10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2010; 6(5):407-418. doi:10.7150/ijbs.6.407 This issue Cite
Research Paper
Subtle evolutionary changes in the distribution of N-glycosylation sequons in the HIV-1 envelope glycoprotein 120
Aarhus University, Department of Genetics and Biotechnology, Forsøgsvej 1, Slagelse 4200, Denmark
Received 2010-3-19; Accepted 2010-7-14; Published 2010-7-21
Abstract
Many viruses are known to undergo rapid evolutionary changes under selective pressures. The HIV-1 envelope glycoprotein 120 (gp120) shows extreme selection for NXS/T sequons, the potential sites of N-glycosylation. Although the average number of sequons in gp120 appears to be relatively stable in the recent past, even slight changes in the distribution of sequons may potentially play crucial roles in protein interaction and viral infection. This study tracked the prevalence and distribution of NXS/T sequons in gp120 over a period of 29 years (from 1981 to 2009). The gp120 showed location specific distribution of sequons with higher density in the outer domain of the molecule. The NXT sequon density decreased in the outer domain (despite the increase in the sequon specific amino acid threonine), but increased in the inner domain. By contrast, the NXS sequon density increased specifically in the outer domain. Related changes were also seen in the distribution probabilities of sequons in two domains. The results indicate that the gp120, chiefly in subtype B, is redistributing NXS/T sequons within the molecule with specific selection for NXS sequons. The subtle evolution of sequons in gp120 may have implications in viral resistance and infection.
Keywords: Molecular evolution, Glycoprotein, Human immunodeficiency virus, N-glycosylation sequons.
Introduction
Protein N-glycosylation is an important co-translational modification process wherein short sugar chains are covalently attached to the amide group of asparagine (N) residue in the amino acid chain [1]. N-glycosylation affects a number of properties of proteins such as solubility, stability and turnover, secretion, protease resistance, protein-protein interaction/recognition and immunogenicity, and hence has an immense biological importance [1-3]. Although asparagine occurs frequently in the protein chain, N-glycosylation requires asparagine to be present in special motifs - NXS/T sequons (where X is any amino acid except proline which is avoided due to conformational hindrance and the third residue is either serine or threonine). Further, N-glycosylation occurs only on some sequons found in membrane-bound or secretory proteins which are exposed to the enzyme oligosaccharyltransferase in the lumen of endoplasmic reticulum [1-6].
The human immunodeficiency virus-1 (HIV-1) envelope glycoprotein 120 (gp120) occurs as a trimeric complex, with each monomer in non-covalent association with gp41 (a transmembrane viral envelope glycoprotein anchor) and interacts with its primary receptor - CD4 glycoprotein of the host T-lymphocyte [7-9]. The gp120 contains an average of 26 NXS/T sequons, and therefore can be extensively N-glycosylated. As much as 55% of the molecular mass of the gp120 is contributed by carbohydrates as a result of N-glycosylation of most of its potential N-glycosylation sequons and therefore it is one of the most heavily glycosylated molecules in nature [10-12]. The HIV-1 pathogenesis, to a large extent, has been attributed to the structural plasticity of the gp120, and in particular, to the variability of N-glycosylation [7, 9, 13]. For example, inhibition studies have demonstrated that proper N-glycosylation of gp120 is critical for the infectivity of the virus [14]. Mutation studies have indicated that N-glycosylation around the CD4 binding site of the gp120 is required for high affinity receptor interaction [10, 15]. It has been suggested that the HIV uses this 'glycan shield' on gp120 for the purpose of host immune evasion [16].
Therefore, it may be hypothesized that under selection pressures, the gp120 must undergo rapid changes with respect to its N-glycosylation sequons [17]. For instance, under selection pressure by mannose-specific lectins, resistant HIV-1 strains showed marked depletion (up to eight out of 22) of the N-glycosylation sequons in gp120 regions distant from interacting sites [18]. Fluctuations in the variable regions of the gp120 itself has been shown to change the N-glycosylation sequons in the early stages of viral infection [19]. Likewise, it is reasonable to expect a long-term trend in the number of N-glycosylation sequons in gp120 under the selection pressure from the host immune system or anti-retroviral drugs since the passage of HIV to human hosts [16, 20].
To data, there are only a few studies which attempted long-term tracking of N-glycosylation sequons. For example, while hemagglutinin of human influenza virus A/H3N2 was found to accumulate the sequons over time, the gp120 of HIV-1 showed only variations in the number and location of sequons, but not any particular trend over time [21]. Similarly, a recent study attributed the abundance of N-glycosylation sequons in the gp120 to Darwinian selection, but found no significant change in the overall number of sequons since the HIV-1 moved from primates to humans [22]. With this information in mind, we sought to answer a different set of questions. Is there any location specific variability in the number and/or distribution of N-glycosylation sequons in the gp120? And what direction its evolution has been taking since the viral passage to human hosts? Finding answers to these questions may be important and relevant as they may have implications in understanding the gp120 evolution, and viral infectivity and resistance [23].
