Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(7):707-715. doi:10.7150/ijbs.6674 This issue Cite
Research Paper
Expression of a GABAB - Receptor in Olfactory Sensory Neurons of Sensilla trichodea on the Male Antenna of the Moth Heliothis virescens
1. University of Hohenheim, Institute of Physiology, Stuttgart, Germany;
2. Bayer CropScience, Monheim, Germany.
Received 2013-5-13; Accepted 2013-6-23; Published 2013-7-20
Abstract
In the olfactory pathway of Drosophila, a GABAB receptor mediated presynaptic gain control mechanism at the first synapse between olfactory sensory neurons (OSNs) and projection neurons has been suggested to play a critical role in setting the sensitivity and detection range of the sensory system. To approach the question if such a mechanism may be realized in the pheromone recognition system of male moths in this study attempts were made to explore if moth's pheromone-responsive cells express a GABAB- receptor. Employing a combination of genome analysis, RT-PCR experiments and screening of an antennal cDNA library we have identified a cDNA which encodes the GABAB-R1 receptor of Heliothis virescens. Moreover, based on the HvirGABAB-R1 sequence we could predict a GABAB-R1 protein from genome sequences of the silkmoth Bombyx mori. To assess whether HvirGABAB-R1 is expressed in OSNs of male antenna we performed whole-mount in situ hybridization (WM-ISH) experiments. Several HvirGABAB-R1 positive cells were visualized under long sensilla trichodea, known to contain pheromone-responsive OSNs. In parallel it was shown that cells under long trichoid hairs were labelled with pheromone receptor specific probes. In addition, the HvirGABAB-R1 specific probe also labelled several cells under shorter olfactory sensilla, but never stained cells under mechanosensory/gustatory sensilla chaetica. Together, the results indicate that a GABAB receptor is expressed in pheromone-responsive OSNs of H. virescens and suggest a presynaptic gain control mechanism in the axon terminals of these cells.
Keywords: moth, olfaction, GABA, pheromone, in situ hybridization.
Introduction
The use of powerful long-range sex pheromones in combination with a sensitive sexually dimorphic detection and processing system has made moth species valuable models for studying reception and processing of pheromone signals [1-3]. In the tobacco budworm, Heliothis virescens, female moths release a blend of several pheromone components to attract the males [4, 5] and male moths detect the components by narrowly tuned olfactory sensory neurons (OSNs) on the antenna, which project their dendrite into the hair-like sensilla trichodea [6-8]. OSNs responding to the major and the principle minor pheromone component, respectively, project their axons to distinct compartments within the male-specific macro glomerular complex (MGC) in the antennal lobe (AL), the first processing center for pheromone information in the brain [8-10]. Here, the axons synapse onto the dendrites of projection neurons (PNs), which propagate pheromone information into higher brain regions [11]. Processing of pheromone signals in the antennal lobe involves GABAergic local interneurons (LNs), which connect most if not all glomeruli [12-14].
A specific and sensitive detection of female-released sex pheromones puts high requirements to the olfactory system of male moths. First of all, pheromone components have to be detected and discriminated within a background of numerous other odorants (plant volatiles) in the environment. On the molecular level this is supposed to be accomplished by using combinations of specialized pheromone binding proteins (PBPs) in the sensillum lymph and narrowly tuned pheromone receptors (PRs) in the membrane of OSNs [1, 15-18]. A second challenge is posed from the nature of the pheromone signal originating from females. Female moths emit pheromone in packages which are distributed downwind by air turbulences and get more and more dispersed in time and space with increasing distance from the pheromone source [19, 20]. As a consequence male moths orientating up-wind in a pheromone plume towards a calling female are facing highly fluctuating pheromone pulses, which increase in number and intensity when coming nearer to the pheromone source. Therefore, fast and effective mechanisms for setting the sensitivity and regulating the gain of the pheromone detection system are considered to be of extreme importance to prevent the system from saturation and to allow effective pheromone tracing and oriented flight.
Recent studies in Drosophila melanogaster [21, 22] but also in mouse [23] indicate that the synaptic transmission from presynaptic OSNs to second order PNs in the AL is regulated to control the gain and adjust the dynamic range of the olfactory system. Evidence has been provided suggesting that GABA released from local interneurons activates GABA receptors in the presynaptic axon terminals of OSN-axons, leading to presynaptic inhibition and thereby reducing the gain of the synaptic transmission. Genes encoding ionotropic GABAA- and metabotropic GABAB receptor types possibly involved in such processes have been identified by cDNA cloning and predicted from genome sequences in several vertebrates and insect species [24-26]. In Drosophila, evidence has been found indicating that a GABAB receptor type was involved in fine tuning the synaptic gain [21, 27]. Interestingly, a strikingly high level of GABAB receptor protein was observed in the terminals of pheromone (11-cis-vaccenyl acetate) -sensitive OSNs and an RNAi-based knockdown of the GABAB receptor lead to an impaired pheromone-induced behaviour of male fruit flies. These results support the notion that presynaptic gain control mechanisms may play a pivotal role in processing of pheromone signals [21]. Since the sensitivity and dynamic range of the moth pheromone detection system is crucial for an immediate behavioural response and for tracing pheromone plumes we hypothesized that GABA-mediated gain control mechanisms may exist in this system which implies that GABAB receptors are expressed in the olfactory sensory neurons. As a first step, in this study we set out to identify GABAB receptors in Heliothis virescens and to explore whether they are expressed in the sensory cells of the male antenna.
