotic conditions than wild-type (16M), but showed no significant difference with 16M when challenged by H2O2. The mutant also showed increased sensitivity to elevated temperature (42°C) and a reduced survival ratio under acidic conditions compared with 16M. The mutant failed to replicate in cultured murine macrophages and was rapidly cleared from the spleens of experimentally infected BALB/c mice. These findings suggest that these operon products make an important contribution to pathogenesis in mice, probably by allowing B. melitensis to adapt to the harsh environment encountered within host macrophages.
Keywords: Brucella, two-component regulatory system, BMEI0066, virulence, CenR, stress.
Introduction
Brucella melitensis is a facultative intracellular pathogen that causes abortion and infertility in domestic animals and a severe debilitating febrile illness in humans. The mechanisms allowing this highly successful intracellular pathogen to adapt to the harsh intracellular environment of host macrophages are not clearly defined. This bacterium is able to survive and replicate within phagocytic cells. Therefore, it has to withstand a number of harsh conditions, including low pH, hypotonic environments and exposure to oxygen intermediates [1]. It has been reported that Brucella Lon functions as an authentic stress-response protease, and that it is required for wild-type virulence during the initial stage of infection, but not for the establishment and maintenance of chronic infection [2]. Hfq, also known as host factor I (HF-I), has been identified as another contributor to macrophage stress adaptation and virulence in the Brucellae [3]. Despite these studies, the mechanisms by which these intracellular pathogens are able to resist the harsh intracellular environment are not fully understood.
Environmental sensing and adaptive responses in bacteria often involve the concerted action of a two-component regulatory system consisting of sensor and response regulator components. Analysis of the genomes of various Brucella spp. has revealed that they encode only 10-12 two-component regulatory systems. So far, five of the thses two-component regulatory systems have been described in Brucella: the FeuP/FeuQ system, which is similar to the R. leguminosarum FeuP involved in the regulation of iron uptake [4]; the NtrB/NtrC system involved in the regulation of nitrogen metabolism [5]; the BvrR/BvrS system, which is associated with cellular invasion, vacuole maturation and intracellular trafficking [6]; the LOV-HK system, which function as a light-sensing module [7]; and BMEI0066, which our previous studies (using random mariner mutagenesis) identified as a two-component response regulator that is required for virulence [8].
A database search of the corresponding deduced amino acid sequences revealed that Brucella BMEI0066 possesses a high level of homology to a domain of the two-component regulatory system protein and shows significant similarity to the DNA-binding OmpR subfamily of response regulators proteins that includes CheY, OmpR, NtrC, and PhoB. BMEI0066's amino acid sequence displays 65% identity and 79% similarity to that of CenR (cell envelope regulator) in Caulobacter crescentus. CenR plays an important role in controlling cell envelop biogenesis and structure in Caulobacter crescentus and is essential for viability and growth. cenR mutants cannot form colonies on plates and fail to grow in liquid medium [9]. However, there are some differences between BMEI0066 and cenR: i) Brucella BMEI0066 gene shares a promoter with its downstream gene BMEI0067, which is a putative cAMP-dependent protein kinase regulatory subunit. These two genes compose an operon that has been predicted to exist in many bacterial species, such as B. melitensis, Mycobacterium tuberculosis, Agrobacterium tumefaciens, Mesorhizobium loti, Sinorhizobium meliloti, and Leptospira interrogans serovar lai. While in Caulobacter crescentus, cenR is an independent gene that does not share a promoter with other genes; ii) in Caulobacter crescentus CenR and CenK (cell envelope kinase) compose a two-component regulatory system, but in Brucella melitensis there is no protein with high identity to CenK (the highest is BMEI1648, identity: 33%), and no gene was predicted to encode the corresponding sensor component of BMEI0066 in Brucella genome.