Materials and Methods
Sequence acquisition
The HIV-1 envelope gp120 sequences were downloaded from the HIV database at Los Alamos National Laboratory (http://www.hiv.lanl.gov/content/index). Only the sequences with information about the year of sampling were used in this study. Further, sequences containing ambiguous amino acids were discarded. The final set constituted 11333 amino acid (and corresponding nucleotide) sequences from 29 years (from 1981 to 2009). As our main aim was to find the general patterns of sequon variability in the gp120 of HIV-1 [17, 22], we have analyzed the sequences by taking all subtypes together. However, we have also analyzed the sequences from subtypes separately (62% sequences were from subtype B, 24% from subtype C and 14% from rest of the minor subtypes) [21] and results have been presented wherever appropriate.
Prevalence of NXS/T sequons
The number of NXS/T sequons (NXS and NXT, where X is any amino acid except proline) and NPS/T sequences were counted in each amino acid sequence. Overlapping sequons, if any, have been taken as separate sequons. The first half of the sequence was considered as the N-terminal region and the second half as the C-terminal region. When a sequence contained odd number of amino acids, additional amino acid was added to the C-terminal region. The NXS/T sequons were also enumerated in N and C-terminal regions separately. The sequon density was considered as the number of sequons per 100 amino acids. The percentage of N, P, S and T amino acids in the full sequence and in the two regions were computed form their respective frequencies over the total number of amino acids. Regions of gp120 having high probability of sequon occurrence were located (Figure 2) merely by stacking the sequences together after normalization for mean sequence length (510 amino acids).
Prediction of NXS/T sequon numbers
The probabilistic occurrence of NXS/T sequons was computed form the transition of amino acid frequencies by considering protein sequence as a Markov chain [6]. Accordingly, an ideal protein with all 20 amino acids in equal proportions has 0.00475 probability (per amino acid) of containing a NXS/T sequon. Thus, a protein sequence of 400 residues has 1.9 predicted NXS/T sequons. As an example, gp120 sequence (AY247224, year 1981) with 512 residues has 25 NXS/T sequons. But, only six sequons (44/512 * 489/512 * 75/512 * 512 = 6.15) may be predicted for this sequence based on amino acid transitions.
Percentage of A and T nucleotides
The AT content (asparagine is encoded by AT rich codons) has previously been identified as one of the possible evolutionary mechanisms to modulate the sequon numbers in glycoproteins [22]. Thus percentage of adenine and thymine were computed form their respective frequencies in the full, and in the N and C-terminal regions of the corresponding nucleotide sequences.
Computing the distribution probability of sequons
The distribution of NXS/T sequons in amino acid chain was modeled based on balls-in-boxes classical occupancy [24-29]. In brief, given n balls randomly distributed to equal number of boxes with equal probability, the number of possible distributions (empty boxes are allowed and the order of the distribution is ignored) follows the partition number [25]. Similarly, given n number of sequons, the amino acid chain may be subdivided into a maximum of n equal parts. Although the number of possible distributions is equal to the partition number, the given integer number of sequons can attain only one particular distribution at a time and the probability of that particular distribution is given by the formula:
Pr(r1, r2, . . . , rn) = 1/nr * r!/(r1! * r2! * . . . * rn!) * r!/(q0! * q1! * . . . * qn!)
Where, r is the number of sequons in a protein sequence, n is the number of partitions (is equal to r) on the protein sequence, ri is the number of sequons in any given partition, qi is the number of partitions with the same number of sequons and ! is the factorial function [25, 29]. As an example, all the possible distributions and respective distribution probabilities for r = 6 is presented in Table S1. In order to avoid too many decimals, log10 distribution probability was used.
Data analysis
The sequence analyses and data handling were done using programs written in the Python programming language (ver. 2.6, http://www.python.org/). The Biopython (http://biopython.org/) tools were used for sequence-parsing. A Microsoft Excel 2003/2007 spreadsheet was used to visualize the data and SigmaPlot (ver. 11, Systat Software Inc, CA, USA) was used to make the contour maps. A linear regression line was fitted to the scatter plots of mean (± 95% confidence interval) values versus year and Pearson's correlation coefficient (r) was computed between the two variables. Owing to the large sample size, parametric tests were favored and means were declared significant at p < 0.05. A Student's t test was used to see if the difference between the actual and the predicted mean values were significant. A significance test was also performed for the slope using the observed statistic over the estimated standard error of the statistic and the t-value was given wherever appropriate. A Z-test for two proportions or two sample means was used wherever applicable.