Material and Methods
Animals
Heliothis virescens pupae were kindly provided by Bayer CropScience, Frankfurt, Germany. Pupae were sexed and allowed to develop at room temperature. After emergence moths were feed on 10% sucrose solution. Moths not older than four days were used for the experiments.
Reverse transcription (RT-) PCR
Total RNA was prepared from different tissues using TRIzol reagent (Invitrogen) following the recommendation of the supplier. For heads (without appendices), labial palps, thoraces and legs a mixture of male and female tissue was used. Total RNA from antennae was prepared separately for males and females. Poly (A)+ RNA was isolated from total RNA, applying oligo (dT)25 magnetic dynabeads (Dynal, Oslo, Norway) using recommended protocols. Poly (A)+ RNA was transcribed into cDNA as previously described [28] and used in RT-PCR experiments with gene-specific primer pairs. PCR conditions were 1 min 40 s at 94°C, then 21 cycles with 94°C for 30 s, 55°C for 40 s and 72°C for 1 min 30 s, with a decrease of the annealing temperature by 0.5°C per cycle. Subsequently, 19 further cycles at the condition of the last cycling step were performed, followed by incubation for 7 min at 72°C. PCR products were purified using the Geneclean II Kit (MP Biomedicals, LLC, IIIkrich, France), cloned into the pGEM-T plasmid (Promega, Madison, USA) and sequenced using vector or gene specific primers. For analysing the tissue distribution of HvirGABAB-R1 expression, the primer pair 5'-(AGTGGTGGACGTAGCACTGCT-3' and 5'-TTTGACTAGTTCCCGGTAGCG-3') was used. The primer pair (5'-CAACGAAGTTGTAACTCGTG-3' and 5'-TTCTTGGCTAGCGTCCACAT-3') directed against the ubiquitously expressed RL31 gene was applied to check the integrity of the different cDNAs.
Identification of a H. virescens GABAB-R1 sequence
In Drosophila the functional metabotropic GABAB receptor is indicated to be a heterodimer of a GABAB-R1 and GABAB-R2 subunit [27]. In order to assess the expression of a GABAB receptor in H. virescens antennae we have focused on the GABAB-R1 subunit. We used the Drosophila melanogaster GABAB-R1 sequence (DmelGABAB-R1; Acc. Nr.: AF318272) to BLAST a genomic database of H. virescens. This led to short H. virescens sequences, with significant similarities to DmelGABAB-R1, which were used to design a specific primer pair (5'-GGTTTAGTAGTGTGGCGATGC-3' and 5'-GACGCGCGAGTTATCGTCGGC-3'). Applying the primers in RT-PCR with male antennal cDNA allowed to amplify a partial HvirGABAB-R1 sequence which was used to generate a digoxigenin (DIG)-labelled PCR-product employing the PCR DIG DNA labeling mix (Roche, Mannheim, Germany). The labelled PCR product was purified, diluted in hybridization solution (30% formamide, 5x SSC, 0.1% lauroylsarcosine, 0.02% SDS, 2% blocking reagent [Roche], 100 μg/ml denatured herring sperm DNA) and used to screen a cDNA library of the antennae of H. virescens [29]. Briefly, phage DNA was transferred to and immobilized on Hybond-N+ nylon transfer membranes (Amersham Biosciences, Freiburg, Germany) and hybridized to the DIG-labelled probe at 30°C. After hybridization membranes were washed twice for 5 min in 2x SSC, 0.1% SDS at room temperature, followed by three washes in 2x SSC, 0.1% SDS for 20 min each at 30°C. Hybridized probes were detected using anti-DIG AP-conjugated antibodies (Roche) and CSPD (Applied Biosystems, Foster City, CA) as substrate. cDNA inserts from positive phage were isolated, subcloned into the pBluescript II SK+ vector and sequenced. This led to a long cDNA sequence, containing an open reading frame of 1578 bp for a putative HvirGABAB-R1, which overlapped with the HvirGABAB-R1 PCR product. Both sequences assembled to a HvirGABAB-R1 coding sequence of 1815 bp, missing parts of the N-terminus.
For further sequence prolongation, DNA (containing H. virescens cDNAs) was isolated from phage forming the antennal cDNA library. The DNA was used in PCR experiments with a primer pair matching the 5´ end of the partial GABAB-R1 sequence (5'-CTTCCCACCGACCCACCGCTCCCTT-3') and a λ-phage specific sequence (5'-GAGGTGGCTTATGAGTATTTCTTCCAGGG-3') flanking the cDNA insertion sites. Sequencing of a resulting PCR product led to a sequence completing the HvirGABAB-R1 coding sequence to 2418 bp encoding 806 amino acids (aa).
