10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(9):872-886. doi:10.7150/ijbs.7126 This issue Cite
Research Paper
A Novel Zinc Finger Protein 219-like (ZNF219L) is Involved in the Regulation of Collagen Type 2 Alpha 1a (col2a1a) Gene Expression in Zebrafish Notochord
1. Institute of Fisheries Sciences, National Taiwan University, Taipei 106, Taiwan;
2. Institute of Biological Chemistry, Academia Sinica, Taipei 115, Taiwan;
3. Department of Biochemistry, Taipei Medical University, Taipei 110, Taiwan;
4. Institute of Cellular and Organismic Biology, Academia Sinica, Taipei 115, Taiwan.
* These authors contributed equally to this work.
Received 2013-7-9; Accepted 2013-8-24; Published 2013-9-5
Abstract
The notochord is required for body plan patterning in vertebrates, and defects in notochord development during embryogenesis can lead to diseases affecting the adult. It is therefore important to elucidate the gene regulatory mechanism underlying notochord formation. In this study, we cloned the zebrafish zinc finger 219-like (ZNF219L) based on mammalian ZNF219, which contains nine C2H2-type zinc finger domains. Through whole-mount in situ hybridization, we found that znf219L mRNA is mainly expressed in the zebrafish midbrain-hindbrain boundary, hindbrain, and notochord during development. The znf219L morpholino knockdown caused partial abnormal notochord phenotype and reduced expression of endogenous col2a1a in the notochord specifically. In addition, ZNF219L could recognize binding sites with GGGGG motifs and trigger augmented activity of the col2a1a promoter in a luciferase assay. Furthermore, in vitro binding experiments revealed that ZNF219L recognizes the GGGGG motifs in the promoter region of the zebrafish col2a1a gene through its sixth and ninth zinc finger domains. Taken together, our results reveal that ZNF219L is involved in regulating the expression of col2a1a in zebrafish notochord specifically.
Keywords: zinc finger protein 219, notochord, zebrafish, collagen type 2 alpha 1a, transcriptional regulation.
Introduction
The notochord is an important feature of the phylum Chordata [1], and is fully differentiated during early embryogenesis of the anterior-posterior body axis through polarization [2]. The notochord is a center of signaling transmission, and also acts as the axial skeleton and neural tube of the embryo [3]. Notochord structure differentiates from the chordamesoderm (part of the mesoderm) during zebrafish embryo gastrulation [4]. At this stage, notochord progenitor cells differentiate, turn into rod-like vacuolated cells at the midline in the trunk, and grow in concert with somatic segmentation. Notochord structure forms through the cross-linking of collagen, elastin, and other extracellular proteins [5]. Certain notochord cells subsequently contribute to the formation of the nucleus pulposus (NP) of intervertebral discs (IVDs) [3, 6, 7], suggesting that defects in notochord development may be related to IVD development and degeneration [8, 9].
Collagen is one of the proteins required for notochord formation. It is also found in a variety of tissues, including cartilage, bone, blood vessels, and the gut. The proteins of the collagen family are diverse in function, but all share a common structural feature, consisting of at least one triple-helical domain corresponding to a number of (Gly-X-Y)n repeats (X is frequently proline, and Y is often hydroxyproline) [10]. To date, more than 50 types of collagen and collagen-like protein have been identified [11]. Most collagen in the body is type I, while type II is the main component of cartilage. Cartilage is composed of chondroblasts, which produce a large amount of extracellular matrix consisting of type II collagen. Collagen type 2 alpha 1 (col2a1) is a major matrix protein found in cartilage; this protein provides tensile strength and a scaffolding network for proteoglycans [10, 12], and has been found to be conserved from mammals to teleosts. It can be detected in the developing cartilage, notochord, skin, floor plate, brain, heart, and other tissues [13-16]. Certain mutations in the col2a1 gene have been linked to type II collagenopathies, which include Stickler syndrome type 1 [17]. Stickler syndrome is a genetic disorder of connective tissue, characterized by abnormal skeletal growth and density, early onset of osteoarthritis, and retinal detachment, as first described by Stickler [18-20]. Mutations in col2a1, col11a1, col11a2, and col9a1 have been reported to cause type 1 to 4 Stickler syndrome [21, 22]. Haploinsufficiency of col2a1 expression, arising from gene silencing or nonsense mutations, is linked to Stickler syndrome [23]. Thus, sufficient production of col2a1 protein is essential for normal skeletal and cartilage development.