Results
N-glycosylation sequons in HIV-1 gp120
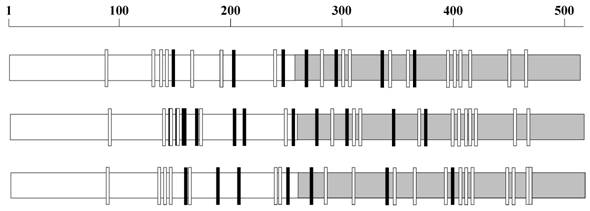
The HIV-1 gp120 has an average of 25.6 (±2.4, standard deviation) N-glycosylation sequons and a mean sequence length of 509.49 (±9.26) amino acids. Although the number and position of sequons are variable in different gp120 sequences (Figure 1), distinct clusters of sequons may be observed. Figure 2 shows the relative probability of finding sequons in the gp120 (n = 11333 sequences). The discrete peaks indicate rather steady occurrence of sequons in particular regions. Further, the NXS and NXT sequon regions are clearly distinguishable. It may be noticed that on or near the protein interaction regions (marked by green arrow-heads), for example, at amino acid position 425-430 (CD4 binding site) [7, 15], the sequon probability is virtually zero. Further, parts of the variable loops (V1/V2 and V3) do not contain any N-glycosylation sites (Figure 2).
Sequon density in gp120
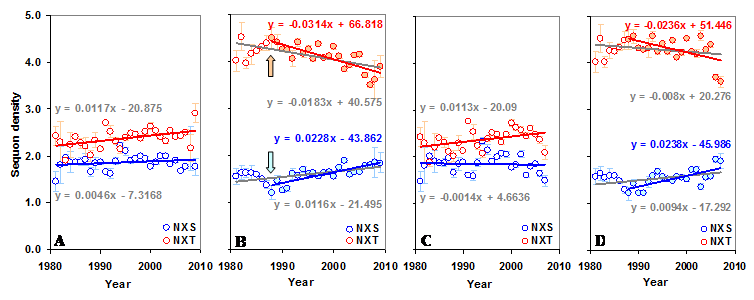
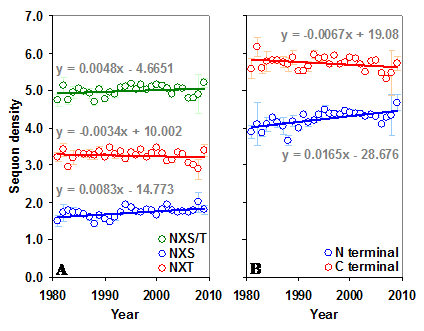
The gp120 has an average sequon density (number of sequons per 100 amino acids) of 5.02 (±0.43) with higher density (5.71±0.58) in the C terminal region compared to the N terminal (4.29±0.59). Further, the C terminal region has much higher NXT sequon density. In contrast, the predicted sequon density in gp120 is nearly 3.84 times lower (Figure S1 A and B). The predicted NXS density is significantly higher (p < 0.05, t = 3.54) in the C terminal region, as is the percentage of serine (5.47% versus 6.06%). There is no significant change in the NXS/T sequon density in the gp120 molecule over time (between 1981 and 2009). However, NXS sequons, when considered alone, show a significant increase (p < 0.05, t = 3.24) (Figure S2 A and B). The sequon density (NXT) is increasing in the N terminal region. In the C terminal region, NXS density is increasing and the NXT density is decreasing (Figure 3 A and B). These changes are much stronger when considered from the year 1988 onwards (arrows in Figure 3 B). Similar trends were observed for sequons in gp120 of HIV-1 subtype B (Figure 3 C and D). Although a comparable pattern was seen in subtype C (sequences available only from 1988 onward) the changes were not significant (data not shown). Inclusion of multiple gp120 sequences, if any, from same patients did not alter the overall pattern of sequon density and distribution.
N-glycosylation sequons in gp120. The N-glycosylation sequons vary in number and distribution in gp120 as exemplified here by three sequences AY247224 from 1981 (top), EU289201 from 1995 (middle) and GU080199 from 2009 (bottom). The NXS sequons are represented as small black rectangles and NXT as white rectangles. The C terminal regions of the sequences are gray shaded.
Occupancy of N-glycosylation sequons. The relative probability of the occurrence of sequons along the gp120 sequence has very distinct peaks. Sequon probability is almost zero on or near the binding site regions. The NXS probability is shown in red and NXT in blue. Binding site regions are pointed by using green arrow-heads. Variable regions of the gp120 sequence are indicated on the top (n = 11333 sequences).
Sequon density in gp120. The N and C terminal regions of gp120 have very different sequon densities. (A) The increase in NXT density over time in the N terminal region is significant (t = 2.88, p < 0.05). (B) The C terminal region has very high NXT density which is significantly decreasing (t = -4.08) over time, but NXS is increasing (t = 3.81). Comparable patterns are observed (C and D) when gp120 of HIV-1 subtype B is considered alone.
AT content and sequon specific amino acids in gp120
The AT content in the N and C terminal regions of the gp120 are significantly different (p < 0.05, 56.46% versus 62.26%). Further, the AT content is decreasing in the whole gp120 molecule over time. This decrease is significant (p < 0.05, t = -2.32) also in the N terminal region, but not in the C terminal region (Figure 4A).