In our previous studies, a mariner transposon insertion mutant of BMEI0066 showed decreased growth ratio when competed with wild-type 16M both in vitro and in vivo [8]. However, transposon insertions might fail to fully eliminate target gene function. Failure to inactivate is common for insertions situated near either end of a gene, but has also been observed with internal insertions [10] (e.g. see [11]). Here we constructed a knock-out mutant of the BMEI0066 gene as well as knock-in (KI) complementation stains to investigate the role of the BMEI0066/ BMEI0067 operon in virulence with a different methodology and its function in stress responses.
Materials and Methods
Bacteria and bacterial cultures. Escherichia coli cultures were routinely grown on Luria-Bertani (Oxoid) plates overnight at 37°C with or without supplemental kanamycin (100 mg/liter), ampicillin (100 mg/liter), or chloramphenicol (30 mg/liter). The wild-type strain and marked deletion strains of B. melitensis were routinely grown on tryptic soy agar (TSA, Bacto) at 37°C in an atmosphere containing 5% (vol/vol) CO2. Brucella melitensis 16M was donated by Dr. Qianni He (Institute of Veterinary Research, Xinjiang Academy of Animal Sciences, China). This strain was used as a virulent control organism and to generate mutants via deletion mutagenesis. All bacterial strains were stored frozen at −80°C in medium supplemented with 15% (vol/vol) glycerol (Table 1). All work with live B. melitensis was performed at a biosafety level 3 facility in China Agricultural Uni versity.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| B. melitensis strains | ||
| 16M | Wild-type | Qianni He's lab |
| ΔBMEI0066::Km | ΔBMEI0066::Km | This work |
| KI | Knock-in complementation of ΔBMEI0066::Km | This work |
| E. coli strains | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ 80lacZΔM15 ΔlacX74 recA1 endA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) nupG | Invitrogen |
| Plasmids | ||
| pBluescript II KS(+) | ColE1, bla, Ampr | Stratagene |
| pEX18Ap | sacB, bla, Ampr | [29] |
| pKD4 | FLP/FRT, Kmr | B. Wanner |
| pBBR1MCS | Broad-host-range plasmid, Cmr | [30] |
| pBBR1-1 | WU281bF- WU282R product(BMEI0066) cloned into pBBR1MCS for complementation assay | This work |
| pBluescript-3.1 | WU0133F- WU0134R / WU0135F- WU0136R cloned into pBluescript | This work |
| pBluescript-3.2 | pBluescript-3a separated by WU0177F- WU0178R (Kan cassette) | This work |
| pEX18Ap-3.1 | WU0133F - WU0136R cloned into pEX18Ap for knock-in complementation | This work |
Recombinant plasmid construction. In order to construct vectors to delete BMEI0066, fragments upstream and downstream of BMEI0066 were amplified by PCR using the primer pairs WU0133F-WU0134R (upstream) and WU0135F-WU0136R (downstream) (Table 2). The reverse primer of the 5′ fragment and the forward primer of the 3′ fragment include approximately 5 to 10 nucleotides complementary to the opposite fragment and a terminal restriction site. The 5′ and 3′ fragments were amplified in separate reactions, gel purified and used as templates for a second round of PCR [12]. The forward primer of the upstream fragment and the reverse primer of the downstream fragment were utilized in a second round of PCR to join the two fragments. The ends of this ligased product were removed by restriction digestion at sites engineered into the primers. The final fragment was gel purified and ligased to pBluescript II KS(+) (Stratagene). To construct the marked deletion plasmid pBluescript-3.2, a kanamycin cassette was inserted between the two fragments following amplification via PCR from the plasmid pKD4 using primers containing a compatible restriction site located within the overlap between the fragments ligased to pBluescript II KS(+). The plasmid pEX18Ap-3.1 containing the WU0133F - WU0136R fragment from the genome of B. melitensis 16M and the counter selectable marker sacB was used for generating a knock-in complementation strain of the mutant ΔBMEI0066::Km. The WU281bF- WU282eR product (BMEI0066 operon) was cloned into pBBR1MCS to construct a complementation plasmid for ΔBMEI0066::Km.