Gene duplication in the zebrafish genome is common [24, 25], and there are two homologues of mammalian col2a1 gene in zebrafish. The col2a1a gene has been well characterized [26], but the expression pattern and function of the col2a1b gene are unclear. Tg(col2a1a:GFP), a transgenic zebrafish line expressing GFP under the control of the col2a1a promoter, was recently generated, and found to express GFP in craniofacial cartilage, ear, notochord, floor plate, hypochord, pectoral fins, and IVD, a pattern similar to that of endogenous col2a1a [27]. Expression of col2a1 in chondrocytes is known to be directly regulated by Sox9 during chondrogenesis in mammals. Sox9 is a member of the vertebrate SoxE family (all of which contain a high-mobility-group (HMG)-box DNA-binding domain), and its effect on col2a1 expression is enhanced by Sox5 and Sox6, which act as cofactors that bind to Sox9 [28-31]. Attempts have been made to identify other transcriptional partners of mammalian Sox9 to better understand the regulatory mechanisms involved in col2a1 activation. However, unlike its mammalian counterpart, zebrafish has two paralogues of the sox9 gene, termed sox9a and sox9b. Previous reports have indicated that the expression patterns of Sox9a and Sox9b are distinct, but overlap in some parts of the brain, craniofacial skeleton, notochord and fins during embryogenesis [32-34]. .Knockdown of Sox9a caused a significant decrease in the GFP signal in craniofacial cartilage, while the GFP signal in notochord remained unchanged [27]. This suggests that factors other than Sox9a are required for expression of col2a1a in zebrafish notochord. Recently, zinc finger protein 219 (ZNF219) was reported to act as a transcriptional partner of Sox9 in the regulation of chondrocyte differentiation in mammals [35-37]. In mice, expression of znf219 mRNA is detected specifically in the developing limb buds, where col2a1 and sox9 are also expressed [37]. ZNF219 has been shown to colocalize with Sox9 in the nucleus. ZNF219 increases activity of the col2a1 gene promoter by upregulating the transcriptional activity of Sox9. This suggests that ZNF219 may affect col2a1 expression by forming a complex with Sox9 [37].
In mammals, znf219 was originally isolated from a human testis cDNA library; it encodes a protein of 723 amino acids with nine individual Cys2His2 (C2H2) zinc finger domains, eight of which are separated-paired [38, 39]. In general, C2H2-type zinc finger proteins can be divided into three groups: triple-C2H2, multiple-adjacent-C2H2, and separated-paired-C2H2. Their functions are diverse, due to their zinc finger structures [40]. There are more than 10,000 types of C2H2 zinc finger domains. It has been reported that classical C2H2 zinc finger proteins are involved in DNA binding, and function in transcriptional regulation. There is considerable variation in zinc finger proteins between different species [38, 39]. Whole genome analysis has been used to predict that there are 216, 837, and 1055 genes encoding C2H2-type zinc finger proteins in the genomes of zebrafish, mouse, and human, respectively [38]. As there are far fewer C2H2 zinc finger proteins in zebrafish than in mammals, it is plausible that zebrafish zinc finger proteins may diverge greatly in terms of their amino acid sequences, and have similar or overlapping functions as compared with their mammalian counterparts.
In this study, we describe theisolation of zebrafish ZNF219-like (ZNF219L) gene, which encodes a novel transcription factor. We demonstrate that ZNF219L recognizes the conserved GGGGG motifs in the col2a1a promoter region, and it regulates col2a1a expression specifically in zebrafish notochord during early stages. Our results indicate that zinc finger domain-containing protein, ZNF219L, might have predominant roles in regulating col2a1a expression in notochord specifically.
Materials and methods
Zebrafish care
Zebrafish embryos were raised at 28.5 °C, and different developmental stages were determined based on criteria described in the Zebrafish Book [41]. All animal procedures were approved by Academia Sinica Institutional Animal Care and Utilization Committee (ASIACUC) (protocol #10-12-114).
