Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2009; 5(6):596-602. doi:10.7150/ijbs.5.596 This issue Cite
Research Paper
Bioinformatics Analysis the Complete Sequences of Cytochrome b of Takydromus sylvaticus and Modeling the Tertiary Structure of Encoded Protein
1. College of Life and Environment Science, Huangshan University, Huangshan, Anhui, 245021, China
2. Bio-electronic Centre of Shanghai University, Shanghai, 200072, China
Received 2009-2-22; Accepted 2009-9-15; Published 2009-9-22
Abstract
Cytochrome b (cyt b) gene complete sequences (1143bp) of Takydromus sylvaticus were sequenced. In order to clarify the phylogenetic position of the Takydromus sylvaticus, we investigated the phylogeny of 15 Takydromus spp. distributed in East-Asia by Maximum Parsimony (MP), Bayesian Inference (BI), and Maximum Likelihood (ML) methods using DNA fragments of cyt b genes. The results supported that the Platyplacopus merged into Takydromus and negated the validity of Platyplacopus. Furthermore, the prediction of tertiary structures of cyt b exhibited the CD loop region contain two short helices forming a hairpin arrangement, namely cd1 and cd2. Thermostability analysis shows that the CD-loop region is unstable thermodynamically and may provide mobility to amino acids located at the heme, and might provide high flexibility to the top of ISP (iron-sulfur protein) and the cavity region of Qo binding site. It suggested that the two short helices of CD loop region of cyt b was a dominating portion for ISP binding site.
Keywords: Takydromus sylvaticus, Cytochrome b (cyt b), Phylogeny, homology modeling, tertiary structure
1. Introduction
Takydromus sylvaticus, is one of the most enigmatic lizards in China, which belongs to Lacortidae, Lacertilia, Squamata, Reptilia. The first discovery of this lacertid species were collected in 1926 from Chung-an County (currently known as the Wuyishan region), Fujian Province, China [1]. In 1929, Pope stated that this species was unpredictably distributed at the type locality and extremely rare or absent elsewhere [2]. However, there were no records of additional specimens or biological observations of this species, until the re-discovery of this species in 2000 from Southern Anhui Province [3]. Further, we described detailed morphological characters of the newly collected specimens of T. sylvaticus [4].
About the taxa of Takydromus sylvaticus, Pope described an independent genus, Apeltonotus, to accommodate both of these species [2], however, he later synonymized this genus with Platyplacopus [5].on the basis of phylogenetic studies using morphological characters, Arnold [6] reduced the rank of Platyplacopus to a subgenus of Takydromus. However, after comparing representative specimens of Platyplacopus and Takydromus, Zhao et al., argued for the validity of Platyplacopus as a full genus on the basis of its distinctness in toe structure from Takydromus [7]. Based on molecular data, Lin et al. and Ota et al. inferred phylogenetic relationships of the genus Takydromus, and negated the validity of Platyplacopus at any taxonomic level [8, 9]. Our previous analyses of 12S rRNA sequences from 14 Takydromus spp. indicated that T. sylvaticus and T. dorsalis are considered to be phylogenetically most closely related to each other, and also do not support the division of Takydromus into the subgenera Platyplacopus and Takydromus [10].
Cytochrome b (cyt b) is one of the cytochromes involved in the electron transport in the respiratory chain of mitochondria. The cyt b gene is the most widely used gene for phylogenetic work, and it is the only cytochrome coded by mitochondrial DNA. In this paper, the complete mitochondrial cyt b gene of T.sylvaticus was sequenced and analyzed based on bioinformatics methods. Moreover, the phylogenetic relationship of Takydromus was reconstructed using cyt b gene data of 15 Takydromus spp., which distributed in East-Asia were collected from the GenBank database. In addition to, the tertiary structure of cyt b encoded protein was predicted using computational program and homology modeling methods.