Assembling of a Bombyx mori GABAB-R1 sequence
In order to identify a GABAB-R1 sequence from the silk moth B. mori (BmorGABAB-R1) we BLAST-searched the available genomic database of B. mori (http://sgp.dna.affrc.go.jp/index.html) with the HvirGABAB-R1 and DmelGABAB-R1 sequences. 14 DNA regions with significant sequence similarity, all positioned on chromosome 15, could be identified. The positions on chromosome 15 of the putative BmorGABAB-R1 exons are; 1 (7008280-7008065), 2 (7004609-7004475), 3 (7001551-7001381), 4 (6995163-6994945), 5 (6986572-6986405), 6 (6982923-6982810), 7 (6981465-6981337), 8 (6978677-6978534), 9 (6975854-6975705), 10 (6974686-6974447), 11 (6971554-6971345), 12 (6969456-6969199), 13 (6966124-6966255) and 14 (6962139-6961930). The identified putative exons were assembled to a continuous DNA strand (Supplementary Material: Figure S1) using the HvirGABAB-R1 and the DmelGABAB-R1 sequence as template. This revealed a putative BmorGABAB-R1 sequence of 2496 bp coding for a protein of 831 aa (Supplementary Material: Figure S2). Blasting the sequence against the NCBI database revealed high sequence similarities in corresponding regions of various GABAB-R1 sequences from other invertebrates and vertebrates, indicating that we assembled a putative BmorGABAB-R1 sequence.
Whole-mount in situ hybridization
To localize the GABAB-R1 expressing cells in the antenna of H. virescens we adapted a whole mount in situ hybridization protocol which was previously used successfully to visualize transcripts of olfactory receptor genes in OSNs of Spodoptera littoralis antennae [30, 31].
Whole-mount in situ hybridization (WM-ISH) was performed in 0.5 ml reaction tubes. For all washes and incubations a volume of 0.5 ml solution were used. Steps were performed at room temperature, if not marked separately. Antennae were dissected from the heads, cut into smaller pieces and directly transferred to fixation solution (4% paraformaldehyde in 0.1 M NaHCO3, pH 9.5). Following incubation over night at 4°C the antennal fragments were washed twice with PBS (phosphate buffered saline = 0.85% NaCl, 1.4 mM KH2PO4, 8 mM Na2HPO4, pH 7.1). Antennal fragments were dehydrated by incubation in methanol, two times for 5 minutes and followed by 1 hour at -20°C. Subsequently rehydration was performed by incubation for 5 min each in Methanol/PBST (ratio 1:1, PBST = PBS + 0.1% Tween 20), Methanol/PBST (3:7) and two times PBST. Fragments were treated again with fixation solution at 4°C for 20 min, washed two times with PBST for 5 min each and incubated with PBST containing 50 µg/ml proteinase K at 37°C for 15 min. After rinsing twice in PBST, antennal fragments were washed for 5 min in PBST. PBST was discarded and fixation solution containing 0.2% glutaraldehyde was added, followed by incubation at 4°C for 20 min. After washing twice in PBST for 5 min each, antennal fragments were treated with 0.1 M triethanolamine containing 0.25% acetic anhydride in water adjusted to pH 8.0 with NaOH for 10 min, followed by two washes for 10 min each in PBST.
Antennal fragments were rinsed with hybridization solution (SOL H: 50% formamide, 5x SSC, 0.1% Tween 20, 0.005% Heparin, 0.1 mg/ml tRNA) and prehybridized in SOL H for at least 4 hours at 65°C. SOL H was removed and replaced by 250 µl fresh SOL H containing the DIG-labelled antisense or sense RNA probe, previously incubated for 10 min at 65°C and at least 10 min on ice. Hybridization was performed overnight at 65°C. Post-hybridization antennal fragments were shortly rinsed and then washed for 1 hour in 2x SSC containing 50% formamide, followed by three washes in 2x SSC, each for 10 min. After a short rinse in PBST antennal fragments were incubated for 1 hour in RNase solution (PBST containing 2 µg/ml RNase A) at 37 °C, followed by a wash in 2x SSC at 37 °C (10 min), 55°C (15 min) and in 0.2x SSC at 55°C (two times 15 min each) and at last in PBST (5 min). Unspecific binding sides were blocked by incubation in blocking solution (BS = 90 mM Tris pH 7.5, 10 mM maleic acid, 150 mM NaCl, 0.03% Triton- X100, 1% blocking-reagent), for at least two hours. BS was removed and replaced by BS containing anti-DIG alkaline phosphatase-conjugated antibody 1:4000 (Roche, Mannheim, Germany). After incubation overnight at 4°C antennal fragments were shortly rinsed twice in BS, washed three times in BS each for 30 min and further washed in BS containing 1 mM Levamisol for 30 min. Followed by three washes in 100 mM Tris pH 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1% Tween 20, 1 mM Levamisol each for 10 min were performed and antennal fragments were rinsed in DAP-buffer (100 mM Tris pH 9.5, 100 mM NaCl, 50 mM MgCl2). Subsequently signals were visualized using NBT (nitroblue tetrazolium) and BCIP (5-brom-4-chlor-3-indolyl phosphate). After signal visualization antennal fragments were washed three times in PBS for 5 min each and incubated in fixation solution for 20 min at 4 °C. After two additional washes in PBS for 5 min each, antennal fragments were embedded into Tissue-Tek O.C.T. Compound (Sakura Finetek Europe, Zoeterwoude, The Netherlands) and 12 µm slices were prepared using a cryostat. Finally, slices were mounted with mowiol (13% mowiol 4-88, 33% glycerin, 130 mM Tris, pH 8.5) and a cover slip. Pictures were made using an Axioskop 2 Mot (Zeiss, Jena, Germany) equipped with an AxioCam MRc5 and the AxioVision LE 4.3 software.