Primers used in this work
| Primer name | Sequence (restriction enzyme) | Fragment |
|---|---|---|
| WU0133F | 5' GGGGTACCAGTTGCACCGGGCGGTTAT 3'(KpnI) | BMEI0066 upstream |
| WU0134R | 5' GGAATTCGCCTGTCCTCGTTACTCTTTGATTT 3'(EcoRI) | BMEI0066 upstream |
| WU0135F | 5' GGAATTCTGGACAGCGCCCGATGCAGGTGTAT 3'(EcoRI) | BMEI0066 downstream |
| WU0136R | 5' CGGGATCCGAAGATCAGTCGCGGTTTGC 3'(BamHI) | BMEI0066 downstream |
| WU0281bF | 5' AACTGCAGTCTTTCGAGATCGGGCA 3'(PstI) | BMEI0066/BMEI0067 operon upstream |
| WU0282R | 5' CGGGATCCGCAACTGTTCGGGCGTGA 3'(BamHI) | BMEI0066 downstream |
| WU0282eR | 5' CGGGATCCAGGATTGGGATGCCGCCGA 3'(BamHI) | BMEI0066/BMEI0067 operon downstream |
| WU0177F | 5' GGAATTCCACGTCTTGAGCGATTGTGTAG 3'(EcoRI) | Kan cassette |
| WU0178R | 5' GGAATTCCGGAAAACGATTCCGAAGCCC 3'(EcoRI) | Kan cassette |
Selection of marked deletion mutants and knock-in complementation strains. Marked deletion mutants were created via allelic exchange following electroporation of the marked plasmid into B. melitensis. Bacteria were grown as described above and pelleted via centrifugation at 1,700 × g for 15 min at 4°C. All subsequent steps were performed on ice or at 4°C. The cell pellet was washed three times with ice-cold sterile water under the same conditions. After the final wash, the cells were resuspended in 1 ml sterile water. The bacterial cell suspension was used in each electroporation with approximately 1 μg DNA in a pre-chilled 1-mm gap cuvette (Bio-Rad) and shocked in a MicroPulser Electroporation Apparatus (Bio-Rad) set at the “Agr" program. SOC-B (6% [wt/vol] tryptic soy broth, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose) medium was immediately added to the cuvette, transferred to microcentrifuge tubes, and incubated overnight at 37°C with agitation [13]. After that, the entire culture was plated onto TSA containing kanamycin. Colonies were replica plated onto TSA containing kanamycin and onto plates containing ampicillin. Marked deletion mutants from allelic exchange should be kanamycin resistant (Kmr) and ampicillin sensitive (Amps) [14]. Verification of mutant genotypes was obtained via PCR to ensure that the gene of interest was deleted and the kanamycin cassette was retained. For construction of a KI complementation strain, pEX18Ap-3.1 was electroporated into ΔBMEI0066::Km and then cells were plated onto TSA containing ampicillin to select for the first homologous recombination, i.e., a cointegration. Colonies were cultured in TSB without antibiotics for 24 h at 37°C, with subsequent plating onto sucrose-containing medium (TSA containing 6% [wt/vol] sucrose, without antibiotic). Colonies that grew on sucrose-containing medium (Sucr) but were sensitive to ampicillin (Amps) and kanamycin (Kms) were picked up as KI complementation strains and verified by PCR.
Cell culture. RAW 264.7 murine macrophages were grown in Dulbecco's modified Eagle's medium (DMEM) with 15% (vol/vol) fetal bovine serum, 1 mM L-glutamine and 1 mM nonessential amino acids. In invasion and replication experiments, 2.5 × 105 cells were seeded into each well of a 24-well plate. The cells were cultured overnight before infection.