2. Materials and Methods
2.1. Samples and DNA Sequencing
Specimens of T. sylvaticus were collected in a recovering subtropical forest and adjacent bamboo plantation (29.45 o N, 118.15 o E) near the Lingnan Nature Reserve, Xiuning County, Anhui Province, China. The reserve is located west of the Bajishan Range that lies on the border of Zhejiang, Jiangxi, and Anhui Province. The genomic DNA extracted from specimens using standard method.
The primers used for Polymerase chain reaction (PCR) amplification were as follows: L1: 5' accaccgttgttttcaactac 3'; H1:5' taggctacaaggactcgagtc 3'. It can amplify a fragment of cyt b gene region approximately 1200 bps in length. Purified PCR products were sequenced from both directions with an ABI-377 automated DNA sequencer. Acquired sequences were blasted against the GenBank database to verify that required sequences had been amplified.
2.2. Phylogenetic Analyses
Cyt b sequences of T. sylvaticus were initially assembled with the SeqEdit package (ABI, USA), and the cyt b gene data of other 14 Takydromus spp. were collected from the GenBank database (Table 1). The cyt b genes of Eremias velox (accession number, AF206549) and Lacerta vivipara (accession number, U69834) were used as outgroup in phylogenetic analyses. Alignments were first conducted using ClustalX v2.03 software [11] with default parameters, and subsequently verified manually. Considering majority sequences were incomplete, 656 bps consensus sequences of cyt b genes of 15 Takydromus spp. were selected for alignment analyses.
The localities of the samples and the origins of cyt b gene data in the study
| Species | Origin | Available length (bps) | Accession number |
|---|---|---|---|
| T. amurensis | Lin, 2006 | 702 | AY248458 (GenBank) |
| T.tachydromoides | Kumazawa, 2007 | 1143 | AB080237 (GenBank) |
| T.hsuehshanesis | Lin, 2006 | 702 | AY248482 (GenBank) |
| T.formosanus | Lin, 2006 | 702 | AY248521 (GenBank) |
| T.stejnegeri | Lin, 2006 | 684 | AY248476 (GenBank) |
| T.septentrionalis | Lin, 2006 | 681 | AY248469 (GenBank) |
| T.sexlineatus | Fu, 2002 | 1043 | AF206533 (GenBank) |
| T.toyamai | Lin, 2006 | 702 | AY248480 (GenBank) |
| T.smaragdinus | Lin, 2006 | 702 | AY248473 (GenBank) |
| T.sauteri | Lin, 2006 | 702 | AY248467 (GenBank) |
| T.intermedius | Lin, 2006 | 702 | AY248462 (GenBank) |
| T.keuhnei | Lin, 2006 | 684 | AY248463 (GenBank) |
| T.dorsalis | Lin, 2006 | 699 | AY248460(GenBank) |
| T.wolteri T.sylvaticus | Lin, 2006 Chen, 2008 | 660 1143 | AY248481 (GenBank) EF495176 (This paper) |
The sequence data were analyzed using maximum parsimony (MP) and maximum likelihood (ML) implemented in PAUP* 4.0 b10 (Swofford, 2003). Heuristic MP Searches were executed in 1000 random addition replicates with all characters unordered and equally weighted, and using tree bisection reconnection (TBR) branch-swapping. Bootstrap branch proportions (BBP) were calculated with 1000 MP replicates. Based on the Akaike Information Criterion (AIC), the GTR+I+G model was selected for maximum likelihood (ML) analyses by Modeltest 3.7 [12]. Heuristic Searches were executed in 10 replicates with the GTR+I+G model, and using TBR branch swapping. BBP values were calculated with 10 ML replicates [13].
Bayesian inference (BI) was carried out using MrBayes 3.1 [14]. The Bayesian posterior probabilities (BPP) used models estimated with Modeltest 3.7 under the AIC. Two separate runs were performed with four Markov chains. Each run was conducted with 1,000,000 generations and sampled every 100 generations. When the log-likelihood scores were found to stabilize, a consensus tree was calculated after omitting the first 25% trees as burn-in [13].
