Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2011; 7(1):41-52. doi:10.7150/ijbs.7.41 This issue Cite
Review
Thiamin (Vitamin B1) Biosynthesis and Regulation: A Rich Source of Antimicrobial Drug Targets?
1. Institute of Modern Biopharmaceuticals, State Key Laboratory Breeding Base of Eco-Enviroment and Bio-Resource of the Three Gorges Area, School of Life Sciences, Southwest University, Beibei Chongqing, 400715, China;
2. State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Sciences, Fudan University, Shanghai, 200433, China.
Received 2010-10-19; Accepted 2011-1-5; Published 2011-1-9
Abstract
Drug resistance of pathogens has necessitated the identification of novel targets for antibiotics. Thiamin (vitamin B1) is an essential cofactor for all organisms in its active form thiamin diphosphate (ThDP). Therefore, its metabolic pathways might be one largely untapped source of antibiotics targets. This review describes bacterial thiamin biosynthetic, salvage, and transport pathways. Essential thiamin synthetic enzymes such as Dxs and ThiE are proposed as promising drug targets. The regulation mechanism of thiamin biosynthesis by ThDP riboswitch is also discussed. As drug targets of existing antimicrobial compound pyrithiamin, the ThDP riboswitch might serves as alternative targets for more antibiotics.
Keywords: thiamin biosynthesis, riboswitch, Mycobacterium tuberculosis, drug targets
Introduction
Inexorable increase of drug resistant pathogens has exacerbated the dire outcome of recalcitrant infectious diseases due to limited effective antibiotics. A notorious case of point is the drug resistance of Mycobacterium tuberculosis, the causative agent of tuberculosis (TB). TB is a leading cause of death worldwide: about two million deaths and 9.4 million prevalent cases (300, 000 multidrug resistant cases included) were estimated in 2008 alone [World Health Organization, 2009]. To tackle infections diseases, especially the drug resistance infection, it is urgent to find novel antibiotics against unexplored drug targets.
To maximally potential toxic and side-effect, an ideal antimicrobial compound should target pathogen unique pathways or molecules. Biosynthetic pathways of several cofactors have been proposed accordingly as encouraging antibiotic targets such as the pantothenate kinase involving in coenzyme A biosynthesis, the lumazine synthase (LS) of riboflavin biosynthesis, and the NMN/NaMN adenylytranferase (NMNAT), NAD synthetase (NADS), NAD kinase (NADK) involving in the NAD (P) biosynthesis pathway [1-3].
Thiamin (Vitamin B1) is indispensable for the activity of the carbohydrate and branched-chain amino acid metabolic enzymes in its active form thiamin diphosphate (ThDP). Therefore it is an essential cofactor for all organisms [4-6]. Most bacteria, as well as fungi and plants, could produce thiamin de novo, but mammals depend solely on the dietary uptake. Recently, the thiamin metabolism pathway of Plasmodium falciparum, the pathogenic agent of tropical malaria, has been suggested as a source of drug targets recently [7]. It is logical that the thiamin biosynthetic enzymes and underlying regulation network might be suitable drug targets for the development of novel antimicrobial agents.
ThDP-dependent enzymes are crucial for pathogens
ThDP-dependent enzymes such as pyruvate dehydrogenase (PDH), 2-oxoglutarate dehydrogenase (OGDH), transketolase (TK), and acetohydroxyacid synthase (AHAS), are involved in various enzymatic reactions and multiple pathways. PDH bridges the glycolysis and the citric acid cycle via acetyl-CoA. ODGH catalyzes a rate-limiting step of the citric acid cycle. TK plays a crucial role in the pentose phosphate pathway and AHAS participates in the branched-chain amino acid biosynthesis. These ThDP-dependent enzymes are essential for many crucial pathways and are conserved among pathogens including M. tuberculosis (Table 1).
The distribution of several ThDP-dependent enzymes in M. tuberculosis H37Rv. Abbreviations: AHAS, acetohydroxyacid synthase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; InPDC, indole-3-pyruvate decarboxylase; PDC, pyruvate decarboxylase; OGDH, 2-oxoglutarate dehydrogenase; PDH, pyruvate dehydrogenase; TK, transketolase.
| EC | Enzyme | ORF | Identity with human homology (amino acid) | Pathway | Known inhibitors | References |
|---|---|---|---|---|---|---|
| 2.2.1.6 | AHAS | Rv3003c Rv3509c | N.A | Branched-chain amino acid biosynthesis | Metsulfuron-methyl, sulfonylureas, imidazolines, pyrimidinythiobenzoates, phthalazin-1 (2H) -one, KHG20612 | [8] [9, 10] |
| 4.1.1.47 | Dxs | Rv2682c | N.A | Thiamin, nonmevalonate and pyridoxal biosynthesis | Bacimethrin, Clomazone, 2-Methyl-3- (4-fluorophenyl) -5- (4-methoxy-phenyl) -4H- pyrazolo[1, 5-a]pyrimidin-7-one, fluoropyruvate | [11, 12] [13] |
| 4.1.1.74 /4.1.1.1 | InPDC /PDC | Rv0853c | 24% (117/497) | Tryptophan catabolism /Glycolysis | N. A | N.A |
| 1.2.4.2 | OGDH | N.A* | N.A | Citric acid cycle | 3-Deaza ThDP, Bacimethrin | [11, 12, 14] |
| 1.2.4.1 | PDH | Rv2497c, Rv2496c | 39% (139/358) | Pyruvate metabolism | Acetylphosphinate, Thiamin thiazolone diphosphate (ThTDP), Thiamin thiothiazolone diphosphate (ThTTDP), Methylacetylphosphonate (PLThDP) | [15-17] |
| 2.2.1.1 | TK | Rv1449c | 25% (162/650) | Pentose-phosphate pathway | Oxythiamin, N3'-pyridyl thiamin, bacimethrin | [11, 12, 18, 19] |
N.A: Not applicable; *M. tuberculosis is identified to be lack of OGDH[90].