Sequencing and sequence analysis
Sequencing was performed on an ABI310 sequencing system using vector and cDNA derived primers and the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). Sequence analysis were made using Chromas Lite 2.01 (http://technelysium.com.au). For further analysis also Genamics Expression (http://genamics.com/expression/index.htm) was used. For prediction of transmembrane domains and the coiled-coil domain the PRED-TMR program was utilized (http://athina.biol.uoa.gr/PRED-TMR/input.html). The MEGA 5.05 [32] software was applied to calculate a neighbor joining tree based on a ClustAL alignment of the amino acid sequences, indicated in the figure legend.
Results
Cloning of a GABAB-R1 sequence from the antennae of H. virescens
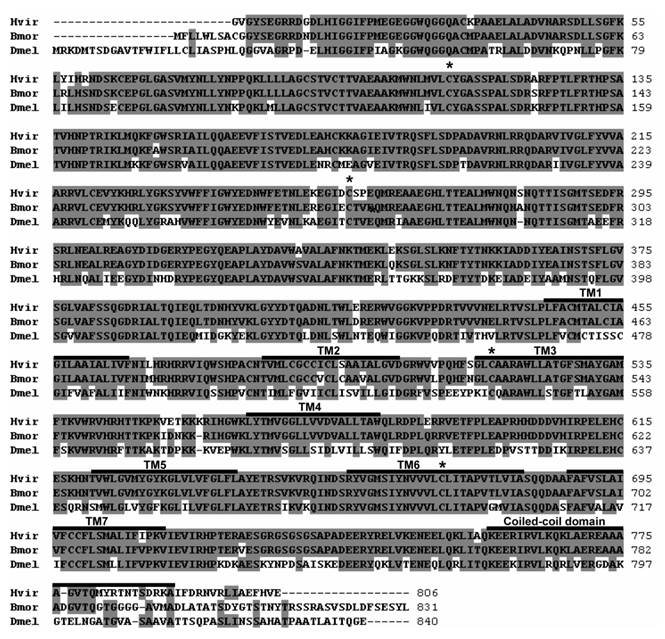
In order to identify GABAB receptors of Heliothis virescens we performed bioinformatic searches in a genomic database of the moth. BLAST analysis with the Drosophila melanogaster GABAB-R1 [27] revealed short genomic DNA sequences which were used to design specific primers for RT-PCR experiments with male antennal cDNA from H. virescens. This led to a PCR-product with high sequence identity to DmelGABAB-R1, which could be prolonged by screening and PCR-based approaches employing an antennal cDNA library. In this way we obtained a putative HvirGABAB-R1 cDNA sequence encoding 806 amino acids (aa). A start codon was not found at the 5' end (Fig. 1). Comparing the HvirGABAB-R1 sequence to DmelGABAB-R1 showed that the N-terminus of the fruit fly sequence was 25 aa longer (Fig. 1). The overall sequence identity between the two sequences was 71.4 %. Blasting the H. virescens sequence in the NCBI data base showed high sequence similarities with GABAB-R1 sequences from various other insect species.
Sequence analysis and comparison
To get further sequence information about GABAB-R1 proteins in moths and to evaluate the length of the missing N-terminus of the HvirGABAB-R1 sequence, attempts were made to assemble a GABAB-R1 sequence from the Bombyx mori genome. Using the HvirGABAB-R1 and DmelGABAB-R1 sequences in BLAST-searches of the genome identified regions with high sequence similarity. Assembling of the predicted exon regions led to a putative BmorGABAB-R1 coding sequence comprising 2493 bp, which started with a start codon and was flanked by a stop codon. The encoded 831 aa long BmorGABAB-R1 shared 92.7 % and 70.6 % aa sequence identity with HvirGABAB-R1 and DmelGABAB-R1, respectively. Most diverse regions in the aa sequence are found at the N- and C-terminal end of the sequence, as found also between Drosophila and human GABAB-R1 [27].
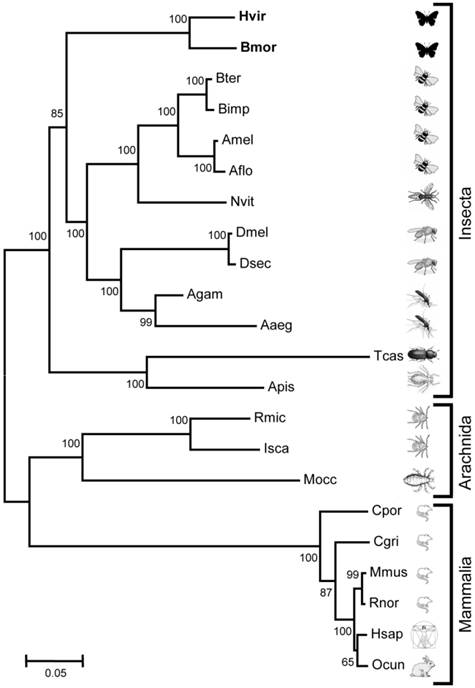
Comparing the N-terminus of the predicted Hvir- and BmorGABAB-R1 revealed that the B. mori sequence is only eight aa longer (Fig. 1). At the C-terminus DmelGABAB-R1 and BmorGABAB-R1 are 12-18 aa longer than HvirGABAB-R1. As found typical for GABAB-R1 sequences from various species [27, 33], both moths GABAB-R1 sequences comprise the typical 7 transmembrane domains, contain several highly conserved cysteins and display a coiled-coil domain at the C-terminal region (Fig. 1). A phylogenetic comparison of the two moth GABAB-R1 sequences with GABAB-R1 sequences from various invertebrate and vertebrate species (Fig. 2) revealed order-specific and class-specific clustering of GABAB-R1 sequences. In a neighbor joining tree, the two Lepidopteran GABAB-R1 sequences form a separated branch, which is most closely related to Hymenopteran and Dipteran GABAB-R1 sequences and more distant to sequences from Coleoptera and Hemiptera.