2.3. Modeling of Cyt b Tertiary Structure
In order to select suitable templates for cyt b protein, the Automated Mode program (http://swissmodel.expasy.org/workspace/index.) of the SWISS-MODEL server were used to research the optimization model. Lastly, we obtained chicken's cyt b tertiary structure (accession number 3h1jC) [15] as a suitable template, and the sequence identity is 73.16%. The alignment results are as for figure 1.
Modeling of cyt b tertiary structure was performed using the programs Swiss-PdbViewer (http://www.expasy.ch/spdbv/). The templates are thereby weighted by their sequence similarity to the target sequence, and steric conflicts between amino acid side chains were removed by manual adjustment. The best-matching loop template is selected using a scoring scheme, which accounts for force field energy, steric hindrance and favorable interactions like hydrogen bond formation. The reconstruction of the model side chains is based on the weighted positions of corresponding residues in the template structures. Starting with conserved residues, the model side chains are built by iso-sterically replacing template structure side chains [16]. The modeling structure was optimized using Insight II software package, and reliability analysis of templates was performed using the programs VMD (version 1.8.6) to calculate the RMS between two molecules.
The alignment results of cyt b protein sequences between the target and templates

3. Results
3.1. Sequence Variation
The complete cyt b gene sequences of T. sylvaicus were 1143bps, and encoded 380 amino-acids sequences. Newly obtained sequences were submitted to GenBank (accession number EF495176). The combined dataset included 295 variable sites and 241 potentially parsimony-informative sites over a total of 656 alignment nucleotide sites.
3.2. Phylogenetic Analyses
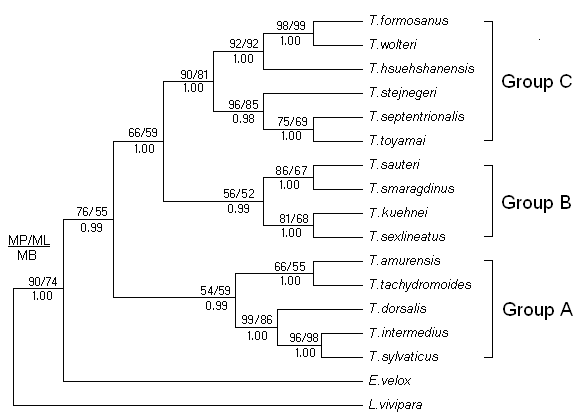
Based on the E.velox and L.vivipara as outgroups, molecular phylogenetic trees were reconstructed using MP method, which were virtually identical to the trees recovered in ML and BI analyses (Figure 2). The analyses found that the phylogenetic trees were consistent, and approximated three major species groups were defined for 15 species lacertids. Group A contained T.amurensis, T.tachydromoides, T.dorsalis, T.intermedius, and T.sylvaticus, Group B contained T.sauteri, T.smaragdinus, T.keuhnei, and T.sexlineatus. Group C contained the remaining species, including T.formosanus, T.wolteri, T.hsuehshanesis, T.toyamai, T.septentrionalis, and T.stejnegeri.
Furthermore, we noticed that the T. sylvaticus and T.intermedius formed sister relatives (96, 98, and 100% support for MP, ML bootstrap proportions and Bayesian posterior probability, respectively). Then, the T.dorsalis was closed to this sister groups with strongly supported values (99, 86 and 100% support). It indicated that they were the closest relatives among all the species used for the phylogenetic reconstruction.
Bayesian inference tree derived from partial cyt b sequences of the lacertids. Numbers above branches are bootstrap support for MP (1000 replicates) / ML (10 replicates) analyses (>50 retained), and numbers below branches indicate Bayesian posterior probabilities (>90% retained). Based on the topological structure of phylogenetic trees, 15 species lacertids were defined approximated three major species groups (A, B and C).