The overall percentage of T and P amino acids are higher in the N terminal part of the gp120, while N and S are higher in the C terminal part. The percentage of sequon specific amino acids N, S and T have not changed much over time in the whole gp120 molecule (Figure S3 B). However, when considered separately in the two regions, N and T amino acids show interesting trend. The percentage of N amino acid is increasing and T is decreasing in the N terminal region (Figure 4B). This trend is quite reversed in the C terminal region (Figure 4C). The proline is significantly increasing in the whole gp120 and in the C terminal region (Figure 4B and 4C).
Distribution of sequons in gp120
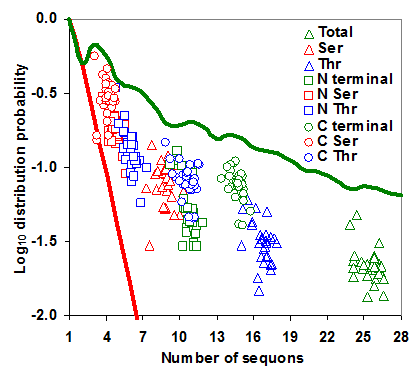
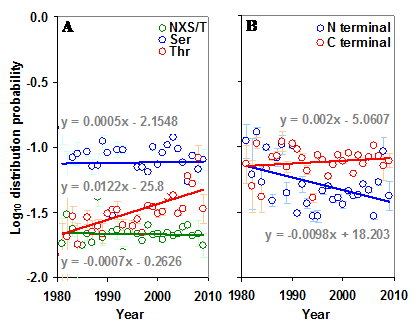
The distribution of sequons in gp120 was modeled based on classical occupancy [25, 29]. The calculation of log10 distribution probability is exemplified in table S1. It may be observed from Figure 5 that the distribution probabilities of sequons in the gp120 or in its two regions form very distinct clusters and are much closer to the maximum distribution probability line, indicating the probabilistically simpler near-random distribution of sequons. The overall distribution of NXS/T sequons in the gp120 has remained same over time. However, NXT distribution, when considered alone has been increasing significantly (p < 0.05, t = 4.28). The distribution of sequons is changing in the N terminal part of the gp120 as evidenced by the decreasing distribution probability (Figure S4 A and B). This is not significant when NXS and NXT sequons are considered separately. But in the C terminal region, the NXT sequon distribution probability is increasing, indicating a change towards the probabilistically simpler distribution over time (Figure 6 A and B). Similar trends were observed for the distribution of sequons in gp120 of HIV-1 subtype B (Figure 6 C and D).
Figure 7 shows the relative probability of finding sequons in the gp120 over time. It is clear that the probability of finding sequons is not random over gp120 or over time, but very is distinct. The two regions of the gp120 have different probabilities of NXS and NXT sequons. It may be noted that there are clear indications of emergence of new sequons (marked by black ovals) both NXS (for instance near amino acid positions 400 and 450) and NXT in the recent years (Figure 7 A and B). The directed disappearance of sequons is not so obvious due to overall high probability of sequons in much of the gp120 molecule.
Percentage of AT and sequon specific amino acids in gp120. (A) The AT content is significantly lower in the C terminal region and decreasing (t = -2.32) over time. (B) The percentage of N and T amino acids are changing significantly (t = 3.56 for N and t = -3.17 for T) in the N terminal region of gp120. (C) The direction of this trend is reversed (t = -2.13 for N and t = 6.33 for T) in the C terminal region.
Distribution probability of sequons in gp120. The log10 distribution probabilities of sequons form distinct clusters and are close to optimal maximum distribution probabilities. The even distribution probability is indicated by red line and the maximum distribution probability by green line.
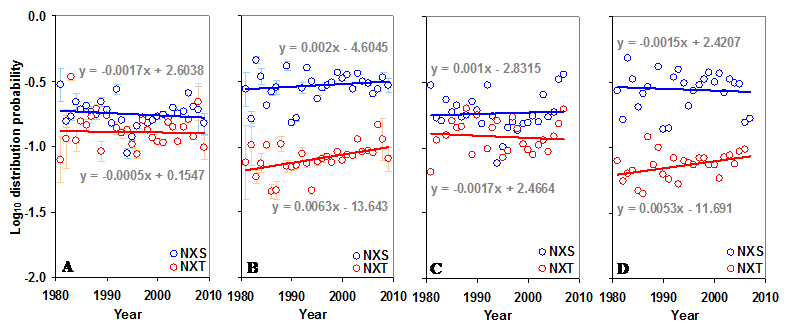
Distribution of sequons in gp120. (A) The log10 distribution probabilities of NXS and NXT have remained same over time in the N terminal region. (B) The log10 distribution probability of NXT is increasing (t = 2.70) over time in the C terminal region. Comparable patterns are observed (C and D) when gp120 of HIV-1 subtype B is considered alone.