Alignment of the HvirGABAB-R1, BmorGABAB-R1 and DmelGABAB-R1 amino acid sequences. Identical amino acid residues in at least two sequences are shaded in grey. Numbers at the right refer to the position of the last residue in a line. Positions of seven putative transmembrane domains (TM1 - TM7), several conserved cysteins and a coiled-coil domain are indicated.
Relationship of HvirGABAB-R1 and BmorGABAB-R1 to GABAB-R1 sequences from species belonging to different classes and orders. Neighbor joining tree based on a ClustAL aligment of amino acid sequences. Bootstrap support values are based on 1000 replicates; only support values above 60% are shown; branch lengths are proportional. Hvir = Heliothis virescens (this study), Bmor = Bombyx mori (this study). Other sequences are from (accession numbers in brackets): Bter = Bombus terrestris (XM_003394282.1), Bimp = Bombus impatiens (XM_003490756.1), Amel = Apis mellifera (XM_392294.4), Aflo = Apis florea (XM_003698600.1), Nvit = Nasonia vitripennis (XM_001605233.2), Dmel = Drosophila melanogaster (AF318272.1), Dsec = Drosophila sechellia (XM_002035835.1), Agam = Anopheles gambiae (XM_319474.3), Aaeg = Aedes aegypti (XM_001652657.1), Tcas = Tribolium castaneum (XM_964268.2), Apis = Acyrthosiphon pisum (XM_001952406.2), Rmic = Rhipicephalus microplus (JN974907.1), Isca = Ixodes scapularis (XM_002406043.1), Mocc = Metaseiulus occidentalis (XM_003747475.1), Cpor = Cavia porcellus (XM_003461152.1), Cgri = Cricetulus griseus (XM_003507407.1), Mmus = Mus musculus (BC056990.1), Rnor = Rattus norvegicus (NM_031028.3), Hsap = Homo sapiens (AK223619.1), Ocun = Oryctolagus cuniculus (XM_002714338.1).
Tissue distribution of GABAB-R1 expression
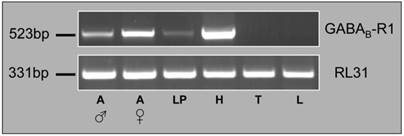
To explore the expression of HvirGABAB-R1 in the antennae and other tissues of the moth we conducted RT-PCR experiments with specific primers and cDNAs from antennae, labial palps, heads (without appendices), thoraces and legs. Male and female antennae were analyzed separately, while for the other body parts a mixture of male and female cDNAs were probed. First, the integrity of the cDNA preparation was controlled by performing PCR reactions with primers for the ubiquitously expressed RL31 gene (Fig. 3). A DNA band of similar intensity was obtained for all cDNAs tested, indicating a similar quality and quantity of the cDNA templates in each preparation. With HvirGABAB-R1 specific primers, respective transcripts were detected in antennal tissue of both sexes as well as in maxillary palps and head tissue, but not in thoraces and legs (Fig. 3). Compared to antennae the differences in band intensities indicate higher GABAB-R1 transcript levels in head and much lower amounts in labial palps. With regard to transcript levels in the antennae of the two sexes, PCR band intensities suggest higher GABAB-R1 expression in females, compared to males.
Expression of HvirGABAB-R1 in different moth tissues. RT-PCR using cDNAs from the tissues indicated and a primer pair matching either HvirGABAB-R1 or the ubiquitously expressed RL31. Male antennae (A ♂), female antennae (A ♀), labial palps (LP), head without appendices (H), thorax (T) and legs (L). The size of RT-PCR amplification products is indicated on the left.
Expression of GABAB-R1 within the antennae
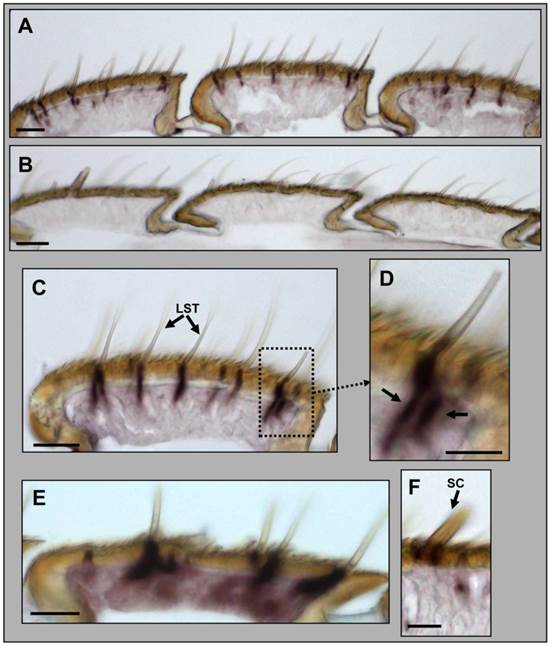
In order to localize the GABAB-R1 expressing cells in the antenna of male H. virescens we adapted a whole mount in situ hybridization protocol, using a DIG-labelled HvirGABAB-R1 antisense RNA probe and color visualization of cells bearing specific transcripts. With the HvirGABAB-R1 specific probe we found stained cells under long and shorter trichoid sensilla (Fig. 4A). For control we applied a DIG-labelled GABAB-R1 sense RNA probe, which gave no hybridization signals (Fig. 4B), confirming the specificity of the signals obtained with the antisense RNA probe. At higher magnification (Fig. 4C and 4D) staining can be assigned to single long trichoid hairs, which have been reported to generally contain two olfactory sensory neurons [6, 34]. In agreement with this number of OSNs, on the slices that were made from antennae after WM-ISH, regularly two labelled cells in close vicinity to each other were visible, indicating co-localization in the same sensillum. In experiments using a probe for the pheromone receptor HR13 single cells were labelled under many but not all long trichoid hairs (Fig. 4E), thus confirming and extending previous results [16, 35]. Cells under sensilla chaetica, which contain mechanosensory and gustatory sensory neurons [36], were not labelled (Fig. 4F); this result indicates that the expression of GABAB-R1 is restricted to olfactory sensory neurons.