3.3. Tertiary Structure Analyses
Firstly, the 300 steps energy minimization calculation to optimize the modeling structure of cyt b protein, including 100 steps steepest descent method and 200 steps conjugate gradient method [17, 18]. Then, in order to solve the problem of local potential barrier, 50ps normal temperature (300K) molecular dynamics (MD) optimization were performed using Insight II package. Moreover, the CA (carbon alpha) congruence between the template structures (cyt b) and the crystallogram of 3h1jC were performed based on VMD program, and the average RMS=0.15nm, it indicates that the modeling structures were reliable for the predicted tertiary structures of cyt b.
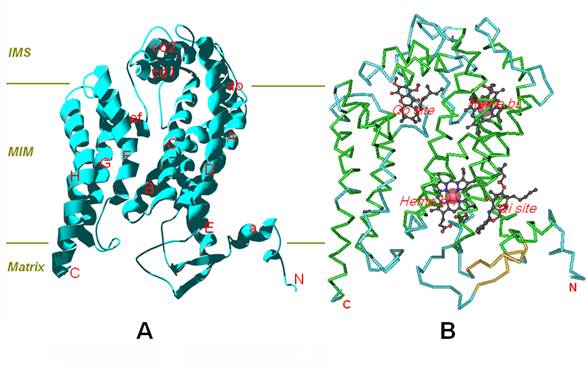
The predicted result shows that the cyt b protein of T.sylvaticus spans the mitochondrial membrane with eight transmembrane (TM) helices, named sequentially from A to H, with both the N- and the C-terminus are located in the mitochondrial matrix. In the tertiary structure, the eight helices are arranged in two helical bundles, one consisting of helices A-E and the other of helices F-H (Figure 3-A). Pairs of transmembrane (TM) helices (A to H) were connected by seven extra-membrane loops, including four long loops (AB, CD, DE, EF) and three short loops (BC, FG, GH). Among the four large loops, the AB and EF loop each contain one helix, namely, ab and ef, the CD loop has two short helices forming a hairpin arrangement, namely, cd1 and cd2. In these four large loops, the DE loop is on the matrix side, while the remaining three long loops are on the intermembrane space (IMS) side. In addition, the amphipathic surface helix-a was located near closed N-terminal.
Furthermore, Cytochrome b contains two bound hemes and two ubiquinol/ubiquinone (Qo/Qi) binding sites (Figure 3-B). The two haems (bL and bH) are incorporated into the first helical bundle, and the axial ligands for haem bL are His84 and His183, His98 and His197 for haem bH, these four histidines are high conserved in the sequences of cyt b [19]. The quinol oxidation (Qo) pocket is located near the IMS side of the membrane surface, and the quinone reduction (Qi) pocket near the matrix side of the membrane with its entrance open to the center of the membrane bilayer. In particular, the cavity of Qi pocket region dives nearly vertically toward the matrix side of the membrane and is surrounded by TM helices A, D and E, and the amphipathic surface helix a.
(A) Ribbon presentation of the cyt b structure of T.sylvaticus. (B) The two bound hemes (bL and bH) and two ubiquinol/ubiquinone (Qo and Qi) binding sites. The structure is oriented with the matrix side at the bottom of the figure, the intermembrane space (IMS) on the top, and the mitochondrial inner membrane (MIM) in the middle, which is roughly delineated with the two parallel lines.
4. Discussions
4.1. Phylogeny of Takydromus
Although use of cytochrome b has some pitfalls, the cyt b gene still is the most widely used gene for phylogenetic analysis for taxonomic, especially are good tools for studying phylogenetics of closely related species [20]. Within species, control region sequences usually are a better choice, because more relaxed structural and functional constraints lead to a faster average substitution rate. In this study, the molecular phylogenetic trees were reconstructed based on cyt b data of Takydromus. The result supported that the Platyplacopus merged into Takydromus and negated the validity of Platyplacopus. It is also consistent with the opinions of Arnold et al. [21, 22] and Lin et al. [8, 23], whom supported the combination of subgenera Platyplacopus and Takydromus into Takydromus.