Macrophage assay. To investigate the role of the operon in intracellular survival, we evaluated the multiplication of B. melitensis 16M and its mutants in RAW 264.7 murine macrophages. This assay was performed as previously reported with modifications[15]. The number of viable bacteria was determined at 1, 6, 12, 24 and 48 h p.i. Monolayers of cells were cultured in 24-well plates and infected with the brucella parental train and its mutants at a multiplicity of infection (MOI) of 10 CFU per cell. To synchronize the infection, the infected plates were centrifuged at 200 × g for 5 min at room temperature, followed by a 20-min incubation at 37°C in an atmosphere containing 5% (vol/vol) CO2. The cells were washed with PBS three times and DMEM containing gentamicin (50 μg/ml) was added to kill extracellular bacteria. To evaluate macrophage invasion, the cells were incubated for 1 h at 37°C. The monolayers were washed with DMEM to remove gentamicin, and then the cells were lysed with 0.5 ml of 0.5% (vol/vol) Tween 20 in sterile water. The CFUs per well was determined by plating dilutions on TSA plates or TSA plates supplemented with 100 μg of kanamycin per ml. The percent bacterial uptake (or invasion) was calculated as the number of bacteria recovered divided by the number of bacteria inoculated into each well. To assess intracellular growth of the bacteria, the infected cells were lysed and CFUs were determined as described above at selected time points post-infection (p.i.). All invasion and survival assays were performed with duplicate wells, and the results presented represent the averages from at least three separate experiments.
Clearance of mutants from mice. Survival or persistence of the mutants in vivo was evaluated following intraperitoneal inoculation of groups (5 mice/group) of 4- to 6-week-old female BALB/c mice with 0.1 ml 1 × 104 to 1 × 105 CFU/ml of the marked deletion mutant or its parental strain, respectively. Mice were euthanized via carbon dioxide asphyxiation at various times post-infection depending on the anticipated clearance rate of the mutant. At each time point, spleens were collected and weighed, homogenized in 1 ml PBS, serially diluted, and 20-μl aliquots of all dilutions were plated onto TSA. Recovered bacteria were enumerated to evaluate the persistence of each strain. (The experimental research involving animals was approved by the Beijing Administration Office of Laboratory Animals.)
Stress response assays. The acid challenge assay was performed as previously reported with the following modifications [16]. For stationary phase, cells were grown for 48 h in 10 ml of TSB medium (pH7.3; 37°C; shaking) with an initial density of 1 × 107 CFU/ml. A 500 μl sample of each strain to be tested was harvested by centrifugation, washed in an equal volume of pH 4.4 TSB (for adapted cultures), resuspended in TSB (500 μl) at the same pH and incubated for 2 h. Adapted cultures were washed and resuspended in pH 3.4 TSB for challenge and incubated for another 2 h. CFUs were determined following dilution and plating on TSA. Percent survival was calculated by dividing the CFUs at 2 h post-acid challenge by the CFUs prior to acid challenge and multiplying by 100.
To compared the growth of the wild-type strain, the mutants, and the complemented strain in hyperosmotic conditions and at high temperature (42°C), the TSB medium was made hypertonic by adding NaCl up to 250 mM, as described previously [17]. Cells were grown in TSB medium at 37°C (control), TSB medium supplemented with 250 mM NaCl at 37°C (hyper-osmotic), and TSB medium at 42°C (high temperature) with the same initial density of 1 × 106 CFU/ml. CFUs were determined at 0 h, 24 h and 48 h. Growth ratio was calculated by dividing the CFUs of 16M by the CFUs of the other strains at the corresponding conditions and time points.
Oxidation resistance assays were conducted according to procedures described previously with a few modifications [18, 19]. In one assay, cells were grown for 48 h in 10 ml of TSB medium (pH 7.3; 37°C; shaking) with an initial density of 1 × 107 CFU/ml and then adjusted to a concentration of 5 × 105 CFU/ml (low cell density). One-milliliter of the bacterial cell suspension was exposed to H2O2 at final concentrations of 1 mM, 2.5 mM, and 5 mM. After exposure for 1 h in a 37°C shaking incubator, the cell suspensions were washed with TSB and serially diluted in phosphate-buffered saline and plated onto TSA. These plates were incubated for 72 h at 37°C with 5% CO2, and the CFUs were enumerated. In another assay, 100 μl of each cell suspension was seeded on a TSA plate and a 5.5 mm sterile filter paper disk was placed in the center of each plate. Ten microliters of either a 3% or a 30% solution of H2O2 was placed onto each disk and the plates were incubated at 37°C with 5% CO2. After 72 h of incubation, the zones of inhibition around each disk were measured.