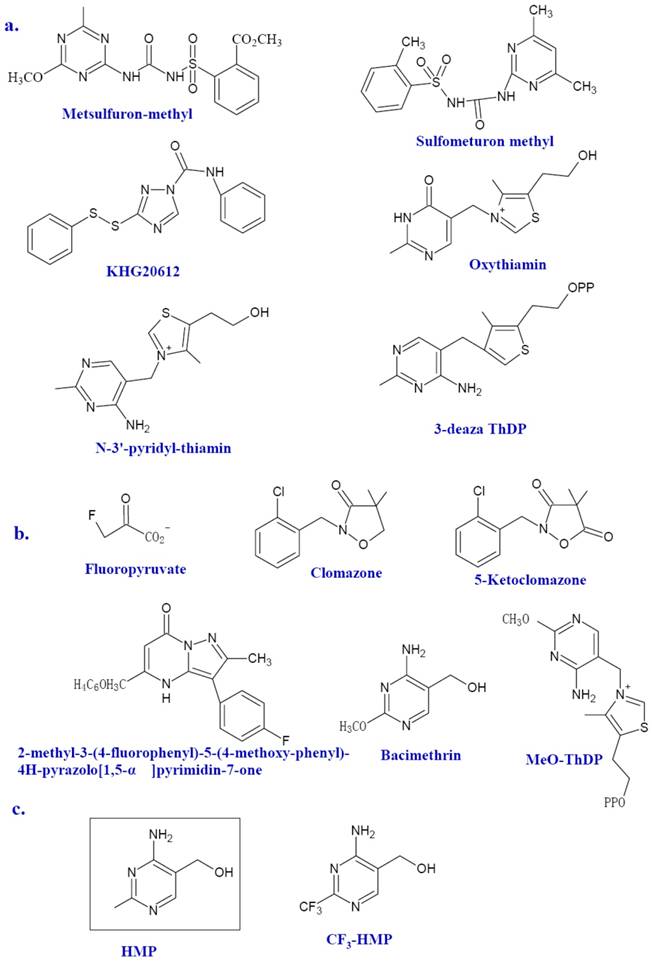
However, few ThDP-dependent enzymes are microbe specific. Therefore, it is no wonder that no clinical novel antibiotics emerged from their inhibitors were reported [20]. One ThDP-dependent enzyme recently in the spotlight is AHAS. AHAS, a ThDP and FAD dependent enzyme, is involved in the synthesis of branched-chain amino acids (BCAAs) in plants, algae, fungi, bacteria, and archaea, but absent in animals. As the first common enzyme in the BCAA biosynthetic pathway, AHAS is the potential targets for herbicides, fungicides, and antimicrobial agents. In fact, many AHAS inhibitors, such as metsulfuron-methyl (Figure 3a), sulfonylureas (Figure 3a), imidazolines, pyrimidinythiobenzoates and phthalazin-1 (2H) -one, have been developed as herbicides [9]. Metsulfuron-methyl can inhibit the activity of AHAS by binding the mouth of the active site and blocking its access to the ThDP [8]. Previous studies of amino acids auxotrophic strains of mycobacteria and the AHAS mutant of Burkholderia pseudomallei have shown that microbial AHAS might be a drug target against infectious disease including tuberculosis [21-23]. Herbicide sulfonylureas can inhibit the Mycobacterium avium AHAS [24]. Several efficient inhibitors against AHAS from M. tuberculosis (for exmple, KHG20612, Figure 3a) and Bacillus anthracis are documented [9, 10, 25, 26].
Other ThDP-dependent enzymes shared by bacteria and animals still deserves considerable attention since there might be huge discrepancy between microbial enzymes and human counterpart in the sequence and higher structures. TK from P. falciparum has been reported as a novel target to treat malarial infection[27]. TK in human beings is also a promising drug target for the treatment of cancer since the suppress activity of TK against tumor cell is much more profound than that against normal cells. Many effective TK inhibitors have been identified such as the oxythiamin and N3'-pyridyl thiamin (Figure 3a) [18, 19]. 3-deazathizmin diphosphate (3-deaza ThDP) is one of the most potent irreversible inhibitors of ThDP-dependent enzymes. The only difference between this compound and ThDP is that the N-3 atom of ThDP has been replaced by a carbon atom resulting in the formation of a neutral thiophene ring in place of the thiazolium ring [14]. This neutral thiophene ring endows 3-deaza ThDP more hydrophobility than that of ThDP thereby stronger interactions with the active site of ThDP dependent enzymes. In fact, based on the enzymatic studies of pyruvate decarboxylase from Zymomonas mobilis, 3-deaza ThDP binds this enzyme more rapidly and tightly compared with ThDP, ie, with a rate of approximately seven-fold faster and 2, 5000 fold more tighter [14]. Similar results are also described in the binding with the E1 subunit of OGDH from E. coli, which showed a binding rate of 70-fold faster and 500-fold tighter than that of ThDP [14].
Bacterial thiamin biosynthesis
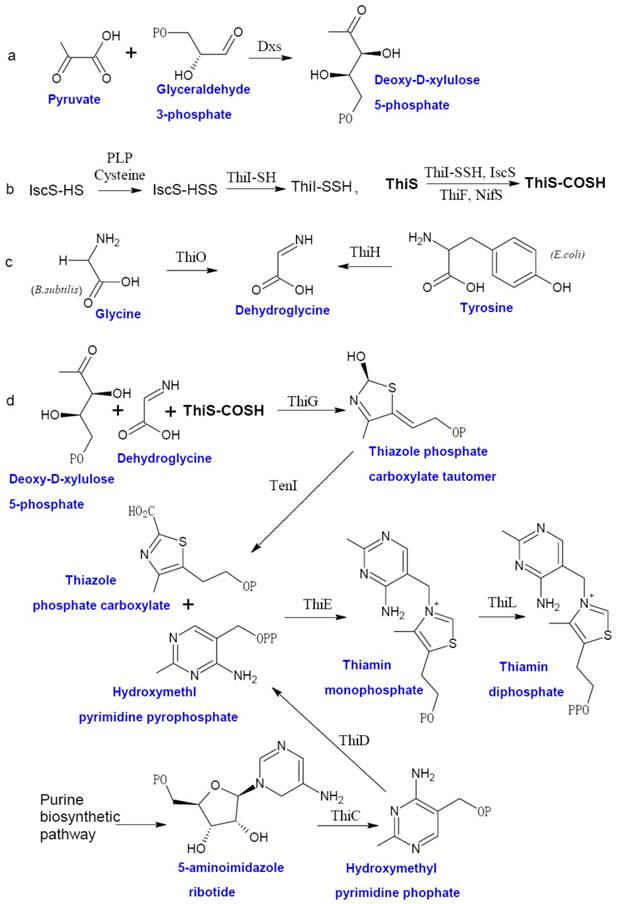
Prokaryotic thiamin biosynthesis is well-documented, which involves the formation of thiazole moiety (4-methyl-5-β-hydroxyethyl thiazole phosphate or THZ-P) and pyrimidine moiety (4-amino-5-hydroxymethyl-2-methyl-pyrimidine pyrophosphate or HMP-PP) separately [5]. Thiamin biosynthetic pathway of facultative anaerobe E. coli and aerobe Bucillus subtilis are the two best studied examples (Figure 1). In E. coli, THZ-P is derived from an oxidative condensation of tyrosine, cysteine, and 1-deoxy-D-xylulose 5-phosphate (DXP). Seven genes (thiF, thiS, thiG, thiH, thiI, iscS, and Dxs) are involved in this process [28-35]. B. subtilis THZ-P biosynthesis is different from E. coli since thiazole synthase (ThiH) is replaced by glycine oxidase (ThiO) [36], which utilizes glycine instead of tyrosine to form dehydroglycine to provide the C2-N3 unit for THZ-P.