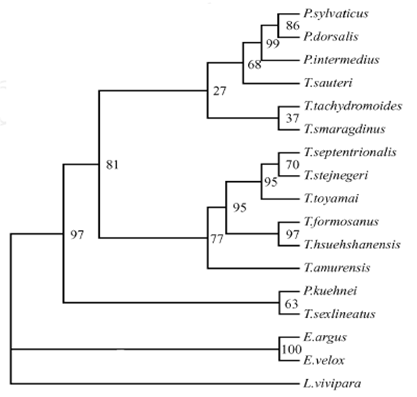
In the trees, the T.dorsalis was closed to the sister groups of T. sylvaticus and T.intermedius with strongly support values (figure 2), and shows the relationships of these species are much closer. However, in our previous study, we found that the T. sylvaticus and T.dorsalis formed sister groups, and then closed to T.intermedius, which based on 12S rRNA gene for phylogenetic analyses [10] (figure 4). It shows the more relationships of these three lacertids are unclarity, although the T.sylvaticus, T.dorsalis, and T.intermedius always clustering in a branch. This phenomenon will be investigated by further experiments and data.
MP tree based on 12S rRNA gene sequences data. Numbers at node are bootstrap values (1000 replicates) from MP method [10]
4.2. Analysis the Tertiary Structure of Cyt b Proteins of T.sylvaticus
The mitochondrial cytochrome b is a membrane protein and a central component of cellular energy conservation machinery in different organisms, and concomitantly translocates protons across membranes for ATP synthesis and various cellular processes [24, 25]. In this study, homology modeling method and computer programs were performed to predict the tertiary structure of cyt b proteins of T.sylvaticus. 3D structures show the cyt b contains eight TM helices connected by seven loops with intra-membrane or extra-membrane domains. Furthermore, cyt b contains two bound hemes and two ubiquinol/ubiquinone binding sites. Among the seven loops, four long loops (AB, CD, DE and EF) are prominent for their special positions: three loops on the intermembrane space (IMS) side and only one loop (DE) on the matrix side. This phenomenon indicates that these four large loops are perhaps most important to the function of cyt b, as they are the primary participants in the formation of the quinol oxidation (Qo) and quinone reduction (Qi) site. Moreover, among the sequence of Qi site residues, His202, Lys228, and Asp229 are highly conserved (Figure 3-A). Gao et al. demonstrated the His202 binds to the carbonyl oxygen of the bound ubiquinone through a water molecule; Asp229 interacts with the other carbonyl oxygen via another water molecule that was stabilized by Lys228. They proposed a two-water molecule mediated four- step mechanism for quinone reduction and protonation at the Qi site [19]. Hacker et al. also identified the mutations of these three residues led to a defective phenotype in heme bH reoxidation, and significantly reduced the rate of quinone reduction, respectively [26]. This suggests that these three residues play crucial roles in Qi binding site.
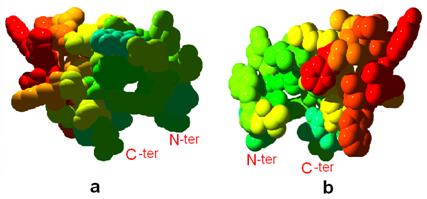
Furthermore, we noticed that the CD loop contains two short helices, including cd1 and cd2. Thermostability analysis shows that the structure of CD-loop region is unstable thermodynamically and may provide mobility to amino acids located at the heme (Figure 5), and might provide high flexibility to the top of ISP (iron-sulfur protein) and the cavity region of Qo binding site. Esser et al. analyzed eight structures of mitochondrial bc1 with bound on Qo site inhibitors, and indicated that the presence of inhibitors causes a bidirectional repositioning of the cd1 helix in the cyt b, and especially, as the cd1 helix forms a major part of the ISP binding crater, any positional shift of this helix modulates the ability of cytochrome b to bind ISP [27]. Zhu et al. also determined the presteady-state kinetics of ISP and cytochrome bL reduction by ubiquinol, and postulated that the iron-sulfur protein accepts the first electron from ubiquinol to generate ubisemiquinone anion to reduce heme bL [28]. On the basis of this mentioned, we speculated the CD loop region is a dominating portion for ISP binding site, and the existence of instability in the CD loop could affect the function of these two short helices. This hypothesis will be investigated by further experiments.