Results and Discussion
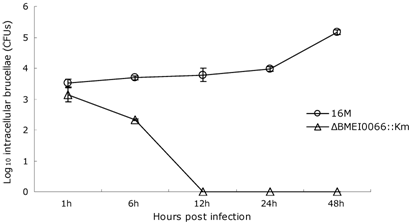
The ΔBMEI0066::Km strain fails to survive inside RAW 264.7 murine macrophages. To assess the ΔBMEI0066::Km mutant's attenuation, RAW 264.7 macrophages were infected with the ΔBMEI0066::Km mutant and with a wild-type strain. CFUs were determined at five time points: 1 h, 6 h, 12 h, 24 h and 48 h post-infection (Fig. 1). At 1 h post-infection, the macrophages contained equivalent bacterial loads, which indicated no significant variation in the ability of the bacteria to invade macrophages. However, at 12 h the ΔBMEI0066::Km strain was cleared from the macrophages. These results show that the ΔBMEI0066::Km mutant has no ability to persist and replicate in RAW 264.7 macrophages, indicating that the ΔBMEI0066/BMEI0067 operon plays an indispensible role for this bacterium to survive inside macrophages.
Multiplication of B. melitensis 16M (○) and ΔBMEI0066::Km (Δ) in RAW 264.7 murine macrophages. Values represent means of three experiments performed in duplicate, and error bars indicate SD.
The BMEI0066 mutant is attenuated in BALB/c mice. To evaluate the ΔBMEI0066::Km mutant's virulence in vivo, BALB/c mice were infected i.p. with 1 × 104 to 1 × 105 CFU/ml of ΔBMEI0066::Km and B. Melitensis 16M. Compared to the parental strain, splenic CFUs in ΔBMEI0066::Km-infected mice were significantly lower at 2 weeks p.i. At week 4, splenic CFUs could not be detected in four of the five ΔBMEI0066::Km-infected mice (Table. 3). This result shows that the ΔBMEI0066::Km strain is highly attenuated in vivo, which is consistent with the results of the in vitro macrophage assay. Compared to the transposon insertion mutant of BMEI0066 in our previous studies, the knock-out mutant ΔBMEI0066::Km showed lower survival capability both in vitro and in vivo.
Mouse spleen colonization by 16M and ΔBMEI0066::Km. Two groups of mice were inoculated i.p. with 1 × 104 to 1 × 105 CFU/ml of the indicated strains. At 2 and 4 weeks p.i., five mice per group were killed, and spleens were removed aseptically, weighed, and processed for bacteriological analysis. Values are means and standard deviations from five spleens homogenized independently in PBS, diluted, and plated in duplicate to determine the CFU per spleen.
| Strain used to infect BALB/c mice | Log CFU/spleen at the indicated week post-infection | |
|---|---|---|
| 2 wk | 4 wk | |
| 16M | 5.5±0.42 | 5.4±0.45 |
| ΔBMEI0066::Km | 2.1±0.21 | < 2 |
BMEI0066 mutants are highly sensitive to environmental stresses. The characteristics of the ΔBMEI0066::Km strain in macrophages and the mouse model promoted us to study its mechanisms. Since BMEI0066 belongs to a two-component regulatory system, and these systems play important roles in environmental sensing and adaptive responses in bacteria, the survival ability of the mutant strain under various stress conditions was tested. Within host macrophages, the pH of phagocytic vacuoles has been shown to decrease rapidly to 4.0 to 4.5 [20]. Early acidification of vacuolar compartments has been shown to be essential for the intracellular survival of Brucella [20, 21], which indicates that it has to adapt itself to the low-pH environment. To determine whether sensitivity to reduced pH may play a role in the attenuation of the ΔBMEI0066::Km, survival at reduced pH was evaluated. The stationary-phase acid tolerance of ΔBMEI0066::Km was about 10-fold lower than that of the wild-type strain or the KI complementation strain (Fig.2). The BMEI0066 mutant was complemented in trans with either the intact BMEI0066 locus (pBBR1-BMEI0066) or the whole set of B. melitensis BMEI0066-BMEI0067 genes. However both of these plasmid-complementation strains (ΔBMEI0066::Km+p1 and ΔBMEI0066::Km+p4) exhibited approximately 10-fold higher survival than the wild-type strain. It is possible that the presence of extra copies of the genes in the vectors (10-12 copies per cell [22]), by overexpressing the BMEI0066 or the BMEI0066/BMEI0067 operon products, affected the transcription of certain genes. These findings suggested that the attenuation of the ΔBMEI0066::Km mutant was probably due to its reduced acid tolerance.