The HMP-PP is produced from aminoimidazole ribotide (AIR) [28], an intermediate of purine biosynthesis pathway. Hydroxymethyl pyrimidine synthase (ThiC) catalyzes AIR to form hydroxymethl pyrimidine phosphate (HMP-P), which is then phosphorylated to HMP-PP by Hydroxymethyl pyrimidine (phosphate) kinase (ThiD). THZ-P and HMP-PP are coupled to form thiamin monophosphate (ThMP) mediated by thiamin phosphate synthase (ThiE), and thiamin phosphate kinase (ThiL) catalyze a final phosphorylation step to yield ThDP, the active form of thiamin.
Thiamin salvage and transport pathways
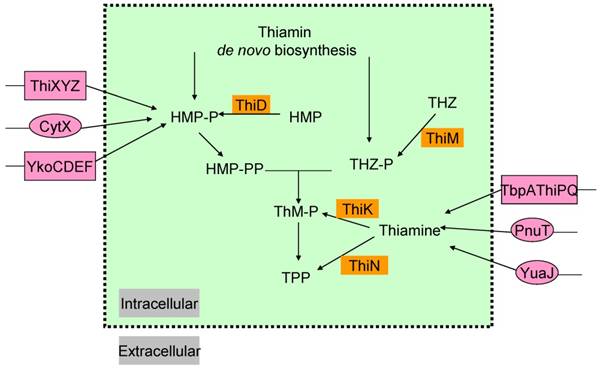
In most microorganisms, thiamin or its components THZ-P and HMP-PP, could all be produced via salvage pathway (Figure 2) [38]. Thiazole alcohol (THZ) can be used to form THZ-P catalyzed by thiazole kinase (ThiM). ThiD is required for the salvage of HMP-PP from pyrimidine alcohol (HMP), while thiamin in bacteria can be converted to ThMP by thiamin kinase (ThiK) in E. coli or to ThDP by thiamin pyrophosphokinase (ThiN) in B. subtilis.
Exogenous thiamin and its components, ThiMP, ThDP, HMP, THZ, could also enter the cell via transporters (Figure 2) [39-41]. Some gram-negative bacteria, such as, E. coli and Salmonella typhimurium, contain an ATP-binding cassette (ABC ) transporter encoded by thiamin-regulated operon, which comprises a periplasmic thiamin-binding protein (TbpA), a transmembrane thiamin channel (ThiP), and an ATPase responsible for active transport (ThiQ) [39]. Gram-positive bacteria use another ABC transporter of operon ykoCDEF. This ABC transporter contains two transmembrane components YkoC and YkoE, an ATPase component YkoD, and a thiamin/HMP-binding protein YkoF [42, 43].
In addition, comparative analysis of thiamin regulatory THI elements (ThDP riboswitches) led to the identification of YuaJ, PnuT, ThiXYZ and CytX as candidate thiamin-related transporters (Figure 2) [42, 44]. It has been identified that ThiXYZ plays an role in the HMP salvage [45]. In this pathway, thiamin produced from various organisms in basic soil is firstly degraded into several components including formyl aminopyrimidine. Secondly, formyl aminopyrimidine binds to the periplasmic component ThiY, and then being transferred into the cell. Thirdly, YlmB mediates deformylation of formyl aminopyrimidine into aminopyrimidine, and fourthly, TenA catalyzes the hydrolysis of aminopyrimidine into hydroxypyrimidine. Finally, ThiD catalyzes two phosphorylation steps to get HMP-PP.
The distribution of thiamin biosynthetic pathway in pathogenic bacteria
Landscapes of thiamin biosynthetic genes or operons of some pathogens outlined via comparative genomics analysis demonstrated some specific thiamin biosynthetic pathways[42]. The salvage and transport pathways are ubiquitous in most bacteria to complement thiamin de novo biosynthesis. This might cripple the value of thiamin biosynthetic enzymes as drug targets since this enable bacteria to obtain available exogenous thiamin. However, for those pathogens lack this salvage pathways and transporters, such as M. tuberculosis, promising targets can be found in the thiamin synthetic pathway [42]. Inhibitors can be designed to block the biosynthesis of thiamin, which will damage the growth and survival of bacteria. Many mechanisms might be used for inhibitors to cross the membrane of bacteria, for example, quinolones penetrate into cell through OmpF porin while chloramphenicol and fluoroquinolones can use Msp porins of Mycobacterium smegmatis [46, 47]. If these pathways exert more fundamental role, such as the indispensability for Listeria monocytogenes replication, this might enhance its drug target value [48].
The biosynthesis of thiamin in bacteria. The thiazole moiety of thiamin is derived from an oxidative condensation of 1-deoxy-D-xylulose 5-phosphate (DXP) (a), cysteine (b), and glycine or tyrosine (c). When the thiazole and pyrimidine moieties are formed, ThiE will coupled them to be thiamin monophosphate and followed by a phosphorylation step to give ThDP (d). Abbreviation: Dxs, 1-deoxy-D-xylulose 5-phosphate synthase; ThiF, adenyltransferase; ThiS, sulfur carrier protein; ThiG, thiazole synthase, ThiO, glycine oxidase; ThiH, thiazole synthase; ThiI, sulfur transferase; ThiC, hydroxymethyl pyrimidine synthase; ThiD, hydroxymethyl pyrimidine (phosphate) kinase; NifS, sulfur donor; TenI, transcriptional regulator TenI; IscS, cysteine desulfurase; ThiE, thiamin phosphate synthase; ThiL, thiamin phosphate kinase. This figure is modified from [37].
The salvage and transport pathways of thiamin in bacteria. Enzymes involved in the salvage of thiamin and its precursors are shown in saffron yellow background. The transport enzymes are shown in magenta background, primary and ATP-dependent transporters are in circles and rectangle, respectively. Abbreviations: ThiK, thiamin kinase; ThiM, thiazole kinase; ThiN, thiamin pyrophosphokinase.