Front (a) and back (b) views shows the thermostability of CD loop region of cyt b. Light blue to red colors show stable to unstable regions, respectively. Tertiary structures are shown in the space-fill mode.
Abbreviations
Cyt b: Cytochrome b; BI: Bayesian inference; ML: Maximum likelihood; MP: Maximum parsimony; ISP: Iron-sulfur protein; PCR: Polymerase chain reaction; MD: Molecular dynamics; TM: Transmembrane; IMS: Intermembrane space; MIM: Mitochondrial inner membrane; Qo/Qi: Ubiquinol/ubiquinone.
Acknowledgements
The studies were supported by the National Science Foundation of China (30870281), the Science Foundation of Huangshan University (2007xkjq026) and the Innovative Foundation of PhDs of Shanghai University.
Conflict of Interest
The authors have declared that no conflict of interest exists.
References
1. Pope CH. Seven New Reptiles from Fukien Province, China. American Museum Novitates.New York. 1928;320:1-2
2. Pope CH. Notes on Reptiles from Fukien and Other Chinese Provinces. Bulletin of the American Museum of Natural History. New York. 1929;58:372-374
3. Xin-sheng T, Pen X. On the rediscovery and the distribution expansion of the Platyplacopus sylvaticus. Chines J Zool. 2002;37:65-66
4. Tang XS, Lu SQ, Chou WH. Description of Male Takydromus sylvaticus (Squamata: Lacertidae) from China, with Notes on Sexual Dimprphism and a Revision of the Morphological Diagnosis of the Species. Zoological Science. 2007;24:496-503
5. Pope CH. The Reptiles of China: Turtles, Crocodilians, Snakes, Lizards. New York: The American Museum of Natural History. 1935:473-476
6. Arnold EN. Towards A Phylogeny and Biogeography of the Lacertidae: Relationships Within An Old-Word Family of Lizards Derived from Morphology. Bulletin of the British Museum ( Natural History) Zoology. 1989;55:209-257
7. Zhao EM, Liu MY. Family Lacertidae Gray, Genus Platyplacopus Boulenger, 1917; In Fauna Sicia, Reptilia Vol 2 Squmata: Lacertilia Ed by Editorial Committee of Fauna Sinca, Academia Sinica. Beijing: Secience Press. 1999:251-257
8. Lin S-M, Wang C-J, Hsu Y-C. et al. Isolation and characterization of 12 tetra-repeated microsatellite loci from the Formosan grass lizard (Takydromus formosanus). Molecular Ecology Notes. 2006;6:57-59
9. Ota H, Honda M, Chen SL. et al. Phylogenetic relationships, taxonomy, character evolution and biogeography of the lacertid lizards of the genus Takydromus (Reptilia: Squamata): a molecular perspective. Biological Journal of the Linnean Society. 2002;76:493-509
10. Tang X-S, Chen Q-L. On the taxonomic status of Platyplacous syslvaticus based on 12S rRNA gene. Acta Zootax Sin. 2006;31:475- 479
11. Larkin MA, Blackshields G, Brown NP. et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947-2948
12. Posada D, Buckley TR. Model selection and model averaging in phylogenetics: advantages of akaike information criterion and bayesian approaches over likelihood ratio tests. Syst Biol. 2004;53:793-808
13. CAI H-X, CHE J, PANG J-F. et al. Paraphyly of Chinese Amolops (Anura, Ranidae) and phylogenetic position of the rare Chinese frog, Amolops tormotus. Zootaxa. 2007;1531:49-55
14. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572-1574