To explore whether the BMEI0066 mutant is sensitive to other stress conditions, oxidation resistance assays, heat growth assays and hypertonic growth assays were performed as described previously [18, 19]. After one-hour exposure to H2O2 at concentrations of 1 mM, 2.5 mM, and 5 mM, the survival rates of wild-type strain were 98.7%, 98.3% and 51.5%, respectively, while the survival rates of BMEI0066 mutant were 97.7%, 98.7% and 48.7%, respectively. Consistently, no significant difference was observed between the mutant and the wild-type strain in the H2O2 disk sensitivity assay, suggesting that the BMEI0066/BMEI0067 operon is not required under oxidative stress. In the optimal temperature environment (37ºC, TSB), each strain showed similar growth characteristics (Fig. 3A). However, at 42°C, the ΔBMEI0066::Km strain yielded significantly fewer CFUs than wild-type both at 24 h and 48 h (about 20-fold and 100-fold less than wild-type at 24 h and 48 h, respectively) (Fig. 3C). In hypertonic conditions, the parent strain yielded slightly more CFUs than ΔBMEI0066::Km at 24 h, but from 24 h to 48 h, the multiplication rate of B. melitensis 16M was 30-fold higher than that of ΔBMEI0066::Km. Although the mutant strain was well restored by the knock-in complementation strategy, the plasmid-based complementation strains ΔBMEI0066::Km+p1 and ΔBMEI0066::Km+p4 could not restore the heat tolerance or the hypertonic adaptation, and they were even more sensitive to these harsh conditions (data not shown). To confirm whether the failure of complementation was caused by the overexpression of the BMEI0066/BMEI0067 operon from the moderate-copy-number vector pBBR1MCS, we introduced the complementation plasmid pBBR1-4 into wild-type B. melitensis 16M to generate 16M+P4. As expected, 16M+P4 demonstrated growth characteristics similar to ΔBMEI0066::Km+p4 (data not shown). It seems that both lack of BMEI0066/BMEI0067 operon products and their overexpression leads to a reduction in hypertonic adaptation. This is not the first report of such kind of misregulation for Brucellae e.g. similar scenario was also observed when complementing B. abortus rpoN mutant with pBBR1MCS-2 (with rpoN) as complementation vector [23].
Mutations in the BMEI0066/BMEI0067 operon result in defective acid tolerance. Cells were grown in TSB (pH 7.3) to stationary phase, washed and resuspended in pH 4.4 TSB for adaptation (2 h), after which the cells were washed and resuspended in pH 3.4 TSB for challenge. Viable counts were taken at 2 h after challenge. Values represent means of three experiments performed in duplicate, and error bars indicate SD. *P < 0.05: BMEI0066 mutant or KI compared to wild-type strain 16M. Statistical analysis was performed with a t-test.
Growth ratio (CFU 16M/CFU mutant) of the mutant strains in TSB at 37°C (a), in TSB supplemented with 250 mM NaCl at 37°C (b), and in TSB at 42°C (c). Values represent means of three experiments performed in duplicate, and error bars indicate SD. *P < 0.05: the growth ratio in hyperosmotic (b) or high temperature condition (c) compared to control condition (a). Statistical analysis was performed with a t-test.