Promising drug targets among thiamin pathway
1-deoxy-D-xylulose 5-phosphate synthase (Dxs)
Dxs catalyses the first step of thiazole biosynthesis by condensation of glyceraldehyde 3-phosphate and pyruvate to produce DXP (Figure 1a) [49]. This step also participates in the biosynthesis of pyridoxol (vitamin B6) and isoprenoids [50, 51]. Intriguingly, ThDP is both the product of Dxp and the cofactor of Dxp. As the rate-limiting enzyme of the mevalonate-independent biosynthesis of isoprenoids, Dxs is conservative and ubiquitous among microbes, including pathogens. Therefore, it is an attractive drug target. Additionally, the resolved Dxs crystal structure from E. coli and Deinococcus radiodurans can further benefit the rational design of inhibitors against it. Dxs contains three domains (I, II, and III), each shares homology with the equivalent domains of TK and the E1 subunit of PDH [52]. Although these enzymes catalyze similar reactions and are all ThDP-dependent, the arrangement of the three domains of Dxs monomer is different from TK and PDH. The active site of Dxs is unique, which is located at the interface of domains I and II of the same monomer, instead of between two monomers as other examples.
Several inhibitors of Dxs have been identified (Figure 3b). Fluoropyruvate is described as a first inhibitor of Dxs, which is supposed to bind covalently to the active site of Dxs [13]. Dxs from Pseudomonas aeruginosa and E. coli were inhibited by fluoropyruvate with an IC50 of 400 μM and 80 μM, respectively [53]. Another known inhibitor of Dxs is 5-ketoclomazone, the metabolite of known commercially available herbicide clomazone [54, 55]. The 5-ketoclomazone was able to inhibit the activity of Dxs from Chlamydomonas with an IC50 value of 0.1 mM [54]. However, clomazone per se has no effect on bacterial growth. This suggests that plants might convert clomazone into active pound 5-ketoclomazone, thereby inhibiting the activity of Dxs[56]. Based on a known transketolase inhibitor, a target-based approach has been used to identify possible M. tuberculosis Dxs inhibitors [57]. A preliminary structure-activity relationship (SAR) of several potential inhibitors against M. tuberculosis was established. So far, 2-methyl-3- (4-fluorophenyl) -5- (4-methoxy-phenyl) -4H-pyrazolo[1, 5-α]pyrimidin-7-one was the most potent Dxs inhibitor (IC50 =10.6μM) despite its somewhat toxicity against mammalian cell [57]. Bacimethrin, an analog of HMP, is the inhibitor of a subset of ThDP-dependent enzymes including α-ketoglutarate dehydrogenase, TK and Dxs. In bacteria, bacimethrin is converted to its active metabolite 2'-methoxy-thiamin diphosphate, which would inhibit thiamin-utilizing enzymes [12]. The last three enzymes of the thiamin synthetic pathway, ThiD, ThiE, and ThiL, are all contributed to this conversion [12]. Therefore, the mutation in any of the three enzymes might alleviate the toxicity of bacimethrin to the cell. It has been tested that bacimethrin could inhibit E. coli cell growth under a low micromolar range of minimum inhibitory concentration (MIC). However, this inhibitory effect can be abrogated by low levels of exogenous thiamin or the HMP moity [11, 12].
The structure of inhibitors. Selected inhibitors of ThDP-dependent enzymes, Dxs and ThiE are shown as a, b, and c, respectively.
Thiamin phosphate synthase (ThiE)
ThiE catalyzes the THZ-P and HMP-PP to yield ThMP. The solved ThiE crystal structures of B. subtilis and M. tuberculosis (http://hdl.handle.net/1813/13623) have similar structure [58]. It is dimer and has an α/β structure with a TIM barrel fold and a core of eight β-strands flanked by eight α-helices. ThiE is a crucial synthase for thiamin. The cellular trace requirement for thiamin enables it to be satisfied by enzymes with weak catalytic activity. The product of yjbQ from E. coli is a thiamin synthase homolog [59]. The yjbQ gene can complement the thiamin auxotrophy of E. coli, Thermotoga, Sulfolobus and Pyrococcus. However, no structural similarity exists between YjbQ and ThiE, which suggested that they might evolve from different origin. ThiE could still serves as an ideal drug target. 4-amino-2-trifluoromethyl-5-hydroxymethylpyrimidine (CF3-HMP) (Figure 3c) is a HMP analog and could inhibit the growth of E. coli. In the bacteria, CF3-HMP is converted by ThiD to CF3-HMP pyrophosphate (CF3-HMP-PP), an analog of HMP-PP, which inhibits the activity of ThiE [60]. Mounting thiamin level either endogenous or exogenous can reverse inhibition.
Other potential targets among thiamin biosynthetic enzymes
Among enzymes involved in thiamin biosynthesis, cysteine desulfurase (IscS) and ThiI also play a role in the 4-thiouridine biosynthetic pathways during the sulfur transfer. ThiH, ThiG, ThiF, and ThiS are genetically identified essential for E. coli THZ-P biosynthesis. ThiO in B. subtilis is essential for THZ-P formation and catalyzes the oxidation of glycine to form glycine imine [61]. ThiC is the only known enzyme to be required for the conversion of AIR to HMP-P in vivo, while ThiD catalyzes an essential ATP-dependent phophorylation of HMP-P to yield HMP-PP. Other thiamin biosynthetic enzymes whose crystal structure have been determined include ThiI (PDB ID: 2C5S), IscS (PDB ID: 1P3W ), ThiF (PDB ID: 1ZUD), ThiS (PDB ID: 1ZUD, 1TYG), ThiO (PDB ID: 1NG3), ThiG (PDB ID: 1TYG), ThiC (PDB ID: 3EPM), ThiD (PDB ID: 1JXI) and ThiL (PDB ID: 3C9T). These structures have shed light on their catalytic mechanisms and will contribute to the rational design of inhibitors.
Essential enzymes of the thiamin biosynthetic pathway are potential antibiotic targets. Inhibitors of these enzymes could block the endogenous thiamin biosynthesis and lead to the depletion of vitamin, thereby damaging the survival and growth of the cell. Transposon site hybridization (Trash) has identified in vitro that thiamin biosynthetic genes thiC, thiD, thiS, thiF, iscS, thiG, thiO and thiL are essential for M. tuberculosis [62]. Further experiments are needed to validate their in vivo essentiality. Inhibitors of established essential gene products might be promising drug leads.