15. Zhang ZL, Huang LS, Shulmeister VM. et al. Electron transfer by domain movement in cytochrome bc(1). Nature. 1998;392:677-684
16. Schwede T, Kopp Jr, Guex N. et al. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Research. 2003;31:3381-3385
17. Ting-Jun H, Yu A, Bing-Gen R. et al. Theoretical Studies on the Crystal Packing Form of MT-II from RatLiver. Chemical J Chinese Universities. 2002;23:271-274
18. Yuan Z, Ze-Sheng L, Miao S. et al. Homology Modeling of Three-dimension Structur e of Hepatitis B Surface Antigen Fragment and Its Ligand Design. Chemical J Chinese Universities. 2005;26:02-105
19. Gao XG, Wen XL, Esser L. et al. Structural basis for the quinone reduction in the bc(1) complex: A comparative analysis of crystal structures of mitochondrial cytochrome bc(1) with bound substrate and inhibitors at the Qi site. Biochemistry. 2003;42:9067-9080
20. Farias IP, Ortà G, Sampaio I. et al. The cytochrome b gene as a phylogenetic marker: the limits of resolution for analyzing relationships among cichlid fishes. J Mol Evol. 2001;53:89-103
21. Arnold EN. Interrelationships and evolution of the east Asian grass lizards, Takydromus (Squamata: Lacertidae). Zool. J. Linn. Soc. 1997;119:267-296
22. Arnold EN, Arribas O, Carranza S. Systematics of the Palaearctic and Oriental lizard tribe Lacertini (Squamata: Lacertidae: Lacertinae), with descriptions of eight new genera. Zootaxa. 2007:3-86
23. Lin SM, Chen CA, Lue KY. Molecular phylogeny and biogeography of the grass lizards genus Takydromus (Reptilia: Lacertidae) of East Asia. Molecular Phylogenetics and Evolution. 2002;22:276-288
24. Trumpower BL, Gennis RB. Energy Transduction by Cytochrome Complexes in Mitochondrial and Bacterial Respiration - the Enzymology of Coupling Electron-Transfer Reactions to Transmembrane Proton Translocation. Annual Review of Biochemistry. 1994;63:675-716
25. Brandt U, Trumpower B. The Protonmotive Q-Cycle in Mitochondria and Bacteria. Critical Reviews in Biochemistry and Molecular Biology. 1994;29:165-197
26. Hacker B, Barquera B, Crofts AR. et al. Characterization of Mutations in the Cytochrome-B Subunit of the Bc(1)-Complex of Rhodobacter-Sphaeroides That Affect the Quinone Reductase Site (Qc). Biochemistry. 1993;32:4403-4410
27. Esser L, Gong X, Yang SQ. et al. Surface-modulated motion switch: Capture and release of iron-sulfur protein in the cytochrome bc(1) complex. PNAS. 2006;103:13045-13050
28. Zhu H, Egawa T, Yeh SR. et al. Simultaneous reduction of iron-sulfur protein and cytochrome b(L) during ubiquinol oxidation in cytochrome bc(1) complex. PNAS. 2007;104:4864-4869
Author contact
![]() Correspondence to: LU Shun-Qing, No.9 Jilingshan Road, College of Life and Environment Science, Huangshan University, Huangshan City, Anhui Province, 245041, China. Tel: +86-559-2546613, Fax: +86-559-2656630, E-mail: cqlw1975edu.cn.
Correspondence to: LU Shun-Qing, No.9 Jilingshan Road, College of Life and Environment Science, Huangshan University, Huangshan City, Anhui Province, 245041, China. Tel: +86-559-2546613, Fax: +86-559-2656630, E-mail: cqlw1975edu.cn.

 Global reach, higher impact
Global reach, higher impact