N-glycosylation sequons in gp120. (A) The contour map shows higher density of NXS in the N terminal region and emergence of NXS in the new locations (marked by black ovals). (B) The density of NXT is higher in the C terminal region. The apparent emergence of NXT in the new location is marked by a black oval.
NPS/T sequences in gp120
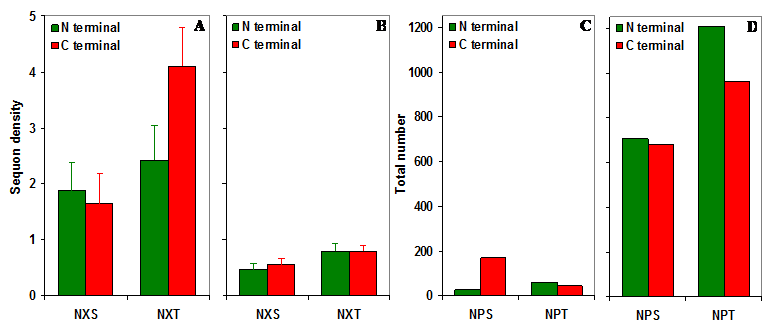
It may be noted that there is a total of 298 NPS/T three amino acid sequences in the entire set of gp120 sequences (n = 11333) studied here. This observation is in stark contrast to the expected number of NPS/T sequences in the gp120 molecule. Based on Markov chain model, the predicted number of NPS/T sequences in 11333 gp120 sequences is 3544 (NPT is significantly higher) which is over 11.89 times higher compared to the observed number of NPS/T sequences (Figure S1 C and D).
Discussion
It has long been appreciated that the N-glycosylation sequons in gp120 are a changing phenotype which provides a mechanism for immune evasion [17, 21, 30]. However, little is known about the exact nature of this change - whether it is a mere fluctuation or a directional trend. In the present study, we attempted to track the number and distribution of sequons in two (N and C terminal) parts of the gp120 molecule over time. The rational for dividing the gp120 sequence into two parts is two fold: First, the gp120 has two parts namely the inner and outer domains [7]. The bulk of the N terminal part of the sequence forms the inner domain and the C terminal part forms the outer domain. The amino acid chain makes a transition from inner to outer domain between sheets β8 and β9, which is almost the mid point of the sequence. Second, it is known that the two domains are unequally glycosylated with a higher proportion occurring in the outer domain [31, 32]. This inequality may either be due to the preferential glycosylation of sequons in the outer domain or simply that it contains many more sequons. It is clear from the present study that sequons are unequally distributed in gp120, with outer domain containing much higher NXT sequon density (Figures 1, 2 and 3). It may be noted that the outer domain is much more exposed to solvent and has a number of regions, which interact with host proteins as compared to the inner domain - much of it is facing gp41 or inner domains of gp120 monomers within the trimeric complex [7]. Therefore, higher sequon density in outer domain might have evolved as a protective mechanism against neutralizing antibodies [16, 32, 33].
In agreement with previous findings [21, 22], the present study found no significant change over time in the number of NXS/T sequons in the whole gp120 molecule. It has been postulated that there may be an upper bound to the number of sequons that can be maintained in the gp120 [21]. The gain or loss of sequons may influence the gp120/virus. For example, the glycan structures are quite bulky (~2000 Da) and therefore, the additional sequons may not provide a selective advantage due to conformational instability or functional redundancy. Similarly, the loss of sequons may have fitness costs, as they lead to failure of infectivity/immune evasion [34]. However, the present study shows a very clear directional change in the density (or number) of sequons in the two domains of gp120. The NXT sequon density is decreasing in the outer domain and the NXS sequon density is increasing. This is in contrary to the norm that the NXT sequons show a much higher Darwinian selection and are often preferentially glycosylated compared to NXS sequons [4-6, 22]. In the current scenario, NXS sequons must be rendering a selective advantage to the outer domain of gp120 molecule. Apparently, the sequons were initially (from 1981 to 1988) changing in directions opposite to the current ones (arrows in Figure 3). This trend is specific for gp120 of HIV-1 subtype B (Figure 3 C and D). The reason for this turnaround is not known, but the overall changes in the sequon density may be the result of antigenic drift since the viral passage to human hosts [8, 16, 18-21]. Directional changes in the sequons were not very obvious in subtype C and other minor subtypes due to narrow sampling period. Results presented here, therefore, may be characteristic of the major subtype B.
The increase in NXS density is significant even when gp120 molecule is considered as a whole. It is interesting to note that there is no net change in the NXT sequon density in the whole gp120 molecule. This is because the decrease in NXT density in outer domain is compensated by significant increase in NXT density in inner domain over the time period between 1981 and 2009. It may be recalled that previous studies failed to notice these intricate trends, instead reported mere fluctuations in the sequon numbers in gp120 over time [21, 22]. This is not surprising because they either considered both the sequons together in the whole gp120 molecule [22] or were too early to detect such a trend in a subset (up to year 2000) of currently available gp120 sequences [21].