The regulation of thiamin metabolism by riboswitches
Riboswitches are highly conserved metabolite-sensing noncoding RNAs which are involved in the regulation of gene expression. They are mostly found in the untranslated regions of mRNAs and can bind to small-molecule metabolites directly, thereby regulating the expression of metabolic-related genes [63-66]. Riboswitches usually act in cis form to control the expression of their downstream genes, but two S-adenosylmethionine (SAM) riboswitches have been demonstrated to be also acted in trans form in addition to their cis-acting upon the downstream genes [67]. Typically riboswitches consist of two domains, ie, the aptamer domain and expression platform. The highly evolutionary conserved aptamer domain is used to recognize a target ligand while the variable expression platform can alter its structure to control genes responsive to metabolite binding [68].
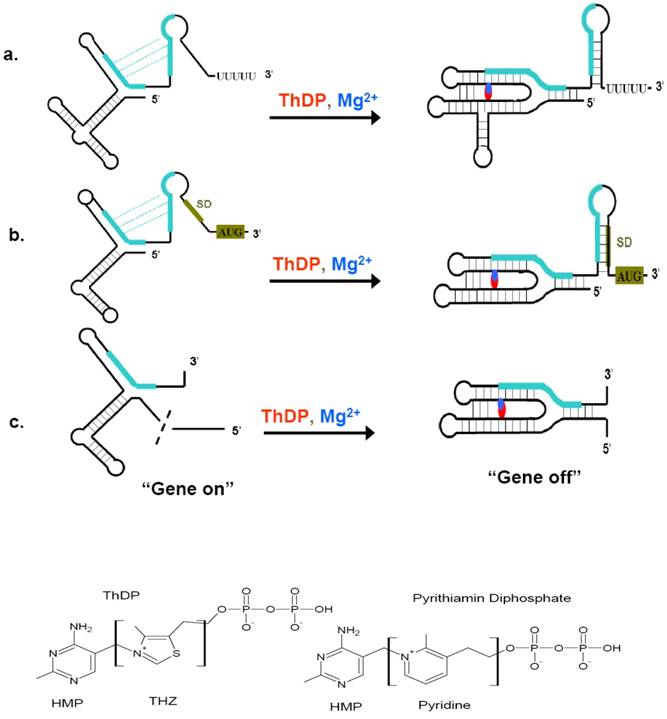
Riboswitches can sense a variety of cellular metabolites whose gene products are always encoded by the riboswitch downstream sequence. These metabolites include coenzymes, amino acids, metal ions, and nucleobases [69]. ThDP riboswitch is one of the most widespread riboswitches. Members of this class have been found in bacteria, archaea, and eukaryotic organisms, regulating the expression of genes involved in the biosynthesis, salvage and transport of thiamin and its precursors [42, 70-75]. ThDP riboswitch also bears the features of other riboswitch. The resolved crystal structure of ThDP riboswitch helps the understanding of their regulation mechanisms. These structures include thiM riboswitch from prokaryote E. coli and thiC riboswitch from plant Arabidopsis thaliana [76, 77]. The high similarities of the two structures demonstrated the riboswitch to be evolutionarily conserved across diverse organisms. ThDP-driven regulation of gene expression is accomplished through interplay of two riboswitch conformations, which permit the expression of thiamin biosynthetic, transport, and salvage genes to be turned “on” or “off” (Figure 4) [78]. When the intracellular concentration of ThDP is relatively high, the aptamer domain of the riboswitch would bind to their cognate ligands and sensing domain adopts a stable metabolite-bound conformation. According to organisms, these conformations would act as transcription terminators or translation inhibitors, even causing mis-splicing of RNA (in eukaryotes) [79]. ThDP riboswith are intrinsic transcription terminator and translation inhibitor. The intrinsic transcription terminator is a stable stem-loop structure followed by several consecutive uridines [80, 81]. When encountered this terminator, the ternary elongation complexes of RNA polymerase would be dissociated rapidly and thus lead to a halt transcription [69]. Therefore, the concentration of ThDP would determine whether to resume the transcription or read through into the following thiamin synthetic genes upon the occupncy of the aptamer domain of riboswitch. Riboswitch can also harness ThDP binding to form a translation inhibitor, thereby regulating the access of mRNA sites near AUG start codons to the Shine-Dalgarno (SD) sequence. As a result, if the intracellular concentration of ThDP is below the threshold, ThDP would dissociate from their riboswitch, leading to the expression of ThDP synthetic genes.
Regulatory mechanism of ThDP riboswitches and the structure of ThDP and Pyrithiamin diphosphate. (Top) When ThDP is at lower concentration during the synthesis of the 5'-UTR, the ThDP riboswitch would not inhibit the expression of ThDP biosynthetic genes (a, b, c, left), then “gene on”. When ThDP is at higher concentration, depending on the the expression platform, the ThDP riboswitch would fold to a transcription terminator (a), a suppressor of translation initiation (b), or a modulator of splicing (c), it is the “gene off” state (a, b, c, right). (Bottom) The structure of ThDP and pyrithiamin diphosphate. The initiation codon and Shine-Dalgarno (SD) sequence are shaded green brown, complementary sequences and their alternate base pairing are shown in green. The red and blue represent ThDP and Mg2+ ions, respectively. This figure is modified from [76, 79].
Riboswitches: ideal drug targets
It has been demonstrated that RNA molecules can fold into intricate three-dimensional structures, which will form pockets for the binding of small compounds. In this sense, the ribosome, tRNA, T box and viral RNAs have been extensively explored as antibacterial or antiviral targets [82]. However, these antimicrobial drugs merely depend on a fortuitous interaction between the compound and its cognate RNA target [83]. In contrast to above RNA, riboswitch represents a novel kind of drug targets due to their high affinity to responsive ligand.
ThDP riboswitch regulate many thiamin metabolism genes in numerous bacteria, rendering them promising drug targets for broad-spectrum antibiotics. Thiamin analog pyrithiamin exemplifies how ThDP riboswitch can act as drug target. The growth of many bacteria and fungi was inhibited by pyrithiamin. Pyrithiamin could be taken up into cell via thiamin transporter and further be phosphorylated into pyrithiamin diphosphate by the enzyme thiamin pyrophosphokinase. Pyrithiamin diphosphate binds ThDP riboswitch at nearly the identical affinity of ThDP, which lead to the expression of riboswitch-regulated thiamin biosynthesis, salvage or transport genes be repressed. Because the limited endogenous thiamin would soon be depleted, the survival or growth of the cell would be prevented in a short time [84]. The sole difference between pyrithiamin diphosphate and ThDP is the central thiazole ring, in which the ThDP thiazole ring was supplanted by a pyridinium ring (Figure 4). Pyrithiamin-resistant mutants of B. subtilis, E. coli, Aspergillus oryzae and Chalmydomonas reinhardtii bearing mutation in the conserved domain of ThDP riboswitch, which unambiguously demonstrated that riboswitch is the drug target for pyrithiamin [70, 74, 84]. Therefore, the wide distribution of ThDP riboswitch in several pathogens including the recalcitrant M. tuberculosis and promiscuous recognition of the central thiazole ring of ThDP by ThDP riboswitches render it possible to find or rationally design ThDP analogs bearing functional groups other than those linking HMP and diphosphate moieties like pyrithiamin [78].