Expression of HvirGABAB-R1 in the antenna of male H. virescens. Whole-mount in situ hybridization (WM-ISH) using a DIG-labelled antisense (A, C and F) and sense (B) RNA probe for HvirGABAB-R1. A, Hybridization signals in three antennal segments under sensilla trichodea. B, No hybridization signals were visualized using a GABAB-R1 sense RNA probe. C, Segment showing several labelled cells under trichoid sensilla. D, Higher magnification of C, showing a sensillum with two labelled cells; marked by arrows. E, WM-ISH using an antisense RNA probe for the pheromone receptor HR13. Single HR13-expressing cells are labelled under long trichoid sensilla. F, No cells are labelled under mechanosensory/gustatory sensilla chaetica. LST = long sensillum trichodeum, SC = sensillum chaeticum. Scale bars: A-E = 20 μm, D and F = 10 µm.
Discussion
In this study, we determined the amino acid sequence of a GABAB-R1 receptor from antennal cDNA of the tobacco budworm Heliothis virescens and predicted an orthologous receptor from the genome database of the silk moth Bombyx mori. In a neighbor joining tree the two sequences form a distinct lepidopteran branch within the insecta lineage. Generally, GABAB-R1 sequences of insect species, which belong to the same animal order were found in separated taxons and GABAB-R1s of the same animal class (Insecta, Arachnida or Mammalia) were most related. Thus, the similarities between GABAB-R1 sequences from different species match perfectly their phylogenetic relationship as indicated in the tree of life (http://tolweb.org/tree). Overall, both moth sequences are highly related to GABAB-R1 sequences of insects from other orders (54-75%) and show significant identities to GABAB-R1 sequences from arachnidan (53-65%) and mammalian species (about 50%). Such a high degree of sequence identity across animal species, orders and classes appears to be a general feature of neurotransmitter receptors, which are tuned to bind specific ligands, like receptors for glutamate or acetylcholine [37, 38]. It is supposed that due to the critical role of such receptors in the physiology of animals, evolution would allow only a limited number of sequence variations and thus keeps the receptor genes under negative selection.
By means of whole mount in situ hybridization with subsequent sectioning of the antenna we revealed expression of GABAB-R1 in cells under long sensilla trichodea, which in general house two sensory cells. Three types of long sensilla hairs have been classified. Among these, type A hairs represent about 80% of the hairs and contain a sensory neuron responsive to the major pheromone component, (Z)-11-hexadecenal (Z11-16:AL) and a second OSN of unknown ligand specificity [6, 39, 40]. In agreement with the antennal representation of Z11-16:AL-responsive sensilla, experiments with a probe specific for HR13 the receptor of the main pheromone component revealed labelling of single cells in many but not all long hairs, thus confirming previous results [35]. With the GABAB-R1-specific probe we have found positive cells under most if not all long sensilla hairs and frequently observed two cells under a single long hair. This suggests that GABAB-R1 is expressed by both OSNs of long trichoid type A hairs. Moreover, based on our data it is suggested that GABAB-R1 is expressed also in OSNs of long sensilla hairs of type B, containing an OSN responsive to the minor pheromone component, (Z)-9-tetradecenal (Z9-14:AL) of H. virescens as well as in type C hairs housing two OSNs sensitive to pheromones used by other species [6, 40]. GABAB-R1-positive cells were also present under shorter trichoid sensilla of male antennae, which in their majority house OSNs responsive to host plant volatiles [10]. This indicates that in male moth the GABAB-R1 receptor is expressed in pheromone-responsive neurons and also in OSNs responding to general odorants.
Based on the results of in situ hybridization it cannot be excluded that GABAB-R1 is also expressed in neurons of the very short sensilla basiconica, representing an additional olfactory sensillum type [34]. However, labelled cells were not found under sensilla chaetica, which house gustatory- and mechanosensory neurons (Jørgensen et al., 2006). Interestingly, in a recent study it was shown that on the tropical wandering spider Cupiennius salei cells of mechanosensilla do express GABAB receptors [41].
Our results also suggest that the GABAB-R1 receptor may also play a role in OSNs of female H. virescens antenna, which possess only short trichoid and basiconic sensilla [34] housing OSNs mainly responding to plant volatiles [42]. In fact, the semi-quantitative RT-PCR data indicate abundant GABAB-R1 transcripts in female antennae compared to males. This may imply a higher expression level; however, one has to consider that the female antenna contains considerable more olfactory sensilla (17000) at higher densities than male antenna (12000) [34]. Therefore, assuming that GABAB-R1 is expressed in most if not all OSNs in these hairs, the disparity of PCR band intensity between sexes may reflect obvious differences in sensilla numbers.