These findings suggest that the BMEI0066/BMEI0067 operon plays an important role in a variety of environmental stresses. As demonstrated for other two-component systems, multiple genes should be under the control of the BMEI0066 operon. It would be interesting to analyse BMEI0066 regulation and BMEI0066 target expression. Two-dimensional gel electrophoresis has proven to be a useful tool for the study of protein expression in general and for identifying stress response proteins in particular, and it has been well applied to the study of protein expression in B. melitensis under different environmental stresses [24, 25, 26]. Therefore by using this technique we can assess the differences in protein expression between a wild-type strain and its BMEI0066 mutant so as to discover the BMEI0066 target genes, and it will permit a further analysis of the BMEI0066-dependent genetic network and will thus contribute to our understanding of the CenR refulator first identified in C. crescentus.
In Caulobacter crescentus, CenR plays a key role in controlling cell envelop biogenesis, and the cenR mutant fails to form colonies on plates and loses the ability to grow in liquid medium [9]. Our data show that B. melitensis BMEI0066 is not essential for viability and BMEI0066 mutants have the same growth phenotype as wild-type both on plates and in liquid medium, indicating a plasticity of the CenR regulation network in C. crescentus and B. melitensis despite the high identity of the amino acid sequences. The plasticity of the regulation network involving CenR might be related to their different lifestyles. B. melitensis is a facultative intracellular pathogen, in which the regulation system has been shaped to fit the interaction of the bacteria with a eukaryotic host over long evolutionary periods. While C. crescentus grows in dilute aquatic environments which lacks the intracellular stress conditions, like low Ph and hypertonic conditions. Comprehensive phylogenomic analyses based on the open reading frames in the genomes of several α-proteobacteria have identified numerous proteins that are unique repertoires of all of its main orders and families including Brucellaceae and Caulobacterales [27]. Hence, there would be some different genes involve in this regulation network which determines the C. crescentus CenR is essential for viability, but the B. melitensis BMEI0066 not.
In conclusion, this study indicates that the B. melitensis BMEI0066/BMEI0067 operon plays an important role in virulence and resistance to stresses such as low pH, hypertonic conditions and high temperature. The rapid clearance of the BMEI0066 mutant in macrophages and mice is possibly due to its lower tolerance of the harsh environment inside the host cells or the high body temperature of fever that usually present in brucellosis [28]. However, the mechanism by which the BMEI0066/BMEI0067 operon promotes virulence and which genes are under its control require further investigation.
Acknowledgements
The authors wish to thank Dr. Jianwu Pei from Texas A&M University, USA for critical review and comments on this manuscript, and Dr. Qianni He from the Institute of Veterinary Research, Xinjiang Academy of Animal Sciences, China for kindly providing us with Brucella melitensis 16M. This work is supported by the Chinese National Key technology R&D program (2006BAD04A05-03).
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. van Furth R. Macrophage-pathogen interactions. Introduction. Immunol Ser. 1994;60:v-vii
2. Robertson G.T. et al. The Brucella abortus Lon functions as a generalized stress response protease and is required for wild-type virulence in BALB/c mice. Mol Microbiol. 2000;35(3):577-88
3. Robertson G.T and Roop R.MJr. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol Microbiol. 1999;34(4):690-700
4. Dorrell N. et al. Identification, cloning and initial characterisation of FeuPQ in Brucella suis: a new sub-family of two-component regulatory systems. FEMS Microbiol Lett. 1998;162(1):143-50
5. Dorrell N. et al. Investigation into the role of the response regulator NtrC in the metabolism and virulence of Brucella suis. Microb Pathog. 1999;27(1):1-11