Screening of novel riboswitch ligand analogs as selective inhibitors of some metabolic pathways also contribute to the exploring of riboswitch as drug targets. Theoretically, any inhibitor of a riboswitch could influence gene expression, regardless of the presence of its ligand [85]. Thus, according to riboswitch crystal structures, novel compounds could be designed as a source of antibiotics. Indeed, the modification of purine derivatives have resulted in the screening of several molecules that inhibit bacterial growth[86]. It has also been identified that a pyrimidine coumpound (PC1) binding guanine riboswitches could kill a subgroup of bacteria [85]. More surprisingly, the fact that PC1 also showed some activity against the infection of Staphlococcus aureus in mouse model suggests it deserves further research to act as an antibiotic candidate. These findings have established the possibility of employing the riboswitch structures to screen novel compounds as candidates for the development of anti-microbial agents.
Discussion and Conclusion
In the search for new drug targets to combat infection, the most desirable strategy is to find compounds targeting pathogen-specific key molecules. The thiamin biosynthetic pathway perfectly matches this criterion. The Dxs and ThiE are proposed as promising enzymatic targets for novel antimicrobial agents. Other enzymes of this pathway could also serve as candidate drug targets.
Riboswitch have been extensively studied as drug targets recently [78, 87-89]. More than 20 riboswitch classes have been identified that respond to a spectrum of metabolites. Each riboswitch aptamer is featured by unique conserved primary and secondary structures [69]. ThDP riboswitch is the target of antimicrobial compound pyrithiamin. Other ThDP analogs like pyrithiamin could also bind to ThDP riboswitch and repress the expression of bacterial thiamin biosynthetic enzymes. The resolved crystal structures of riboswitch could help the designing of their inhibitors. Targeting riboswitch to develop antimicrobial agents has significant advantages. Despite distantly evolved, ubiquitous riboswitches bear conserved primary and secondary structures, which renders them ideal targets for broad-spectrum antibiotics. Moreover, multi-pronged targeting strategy against multiple riboswitches (e.g. there are thiC and thiO riboswitches in M. tuberculosis) can dramatically minimize the possibility of drug resistance.
The challenge is that the trace thiamin requirement of microbes can be readily compensated exogenously or by the activation of some other thiamin synthetic enzymes not yet known (for example, yjbQ). Therefore, the inhibition can be easily invalidated. However, for pathogens such as M. tuberculosis de novo synthesis of thiamin is indispensable due to absence of thiamin salvage or transporters, inhibition can be more effective. Further validation of the essentiality of genes involved in the thiamin metabolism and regulatory may reveal more potential drug targets.
Acknowledgements
This work was supported by the National key infectious disease project (Grant Nos. 2008ZX10003-006 and 2008ZX10003-001), Excellent PhD thesis fellowship of southwest university (Grant Nos. kb2009010, ky2009009 and kb2010017), the Fundamental Research Funds for the Central Universities (Grant No. XDJK2009A003), National Natural Science Foundation (grant No. 81071316) Natural Science Foundation Project of CQ CSTC (Grant No. CSTC, 2010BB5002).
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
1. Spry C, Kirk K, Saliba K.J. Coenzyme A biosynthesis: an antimicrobial drug target. Fems Microbiology Reviews. 2008;32:56-106
2. Mdluli K, Spigelman M. Novel targets for tuberculosis drug discovery. Current Opinion in Pharmacology. 2006;6:459-67
3. Magni G, Di Stefano M, Orsomando G. et al. NAD (P) biosynthesis enzymes as potential targets for selective drug design. Current Medicinal Chemistry. 2009;16:1372-90
4. Settembre E, Begley T.P, Ealick S.E. Structural biology of enzymes of the thiamin biosynthesis pathway. Curr Opin Struct Biol. 2003;13:739-47
5. Begley T.P, Downs D.M, Ealick S.E. et al. Thiamin biosynthesis in prokaryotes. Arch Microbiol. 1999;171:293-300
6. Pohl M, Sprenger G.A, Muller M. A new perspective on thiamine catalysis. Curr Opin Biotechnol. 2004;15:335-42
7. Muller I.B, Hyde J.E, Wrenger C. Vitamin B metabolism in Plasmodium falciparum as a source of drug targets. Trends in Parasitology. 2010;26:35-43
8. McCourt J.A, Pang S.S, King-Scott J. et al. Herbicide-binding sites revealed in the structure of plant acetohydroxyacid synthase. Proc Natl Acad Sci U S A. 2006;103:569-73
9. Kalme S, Pham C.N, Gedi V. et al. Inhibitors of Bacillus anthracis acetohydroxyacid synthase. Enzyme and Microbial Technology. 2008;43:270-275
10. Choi K.J, Yu Y.G, Hahn H.G. et al. Characterization of acetohydroxyacid synthase from Mycobacterium tuberculosis and the identification of its new inhibitor from the screening of a chemical library. FEBS Lett. 2005;579:4903-10
11. Zilles J.L, Croal L.R, Downs D.M. Action of the thiamine antagonist bacimethrin on thiamine biosynthesis. J Bacteriol. 2000;182:5606-10
12. Reddick J.J, Saha S, Lee J. et al. The mechanism of action of bacimethrin, a naturally occurring thiamin antimetabolite. Bioorg Med Chem Lett. 2001;11:2245-8
13. Hirsch A.K.H, Diederich F. The non-mevalonate pathway to isoprenoid biosynthesis: A potential source of new drug targets. Chimia. 2008;62:226-230
14. Mann S, Melero CP, Hawksley D. et al. Inhibition of thiamin diphosphate dependent enzymes by 3-deazathiamin diphosphate. Organic & Biomolecular Chemistry. 2004;2:1732-41
15. Nemeria N.S, Korotchkina L.G, Chakraborty S. et al. Acetylphosphinate is the most potent mechanism-based substrate-like inhibitor of both the human and Escherichia coli pyruvate dehydrogenase components of the pyruvate dehydrogenase complex. Bioorg Chem. 2006;34:362-79