For the GABAB receptors from Drosophila it has been found in heterologous systems that a co-expression of GABAB-R1 and GABAB-R2 is necessary to form a functional GABAB receptor [27]; in addition, the results of RNAi-based kock-down experiments suggest a role of GABAB-R2 in presynaptic gain control [21]. Therefore it is possible that expression of GABAB-R2 is necessary in OSNs of the moths. However, so far a GABAB-R2 receptor subtype of the moth H. virescens has not been identified yet.
In conclusion, we have identified a GABAB-R1 receptor and provided evidence for its expression in pheromone-responsive neurons as well as in OSNs responding to general odors. This finding is in line with synaptic contacts identified between OSNs and GABAergic LNs in insects [43] and a proposed role of a GABAB receptor in the mechanisms underlying presynaptic gain control in the olfactory system of Drosophila [21, 22]. Forthcoming studies will have to show whether the HvirGABAB-R1 protein is in fact localized in the axon terminals of OSNs in the antennal lobe thus corroborating the notion that GABAB receptors may be part of the presynaptic gain control mechanisms important for the recognition of pheromone signals by male moths.
Supplementary Material
Fig.S1 - S2.
Acknowledgements
The authors would like to thank the insect rearing unit at Bayer CropScience AG Frankfurt for providing Heliothis virescens pupae. We are grateful to Heidrun Froß for excellent technical assistance. This work was supported by the Deutsche Forschungsgemeinschaft, SPP1392 by grants to J.K. (KR1786/4-2).
Data deposition
The HvirGABAB-R1 sequence reported in this paper has been deposited in the EMBL database under accession number HG004164
Competing Interests
The authors have declared that no competing interest exists.
References
1. Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373-91
2. Martin JP, Beyerlein A, Dacks AM, Reisenman CE, Riffell JA, Lei H, Hildebrand JG. The neurobiology of insect olfaction: sensory processing in a comparative context. Prog Neurobiol. 2011;95:427-47
3. Blomquist GJ, Vogt RG. Insect pheromone biochemistry and molecular biology. The biosynthesis and detection of pheromones and plant volatiles. London: Elsevier Academic Press. 2003
4. Klun JA, Bierl-Leonhardt BA, Plimmer JR, Sparks AN, Primiani M, Chapman OL, Lepone G, Lee GH. Sex pheromone chemistry of the female tobacco budworm moth, Heliothis virescens. J Chem Ecol. 1980;6:177-83
5. Teal PE, Tumlinson JH, Heath RR. Chemical and behavioral analyses of volatile sex pheromone components released by calling Heliothis virescens (F.) females (Lepidoptera: Noctuidae). J Chem Ecol. 1986;12:107-26
6. Baker TC, Ochieng SA, Cosse AA, Lee SG, Todd JL, Quero C, Vickers NJ. A comparison of responses from olfactory receptor neurons of Heliothis subflexa and Heliothis virescens to components of their sex pheromone. J Comp Physiol A. 2004;190:155-65
7. Mustaparta H. Chemical information processing in the olfactory system of insects. Physiol Rev. 1990;70:199-245
8. Berg BG, Almaas TJ, Bjaalie JG, Mustaparta H. The macroglomerular complex of the antennal lobe in the tobacco budworm moth Heliothis virescens: specified subdivision in four compartments according to information about biologically significant compounds. J Comp Physiol A. 1998;183:669-82
9. Berg BG, Galizia CG, Brandt R, Mustaparta H. Digital atlases of the antennal lobe in two species of tobacco budworm moths, the Oriental Helicoverpa assulta (male) and the American Heliothis virescens (male and female). J Comp Neurol. 2002;446:123-34
10. Hillier NK, Vickers NJ. Physiology and antennal lobe projections of olfactory receptor neurons from sexually isomorphic sensilla on male Heliothis virescens. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2007;193:649-63
11. Ro H, Muller D, Mustaparta H. Anatomical organization of antennal lobe projection neurons in the moth Heliothis virescens. J Comp Neurol. 2007;500:658-75
12. Berg BG, Schachtner J, Homberg U. Gamma-aminobutyric acid immunostaining in the antennal lobe of the moth Heliothis virescens and its colocalization with neuropeptides. Cell Tissue Res. 2009;335:593-605
13. Hoskins SG, Homberg U, Kingan TG, Christensen TA, Hildebrand JG. Immunocytochemistry of GABA in the antennal lobes of the sphinx moth Manduca sexta. Cell Tissue Res. 1986;244:243-52
14. Seki Y, Kanzaki R. Comprehensive morphological identification and GABA immunocytochemistry of antennal lobe local interneurons in Bombyx mori. J Comp Neurol. 2008;506:93-107
15. Grosse-Wilde E, Gohl T, Bouche E, Breer H, Krieger J. Candidate pheromone receptors provide the basis for the response of distinct antennal neurons to pheromonal compounds. Eur J Neurosci. 2007;25:2364-73
16. Gohl T, Krieger J. Immunolocalization of a candidate pheromone receptor in the antenna of the male moth, Heliothis virescens. Invert Neurosci. 2006;6:13-21