The increase in the content of AT (asparagine is encoded by AT rich codons) and sequon specific amino acids were previously been identified as the possible evolutionary mechanisms to modulate the sequon numbers in glycoproteins [22]. However, none of these mechanisms are at work here. The AT content is decreasing in the whole gp120 molecule and in the inner domain, but not in the outer domain. Further, the NXT density in outer domain is decreasing despite significant increase in the T amino acid. This trend is quite reversed in the inner domain (Figure 4). Another interesting mechanism to modulate sequon numbers is through NPS/T [6]. As the proline containing sequences (NPS/T) are not used for glycosylation due to conformational hindrance, sequon numbers can be increased or decreased simply by replacing/removing proline in NXS/T during evolutionary time course. In gp120, this mechanism contributed much to the sequon increase (not during the recent years). It may be noted that as against just 3.8 fold increase in the NXS/T density, there is an 11.9 fold decrease in the number of NPS/T sequences (Figure S1). On the other hand, the actual number of NPS/T sequences was shown to be significantly higher than expected in glycoproteins (such as ABC proteins) which contain very low sequon density [6].
The changes in the number or type of sequons also change the distribution of sequons. However, distribution of sequons can also be changed without changing the number or type of sequons. Due to the unequal number of NXS and NXT sequons in the two domains of gp120, their distributions are obviously 'uneven' in the whole gp120 molecule. It may be noted that there are no studies modeling the distribution of sequons in gp120. The present results show that the distribution probabilities of NXS and/or NXT sequons in gp120 or in its two domains are very close to the maximum distribution probability (Figure 5). That is, the sequons attain a distribution which has a maximum or near-maximum probability of occurrence based on the random mechanism [25, 29]. This is very different from the 'even' distribution (which is simply one out of many possible random distributions) one would expect for a given number of sequons. Poon et al. [17] suggested an 'even' distribution of sequons (in the functional space) on the assumption that glycan groups are bulky and therefore sequons must have been selected to be evenly distributed to avoid conformational hindrance or functional redundancy. By contrast, a non-random distribution of sequons in gp120 was shown to be essential for infectivity of HIV-1 [35]. Here, uniform distribution was inaptly considered as random and hence the term 'non-random' means a distribution which is not even.
As seen by the increasing distribution probability, the re-distribution of NXT sequons in gp120 over time (from 1981 to 2009) can be taking place according to the random process [29]. Similarly, as a result of decreasing density, the distribution of NXT is changing towards a more random one in the outer domain (Figure 6). On the other hand, large change (decreasing probability) in the distribution of NXS/T sequons in inner domain is against random process. That is, there must be a selective advantage for this change and gp120/virus must have an evolutionary mechanism to attain this directed change. Interestingly, as a result of changing number and distribution, gp120 appears to be accumulating sequons in new locations (Figure 7). For example, the emergence of NXS sequons in the outer domain - near amino acid positions 400 and 450, which fall in the variable loop regions V4 and V5, respectively. The position of sequons is important in protein interactions - both antibody avoidance and receptor binding [36-40] and sequons in relatively constant positions tend to be high-mannose type glycans, while sequons in shifting positions tend to have complex type glycans. It is shown that macrophage-derived viruses tend to be more neutralization resistant as their gp120 contains complex glycans on the shifting sequons [41]. The gain or loss of sequons and their distribution in gp120 may give a selective advantage for the virus to evolve against host immune response. For example, it is shown that cumulative depletion of sequons in similar locations affect the infectivity of SIV [34]. The sequons may also vary depending on early/late stages of viral infection and sequence length variability itself has shown to affect the pattern of sequons (by selective addition or elimination) in gp120/viruses within infected individuals [19, 36]. The results presented here show delicate changes in the distribution of sequons in gp120 (chiefly for major subtype B) taking place over a long period since the viral passage to human hosts. However, our results must be viewed in the context of many factors (as mentioned previously) which affect sequons in gp120.
In summary, we have shown that there are directional trends in both the number and distribution of N-glycosylation sites in HIV-1 gp120, with a clear selection for NXS sequons and rearrangement of NXT sequons. These observations indicate that the virus/subtypes must be currently (from 1981 to 2009) experiencing fitness benefits through the changes in the number and type of sequons, as well as their distribution, in the two domains of gp120.
Conflict of Interests
The authors have declared that no conflict of interest exists.