16. Xiong Y, Li Y, He H. et al. Theoretical calculation of the binding free energies for pyruvate dehydrogenase E1 binding with ligands. Bioorg Med Chem Lett. 2007;17:5186-90
17. Nemeria N, Yan Y, Zhang Z. et al. Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by thiamin 2-thiazolone and thiamin 2-thiothiazolone diphosphates. Evidence for reversible tight-binding inhibition. J Biol Chem. 2001;276:45969-78
18. Rais B, Comin B, Puigjaner J. et al. Oxythiamine and dehydroepiandrosterone induce a G1 phase cycle arrest in Ehrlich's tumor cells through inhibition of the pentose cycle. FEBS Lett. 1999;456:113-8
19. Le Huerou Y, Gunawardana I, Thomas A.A. et al. Prodrug thiamine analogs as inhibitors of the enzyme transketolase. Bioorg Med Chem Lett. 2008;18:505-8
20. Agyei-Owusu K, Leeper F.J. Thiamin diphosphate in biological chemistry: analogues of thiamin diphosphate in studies of enzymes and riboswitches. FEBS J. 2009;276:2905-16
21. Bange F.C, Brown A.M, Jacobs W.R.Jr. Leucine auxotrophy restricts growth of Mycobacterium bovis BCG in macrophages. Infect Immun. 1996;64:1794-9
22. Guleria I, Teitelbaum R, McAdam R.A. et al. Auxotrophic vaccines for tuberculosis. Nat Med. 1996;2:334-7
23. Atkins T, Prior R.G, Mack K. et al. A mutant of Burkholderia pseudomallei, auxotrophic in the branched chain amino acid biosynthetic pathway, is attenuated and protective in a murine model of melioidosis. Infect Immun. 2002;70:5290-4
24. Zohar Y, Einav M, Chipman D.M. et al. Acetohydroxyacid synthase from Mycobacterium avium and its inhibition by sulfonylureas and imidazolinones. Biochim Biophys Acta. 2003;1649:97-105
25. Sohn H, Lee K.S, Ko Y.K. et al. In vitro and ex vivo activity of new derivatives of acetohydroxyacid synthase inhibitors against Mycobacterium tuberculosis and non-tuberculous mycobacteria. Int J Antimicrob Agents. 2008;31:567-71
26. Gedi V, Jayaraman K, Kalme S. et al. Evaluation of substituted triazol-1-yl-pyrimidines as inhibitors of Bacillus anthracis acetohydroxyacid synthase. Biochim Biophys Acta. 2010;1804:1369-75
27. Joshi S. Identification of potential P. falciparum transketolase inhibitors: pharmacophore design, in silico screening and docking studies. Journal of Biophysical Chemistry. 2010;01:96-104
28. Lawhorn B.G, Mehl R.A, Begley T.P. Biosynthesis of the thiamin pyrimidine: the reconstitution of a remarkable rearrangement reaction. Organic & Biomolecular Chemistry. 2004;2:2538-46
29. Vander Horn P.B, Backstrom A.D, Stewart V. et al. Structural genes for thiamine biosynthetic enzymes (thiCEFGH) in Escherichia coli K-12. J Bacteriol. 1993;175:982-92
30. Sprenger G.A, Schorken U, Wiegert T. et al. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc Natl Acad Sci U S A. 1997;94:12857-62
31. Taylor S.V, Kelleher N.L, Kinsland C. et al. Thiamin biosynthesis in Escherichia coli. Identification of this thiocarboxylate as the immediate sulfur donor in the thiazole formation. J Biol Chem. 1998;273:16555-60
32. Xi J, Ge Y, Kinsland C. et al. Biosynthesis of the thiazole moiety of thiamin in Escherichia coli: identification of an acyldisulfide-linked protein--protein conjugate that is functionally analogous to the ubiquitin/E1 complex. Proc Natl Acad Sci U S A. 2001;98:8513-8
33. Leonardi R, Fairhurst S.A, Kriek M. et al. Thiamine biosynthesis in Escherichia coli: isolation and initial characterisation of the ThiGH complex. FEBS Lett. 2003;539:95-9
34. Martinez-Gomez N.C, Robers M, Downs D.M. Mutational analysis of ThiH, a member of the radical S-adenosylmethionine (AdoMet) protein superfamily. Journal of Biological Chemistry. 2004;279:40505-40510
35. Kriek M, Martins F, Leonardi R. et al. Thiazole synthase from Escherichia coli: an investigation of the substrates and purified proteins required for activity in vitro. J Biol Chem. 2007;282:17413-23
36. Settembre E.C, Dorrestein P.C, Park J.H. et al. Structural and mechanistic studies on ThiO, a glycine oxidase essential for thiamin biosynthesis in Bacillus subtilis. Biochemistry. 2003;42:2971-81
37. Jurgenson C.T, Begley T.P, Ealick S.E. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem. 2009;78:569-603
38. Melnick J, Lis E, Park J.H. et al. Identification of the two missing bacterial genes involved in thiamine salvage: thiamine pyrophosphokinase and thiamine kinase. J Bacteriol. 2004;186:3660-2
39. Webb E, Claas K, Downs D. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J Biol Chem. 1998;273:8946-50
40. Bellion E, Lash T.D. Transport of 2-methyl-4-amino-5-hydroxymethylpyrimidine by Salmonella typhimurium. Biochim Biophys Acta. 1983;735:337-40
41. Bellion E, Lash T.D, McKellar B.R. Transport of thiamine and 4-methyl-5-hydroxyethylthiazole by Salmonella typhimurium. Biochim Biophys Acta. 1983;735:331-6
42. Rodionov D.A, Vitreschak A.G, Mironov A.A. et al. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J Biol Chem. 2002;277:48949-59
43. Devedjiev Y, Surendranath Y, Derewenda U. et al. The structure and ligand binding properties of the B. subtilis YkoF gene product, a member of a novel family of thiamin/HMP-binding proteins. J Mol Biol. 2004;343:395-406
44. Gelfand M.S, Rodionov D.A. Comparative genomics and functional annotation of bacterial transporters. Physics of Life Reviews. 2008;5:22-49
45. Jenkins A.H, Schyns G, Potot S. et al. A new thiamin salvage pathway. Nat Chem Biol. 2007;3:492-7
46. Hirai K, Aoyama H, Irikura T. et al. Differences in susceptibility to quinolones of outer membrane mutants of Salmonella typhimurium and Escherichia coli. Antimicrob Agents Chemother. 1986;29:535-8
47. Danilchanka O, Pavlenok M, Niederweis M. Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob Agents Chemother. 2008;52:3127-34
48. Schauer K, Stolz J, Scherer S. et al. Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J Bacteriol. 2009;191:2218-27