17. Forstner M, Breer H, Krieger J. A receptor and binding protein interplay in the detection of a distinct pheromone component in the silkmoth Antheraea polyphemus. Int J Biol Sci. 2009;5:745-57
18. Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005;307:1638-42
19. Vickers NJ. Winging it: moth flight behavior and responses of olfactory neurons are shaped by pheromone plume dynamics. Chem Senses. 2006;31:155-66
20. Carde RT, Willis MA. Navigational strategies used by insects to find distant, wind-borne sources of odor. J Chem Ecol. 2008;34:854-66
21. Root CM, Masuyama K, Green DS, Enell LE, Nassel DR, Lee CH, Wang JW. A presynaptic gain control mechanism fine-tunes olfactory behavior. Neuron. 2008;59:311-21
22. Olsen SR, Wilson RI. Lateral presynaptic inhibition mediates gain control in an olfactory circuit. Nature. 2008;452:956-60
23. McGann JP, Pirez N, Gainey MA, Muratore C, Elias AS, Wachowiak M. Odorant representations are modulated by intra- but not interglomerular presynaptic inhibition of olfactory sensory neurons. Neuron. 2005;48:1039-53
24. Bettler B, Kaupmann K, Mosbacher J, Gassmann M. Molecular structure and physiological functions of GABA(B) receptors. Physiol Rev. 2004;84:835-67
25. Gou ZH, Wang X, Wang W. Evolution of neurotransmitter gamma-aminobutyric acid, glutamate and their receptors. Zoolog Res. 2012;33:E75-E81
26. Darlison MG, Pahal I, Thode C. Consequences of the evolution of the GABA(A) receptor gene family. Cell Mol Neurobiol. 2005;25:607-24
27. Mezler M, Muller T, Raming K. Cloning and functional expression of GABA(B) receptors from Drosophila. Eur J Neurosci. 2001;13:477-86
28. Krieger J, Raming K, Dewer YME, Bette S, Conzelmann S, Breer H. A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur J Neurosci. 2002;16:619-28
29. Krieger J, Gaenssle H, Raming K, Breer H. Odorant binding proteins of Heliothis virescens. Insect Biochem Mol Biol. 1993;23:449-56
30. Brigaud I, Montagne N, Monsempes C, Francois MC, Jacquin-Joly E. Identification of an atypical insect olfactory receptor subtype highly conserved within noctuids. FEBS J. 2009;276:6537-47
31. Malpel S, Merlin C, Francois MC, Jacquin-Joly E. Molecular identification and characterization of two new Lepidoptera chemoreceptors belonging to the Drosophila melanogaster OR83b family. Insect Mol Biol. 2008;17:587-96
32. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731-9
33. Kaupmann K, Huggel K, Heid J, Flor PJ, Bischoff S, Mickel SJ, McMaster G, Angst C, Bittiger H, Froestl W, Bettler B. Expression cloning of GABA(B) receptors uncovers similarity to metabotropic glutamate receptors. Nature. 1997;386:239-46
34. Almaas TJ, Mustaparta H. Pheromone reception in tobacco budworm moth, Heliothis virescens. J Chem Ecol. 1990;16:1331-47
35. Krieger J, Grosse-Wilde E, Gohl T, Dewer YME, Raming K, Breer H. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc Natl Acad Sci U S A. 2004;101:11845-50
36. Jorgensen K, Almaas TJ, Marion-Poll F, Mustaparta H. Electrophysiological characterization of responses from gustatory receptor neurons of sensilla chaetica in the moth Heliothis virescens. Chem Senses. 2007Nov;32(9):863-79
37. Chiu J, DeSalle R, Lam HM, Meisel L, Coruzzi G. Molecular evolution of glutamate receptors: a primitive signaling mechanism that existed before plants and animals diverged. Mol Biol Evol. 1999;16:826-38
38. Tsunoyama K, Gojobori T. Evolution of nicotinic acetylcholine receptor subunits. Mol Biol Evol. 1998;15:518-27
39. Almaas TJ, Mustaparta H. Heliothis virescens: Response characteristics of receptor neurons in sensilla trichodea type 1 and type 2. J Chem Ecol. 1991;17:953-72
40. Berg BG, Tumlinson JH, Mustaparta H. Chemical communication in heliothine moths. IV. Receptor neuron responses to pheromone compounds and formate analogues in the male tobacco budworm moth Heliothis virescens. J Comp Physiol A. 1995;177:527-34
41. Panek I, Meisner S, Torkkeli PH. Distribution and function of GABAB receptors in spider peripheral mechanosensilla. J Neurophysiol. 2003;90:2571-80
42. Hillier NK, Kleineidam C, Vickers NJ. Physiology and glomerular projections of olfactory receptor neurons on the antenna of female Heliothis virescens (Lepidoptera: Noctuidae) responsive to behaviorally relevant odors. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2005;192:199-219
43. Distler PG, Boeckh J. Synaptic connections between identified neuron types in the antennal lobe glomeruli of the cockroach, Periplaneta americana: II. Local multiglomerular interneurons. J Comp Neurol. 1997;383:529-40
Author contact
![]() Corresponding author: Dr. Jürgen Krieger, University of Hohenheim, Institute of Physiology (230), Garbenstrasse 30, 70599 Stuttgart, Germany. e-mail: juergen.kriegerde.
Corresponding author: Dr. Jürgen Krieger, University of Hohenheim, Institute of Physiology (230), Garbenstrasse 30, 70599 Stuttgart, Germany. e-mail: juergen.kriegerde.

 Global reach, higher impact
Global reach, higher impact