References
1. Spiro RG. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiol. 2002;12:43R-56R
2. Dennis JW, Granovsky M, Warren CE. Protein glycosylation in development and disease. Bioessays. 1999;21:412-421
3. Verki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiol. 1993;3:97-130
4. Ben-Dor S, Esterman N, Rubin E, Sharon N. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiol. 2004;14:95-101
5. Petrescu A-J, Milac A-L, Petrescu SM. et al. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiol. 2004;14:103-114
6. Rao RSP, Buus OT, Wollenweber B. Evolutionary pattern of N-glycosylation sequon numbers in eukaryotic ABC protein superfamilies. Bioinf Biol Insights. 2010;4:9-17
7. Kwong PD, Wyatt R, Robinson J. et al. Structure of an HIVgp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648-659
8. Kwong PD, Doyle ML, Casper DJ. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678-682
9. Poumbourios P, Maerz AL, Drummer HE. Functional evolution of the HIV-1 envelope glycoprotein 120 association site of glycoprotein 41. J Biol Chem. 2003;278:42149-42160
10. Botarelli P, Houlden BA, Haigwood NL. et al. N-glycosylation of HIV-gp120 may constrain recognition by T lymphocytes. J Immunol. 1991;9:3128-3132
11. Leonard CK, Spellman MW, Riddle L. et al. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373-10382
12. Zhu X, Borchers C, Bienstock RJ, Tomer KB. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochem. 2000;39:11194-11204
13. Losman B, Bolmstedt A, Schønning K. et al. Protection of neutralization epitopes in the V3 loop of oligomeric human immunodeficiency virus type 1 glycoprotein 120 by N-linked oligosaccharides in the V1 region. AIDS Res Hum Retroviruses. 2001;17:1067-1076
14. Montefiori DC, Robinson WR Jr, Mitchell WM. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1988;85:9248-9252
15. Dirckx L, Lindemann D, Ette R. et al. Mutation of conserved N-glycosylation sites around the CD4-binding site of human immunodeficiency virus type 1 GP120 affects viral infectivity. Virus Res. 1990;18:9-20
16. Wei X, Decker JM, Wang S. et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307-312
17. Poon AFY, Lewis FI, Pond SLK, Frost SWD. Evolutionary interactions between N-linked glycosylation sites in the HIV-1 envelope. PLoS Comput Biol. 2007;3:110-119
18. Balzarini J, Laethem KV, Hatse S. et al. Marked depletion of glycosylation sites in HIV-1 gp120 under selection pressure by the mannose-specific plant lectins of Hippeastrum hybrid and Galanthus nivalis. Mol Pharmacol. 2005;67:1556-1565
19. Novitsky V, Lagakos S, Herzig M. et al. Evolution of proviral gp120 over the first year of HIV-1 subtype C infection. Virol. 2009;383:47-59
20. Gunthard HF, Leigh-Brown AJ, D'Aquila RT. et al. Higher selection pressure from antiretroviral drugs in vivo results in increased evolutionary distance in HIV-1 pol. Virol. 1999;259:154-165
21. Zhang M, Gaschen B, Blay W. et al. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiol. 2004;14:1229-1246
22. Cui J, Smith T, Robbins PW, Samuelson J. Darwinian selection for sites of Asn-linked glycosylation in phylogenetically disparate eukaryotes and viruses. Proc Natl Acad Sci USA. 2009;106:13421-13426
23. Wolk T, Schreiber M. N-Glycans in the gp120 V1/V2 domain of the HIV-1 strain NL4-3 are indispensable for viral infectivity and resistance against antibody neutralization. Med Microbiol Immunol. 2006;195:165-172
24. Ewens WJ, Wilf HS. Computing the distribution of the maximum in balls-and-boxes problems with application to clusters of disease cases. Proc Natl Acad Sci USA. 2007;104:11189-11191
25. Feller W. An introduction to probability theory and its applications. New York, USA: Wiley. 1968
26. Lamboy WF, Moreno-Hagelsieb G. A new method of solution for the occupancy problem and its application to operon size prediction. J Theor Biol. 2004;227:315-322
27. Park CJ. A note on the classical occupancy problem. Ann Math Stat. 1972;43:1698-1701
28. Williamson PP, Mays DP, Asmerom GA, Yang Y. Revisiting the classical occupancy problem. Am stat. 2009;63:356-360
29. Wu G, Yan S-M. Analysis of distribution of amino acids in the primary structure of tumor suppressor p53 family according to the random mechanism. J Mol Model. 2002;8:191-198
30. Alexander S, Elder JH. Carbohydrate dramatically influences immune reactivity of antisera to viral glycoprotein antigens. Science. 1984;226:1328-1330
31. Chen H, Xu X, Jones IM. Immunogenicity of the outer domain of a HIV-1 clade C gp120. Retrovirol. 2007;4:33