49. Schwender J, Seemann M, Lichtenthaler H.K. et al. Biosynthesis of isoprenoids (carotenoids, sterols, prenyl side-chains of chlorophylls and plastoquinone) via a novel pyruvate/glyceraldehyde 3-phosphate non-mevalonate pathway in the green alga Scenedesmus obliquus. Biochem J. 1996;316( Pt 1):73-80
50. Hill R.E, Himmeldirk K, Kennedy I.A. et al. The biogenetic anatomy of vitamin B6. A 13C NMR investigation of the biosynthesis of pyridoxol in Escherichia coli. J Biol Chem. 1996;271:30426-35
51. Lange B.M, Rujan T, Martin W. et al. Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc Natl Acad Sci U S A. 2000;97:13172-7
52. Xiang S, Usunow G, Lange G. et al. Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J Biol Chem. 2007;282:2676-82
53. Altincicek B, Hintz M, Sanderbrand S. et al. Tools for discovery of inhibitors of the 1-deoxy-D-xylulose 5-phosphate (DXP) synthase and DXP reductoisomerase: an approach with enzymes from the pathogenic bacterium Pseudomonas aeruginosa. FEMS Microbiol Lett. 2000;190:329-33
54. Mueller C, Schwender J, Zeidler J. et al. Properties and inhibition of the first two enzymes of the non-mevalonate pathway of isoprenoid biosynthesis. Biochem Soc Trans. 2000;28:792-3
55. Zeidler J, Schwender J, Mueller C. et al. The non-mevalonate isoprenoid biosynthesis of plants as a test system for drugs against malaria and pathogenic bacteria. Biochem Soc Trans. 2000;28:796-8
56. Rodriguez-Concepcion M. The MEP pathway: a new target for the development of herbicides, antibiotics and antimalarial drugs. Curr Pharm Des. 2004;10:2391-400
57. Mao J, Eoh H, He R. et al. Structure-activity relationships of compounds targeting mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate synthase. Bioorg Med Chem Lett. 2008;18:5320-3
58. Chiu H.J, Reddick J.J, Begley T.P. et al. Crystal structure of thiamin phosphate synthase from Bacillus subtilis at 1.25 A resolution. Biochemistry. 1999;38:6460-70
59. Morett E, Saab-Rincon G, Olvera L. et al. Sensitive genome-wide screen for low secondary enzymatic activities: the YjbQ family shows thiamin phosphate synthase activity. J Mol Biol. 2008;376:839-53
60. Reddick J.J, Nicewonger R, Begley T.P. Mechanistic studies on thiamin phosphate synthase: evidence for a dissociative mechanism. Biochemistry. 2001;40:10095-102
61. Park J.H, Dorrestein P.C, Zhai H. et al. Biosynthesis of the thiazole moiety of thiamin pyrophosphate (vitamin B1). Biochemistry. 2003;42:12430-8
62. Sassetti C.M, Boyd D.H, Rubin E.J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77-84
63. Mandal M, Breaker R.R. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451-63
64. Nudler E, Mironov A.S. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11-7
65. Soukup J.K, Soukup G.A. Riboswitches exert genetic control through metabolite-induced conformational change. Curr Opin Struct Biol. 2004;14:344-9
66. Coppins R.L, Hall K.B, Groisman E.A. The intricate world of riboswitches. Curr Opin Microbiol. 2007;10:176-81
67. Loh E, Dussurget O, Gripenland J. et al. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770-9
68. Breaker R.R. Complex riboswitches. Science. 2008;319:1795-7
69. Roth A, Breaker R.R. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305-34
70. Kubodera T, Watanabe M, Yoshiuchi K. et al. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5'-UTR. FEBS Lett. 2003;555:516-20
71. Miranda-Rios J, Navarro M, Soberon M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc Natl Acad Sci U S A. 2001;98:9736-41
72. Sudarsan N, Barrick J.E, Breaker R.R. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644-7
73. Bettendorff L. At the crossroad of thiamine degradation and biosynthesis. Nat Chem Biol. 2007;3:454-5
74. Croft M.T, Moulin M, Webb M.E. et al. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci U S A. 2007;104:20770-5
75. Kazanov M.D, Vitreschak A.G, Gelfand MS. Abundance M.S and functional diversity of riboswitches in microbial communities. BMC Genomics. 2007;8:347
76. Serganov A, Polonskaia A, Phan A.T. et al. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167-71
77. Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208-11
78. Blount K.F, Breaker R.R. Riboswitches as antibacterial drug targets. Nat Biotechnol. 2006;24:1558-64
79. Nudler E. Flipping riboswitches. Cell. 2006;126:19-22
80. Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495-504
81. Yarnell W.S, Roberts J.W. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611-5
82. Thomas J.R, Hergenrother P.J. Targeting RNA with small molecules. Chem Rev. 2008;108:1171-224
83. Monaghan R.L, Barrett J.F. Antibacterial drug discovery--then, now and the genomics future. Biochem Pharmacol. 2006;71:901-9
84. Sudarsan N, Cohen-Chalamish S, Nakamura S. et al. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325-35
85. Mulhbacher J, Brouillette E, Allard M. et al. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 2010;6:e1000865
86. Kim J.N, Blount K.F, Puskarz I. et al. Design and antimicrobial action of purine analogues that bind Guanine riboswitches. ACS Chem Biol. 2009;4:915-27
87. Lea C.R, Piccirilli J.A. 'Turning on' riboswitches to their antibacterial potential. Nat Chem Biol. 2007;3:16-7
88. Blouin S, Mulhbacher J, Penedo J.C. et al. Riboswitches: ancient and promising genetic regulators. Chembiochem. 2009;10:400-16
89. Breaker R.R. Riboswitches: from ancient gene-control systems to modern drug targets. Future Microbiol. 2009;4:771-3
90. Tian J, Bryk R, Itoh M. et al. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: identification of alpha-ketoglutarate decarboxylase. Proc Natl Acad Sci U S A. 2005;102:10670-5
Author contact
![]() Corresponding author: Jianping Xie, E-mail: jianpingxiefudancom; Telephone: 86-23-68367108, Fax: 86-23-68252365
Corresponding author: Jianping Xie, E-mail: jianpingxiefudancom; Telephone: 86-23-68367108, Fax: 86-23-68252365

 Global reach, higher impact
Global reach, higher impact