6. Sola-Landa A. et al. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol Microbiol. 1998;29(1):125-38
7. Swartz T.E. et al. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317(5841):1090-3
8. Wu Q. et al. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. Microbiol BMC. 2006;6:102
9. Skerker J.M. et al. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3(10):e334
10. Salama N.R, Manoil C. Seeking completeness in bacterial mutant hunts. Curr Opin Microbiol. 2006;9(3):307-11
11. Kang Y. et al. Systematic mutagenesis of the Escherichia coli genome. Bacteriol J. 2004;186(15):4921-30
12. Murphy K.C, Campellone K.G, Poteete A.R. PCR-mediated gene replacement in Escherichia coli. Gene. 2000;246(1-2):321-30
13. Lai F, Schurig G.G, Boyle S.M. Electroporation of a suicide plasmid bearing a transposon into Brucella abortus. Microb Pathog. 1990;9(5):363-8
14. Kahl-McDonagh M.M, Ficht T.A. Evaluation of protection afforded by Brucella abortus and Brucella melitensis unmarked deletion mutants exhibiting different rates of clearance in BALB/c mice. Infect Immun. 2006;74(7):4048-57
15. Pei J. et al. Brucella abortus Rough Mutants Induce Macrophage Oncosis That Requires Bacterial Protein Synthesis and Direct Interaction with the Macrophage. Infect Immun. 2006;74(5):2667-75
16. Lee I.S, Slonczewski J.L, Foster J.W. low-pH-inducible A, stationary-phase acid tolerance response in Salmonella typhimurium. Bacteriol J. 1994;176(5):1422-6
17. Hernandez-Castro R. et al. The aquaporin gene aqpX of Brucella abortus is induced in hyperosmotic conditions. Microbiology. 2003;149(Pt 11):3185-92
18. Hornback M.L, Roop R.M. 2nd, The Brucella abortus xthA-1 gene product participates in base excision repair and resistance to oxidative killing but is not required for wild-type virulence in the mouse model. Bacteriol J. 2006;188(4):1295-300
19. Gee J.M. et al. Role of catalase in the virulence of Brucella melitensis in pregnant goats. Vet Microbiol. 2004;102(1-2):111-5
20. Porte F, Liautard J.P, Kohler S. Early acidification of phagosomes containing Brucella suis is essential for intracellular survival in murine macrophages. Infect Immun. 1999;67(8):4041-7
21. Detilleux P.G, Deyoe B.L, Cheville N.F. Effect of endocytic and metabolic inhibitors on the internalization and intracellular growth of Brucella abortus in Vero cells. Am J Vet Res. 1991;52(10):1658-64
22. Elzer P.H. et al. Characterization and genetic complementation of a Brucella abortus high-temperature-requirement A (htrA) deletion mutant. Infect Immun. 1994;62(10):4135-9
23. Iannino F, Ugalde R.A, Inon de Iannino N. Characterization of Brucella abortus sigma factor sigma54 (rpoN): genetic complementation of Sinorhizobium meliloti ntrA mutant. Microb Pathog. 2008;45(5-6):394-402
24. Teixeira-Gomes A.P, Cloeckaert A, Zygmunt M.S. Characterization of heat, oxidative, and acid stress responses in Brucella melitensis. Infect Immun. 2000;68(5):2954-61
25. Teixeira-Gomes A.P. et al. Mapping and identification of Brucella melitensis proteins by two-dimensional electrophoresis and microsequencing. Electrophoresis. 1997;18(1):156-62
26. Wang Y. et al. Comparative proteomics analyses reveal the virB of B. melitensis affects expression of intracellular survival related proteins. PLoS One. 2009;4(4):e5368
27. Gupta R.S, Mok A. Phylogenomics and signature proteins for the alpha proteobacteria and its main groups. Microbiol BMC. 2007;7:106
28. Amirmozafari N. et al. Comparison of heat shock response in Brucella abortus and Brucella melitensis. Pak J Biol Sci. 2008;11(2):188-94
29. Hoang T.T. et al. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212(1):77-86
30. Kovach M.E. et al. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 1994;16(5):800-2
Author contact
![]() Correspondence to: Department of Infectious Disease and Microbiology, College of Veterinary Medicine, China Agricultural University, Beijing 100193. Phone: +86-10- 62733901. E-mail: wuqmedu.cn.
Correspondence to: Department of Infectious Disease and Microbiology, College of Veterinary Medicine, China Agricultural University, Beijing 100193. Phone: +86-10- 62733901. E-mail: wuqmedu.cn.

 Global reach, higher impact
Global reach, higher impact