32. Hotzel I. Conservation of inner domain modules in the surface envelope glycoproteins of an ancient rabbit lentivirus and extant lentiviruses and betaretroviruses. Virol. 2008;372:201-207
33. Doria-Rose NA, Learn GH, Rodrigo AG. et al. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J Virol. 2005;79:11214-11224
34. Ohgimoto S, Shioda T, Mori K. et al. Location-specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J Virol. 1998;72:8365-8370
35. Lee WR, Syu WJ, Du B. et al. Nonrandom distribution of gp120 N-linked glycosylation sites important for infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:2213-2217
36. Belair M, Dovat M, Foley B. et al. The polymorphic nature of HIV type 1 env V4 affects the patterns of potential N-glycosylation sites in proviral DNA at the intrahost level. Aids Res Hum Retroviruses. 2009;25:199-206
37. Delos SE, Burdick MJ, White JM. A single glycosylation site within the receptor-binding domain of the avian sarcoma/leukosis virus glycoprotein is critical for receptor binding. Virol. 2002;294:354-363
38. Walmsley AR, Hooper NM. Distance of sequons to the C-terminus influences the cellular N-glycosylation of the prion protein. Biochem J. 2003;370:351-355
39. Weber ANR, Morse MA, Gay NJ. Four N-linked glycosylation sites in human toll-like receptor 2 cooperate to direct efficient biosynthesis and secretion. J Biol Chem. 2004;279:34589-34594
40. Wojczyk BS, Takahashi N, Levy MT. et al. N-glycosylation at one rabies virus glycoprotein sequon influences N-glycan processing at a distant sequon on the same molecule. Glycobiol. 2005;15:655-666
41. Lin G, Simmons G, Pohlmann S. et al. Differential N-linked glycosylation of human immunodeficiency virus and Ebola virus envelope glycoproteins modulates interactions with DC-SIGN and DC-SIGNR. J Virol. 2003;77:1337-1346
Figures and Tables
NXS/T sequons in gp120. (A) The observed density of NXS/T (where X is not proline) sequons in gp120. (B) The predicted density of NXS/T sequons is about four times lower compared to the observed density. (C) The observed number of NPS/T sequences in gp120. (D) The predicted number of NPS/T sequences is about 12 times higher compared to the observed number.
Sequon density in gp120. (A) The NXS density is increasing (t = 3.24) over time in the whole gp120 molecule. (B) The NXS/T sequon density is increasing (t = 4.53) in the N terminal region.
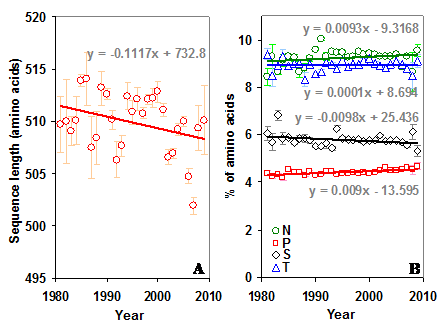
Sequence length and sequon specific amino acids in gp120. (A) There has been no significant change in the length of the sequence over time. (B) The percentage of sequon specific amino acids too remained same over time in gp120. However, proline has been increasing (t = 5.08) over time.
Distribution of sequons in gp120. (A) The log10 distribution probability of NXT is increasing (t = 4.28) over time in the whole gp120 molecule. (B) The log10 distribution probability of sequons in the N terminal region is decreasing (t = -2.57) over time.
Distributions and distribution probabilities.
| Sl. No. | Partition | Count | Distribution probability | Log10 distribution probability | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | ||||
| 1 | S | S | S | S | S | S | 1,1,1,1,1,1 | 0.0154321 | -1.812 |
| 2 | S | S | S | S | SS | 0,1,1,1,1,2 | 0.231481 | -0.635 | |
| 3 | S | S | SS | SS | 0,0,1,1,2,2 | 0.3472222 | -0.459 | ||
| 4 | SS | SS | SS | 0,0,0,2,2,2 | 0.038580 | -1.414 | |||
| 5 | S | S | S | SSS | 0,0,1,1,1,3 | 0.154321 | -0.812 | ||
| 6 | S | SS | SSS | 0,0,0,1,2,3 | 0.154321 | -0.812 | |||
| 7 | SSS | SSS | 0,0,0,0,3,3 | 0.006430 | -2.192 | ||||
| 8 | S | S | SSSS | 0,0,0,1,1,4 | 0.0385803 | -1.414 | |||
| 9 | SS | SSSS | 0,0,0,0,2,4 | 0.009645 | -2.016 | ||||
| 10 | S | SSSSS | 0,0,0,0,1,5 | 0.003858 | -2.414 | ||||
| 11 | SSSSSS | 0,0,0,0,0,6 | 0.000129 | -3.891 | |||||
1Even distribution - the distribution in which all the partitions are occupied.
2Maxumum distribution - the distribution which has the maximum probability of occurrence. Here,
Pr(0,0,1,1,2,2) = 1/66 * 6!/(0! * 0! * 1! * 1! * 2! * 2!) * 6!/(2! * 2! * 2! * 0! * 0! * 0! * 0!).
3As an example, the distribution of NXT sequons in the N-terminal part of the gp120 (EU289201, year 1995) shown in Figure 1 is (0, 0, 1, 4, 0, 1).
Author contact
![]() Corresponding author: Email: raodk; Tel: +45 8999 3569.
Corresponding author: Email: raodk; Tel: +45 8999 3569.

 Global reach, higher impact
Global reach, higher impact