Rapid Amplification of cDNA Ends (RACE)
In order to obtain full-length cDNA, the 5' ends of zebrafish znf219L mRNA were amplified by random amplification of cDNA 5' ends (RACE) PCR using a Marathon cDNA amplification kit (Clontech, Palo Alto, CA, USA) according to the supplier's instructions. In brief, total RNA was isolated from zebrafish kidney or heart by the guanidinium thiocyanate method [42], and poly(A)+ RNA was subsequently prepared. First-strand cDNA was synthesized using a modified poly-T primer and 1 μg of poly (A)+ RNA. Second-strand cDNA was generated using an enzyme cocktail containing RNase H, DNA polymerase, and DNA ligase. Asymmetric adapter primers (AP primers) were then ligated to both ends of the double-stranded cDNA. An aliquot of this zebrafish cDNA collection was diluted 1:100 and subjected to two different anchored PCRs. 5' RACE was first performed with a 27-mer sense primer (AP1) against the adapter and an anti-sense primer against znf219L (5'- GGG GAA GGA ATT GGG GAC TGT GGT CCA GGG ATG GCA TGG AGG GAG GAG ACA G-3'); the second round of PCR was carried out with a nested 23-mer sense primer (AP2) and a nested antisense primer against znf219L (5'-TGG GGG CTC AGA GGA AAG AGC CAG CAT ACA TTC TGG TGG GGA ATC CAT -3'). The PCR products were cloned into the pGEM-T vector (Promega, Madison, WI, USA) and sequenced.
Cloning of full-length and deletion mutations of zebrafish znf219L
The znf219L sequence was submitted to GenBank with the accession no. JX141438. Full-length and deletion variants of the zebrafish znf219L coding sequence were generated by PCR amplification using gene-specific primers, as shown in Table 1. In order to express fusion proteins of ZNF219L (or ZNF219L deletion mutants tagged with HA) in NIH3T3 cells, PCR products of full-length ZNF219L or deletion mutant forms of ZNF219L were ligated into pGEM-T-easy vector (Promega) through TA cloning, and then further sub-cloned into pcDNA3-HA (Invitrogen, Carlsbad, CA, USA) via HindIII and KpnI restriction enzyme sites. The expression vectors were transfected into NIH3T3 using the transfection protocol described in the following section.
Primers used for PCR amplification of cDNA encoding full-length and deletion mutations of zebrafish znf219L.
| Gene | Sequences of forward (F) and reverse (R) primers |
|---|---|
| znf219L | HindIII-F, 5'- AAA AGC TTA TGG ATT CCC CAC CAG AAT GTA TGC TG-3' KpnI-R, 5'- GGG GTA CCC TAT TCC CCC AAG ACA CCT TCC TCC TC-3' |
| znf219L-△1 | HindIII-F, 5'- AAA AGC TTCCA GAA ACT CCC GTC CCA CAA ATG TCT-3' KpnI-R, 5'- GGG GTA CCC TAT TCC CCC AAG ACA CCT TCC TCC TC-3' |
| znf219L-△2 | HindIII-F, 5'- AAA AGC TTCAT CAG CAT CCT TCA TCA C-3' KpnI-R, 5'- GGG GTA CCC TAT TCC CCC AAG ACA CCT TCC TCC TC-3' |
| znf219L-△3 | HindIII-F, 5'- AAA AGC TTA TGG ATT CCC CAC CAG AAT GTA TGC TG-3' KpnI-R, 5'- GGG GTA CCT GGA CAT GCC CAC TCT GGC AC -3' |
| znf219L-△4 | HindIII-F, 5'- AAA AGC TTC GTG AGC AAA GGA ATG CTA TGG CA-3' KpnI-R, 5'- GGG GTA CCC TAT TCC CCC AAG ACA CCT TCC TCC TC-3' |
| znf219L-△5 | HindIII-F, 5'- AAA AGC TTA TGG ATT CCC CAC CAG AAT GTA TGC TG-3' KpnI-R, 5'- GGG GTA CCT GCC ATA GCA TTC CTT TGC TCA CG-3' |
| znf219L-△6 | HindIII-F, 5'- AAA AGC TTA TGG ATT CCC CAC CAG AAT GTA TGC TG-3' KpnI-R, 5'- GGG GTA CCG TTA CCC ATT TCT GCA ACC TG-3' |
Luciferase reporter gene assay
Luciferase reporter gene assay was performed using pGL3 Luciferase Reporter Vectors (Promega), which contained a modified coding region for firefly (Photinus pyralis) luciferase driven by the zebrafish col2a1a -386 proximal promoter and -386 mut (all three zZNF219L binding sites were mutated from GGGGG to GAAAG by PCR mutagenesis). The following primers were used to amplify the zebrafish col2a1a -386 proximal and -386 mut promoter: forward primer: 5'- AAG CCC ACC ACT TGC CAG GAA A-3'; reverse primer: 5'- TTG GAT GTG GAT GTG TTT GGG-3'. The luciferase reporter constructs were transfected into carp fin epithelioid cells. One microgram of promoter DNA and 0.5 mg of pSV-β-galactosidase were co-transfected using the PolyJet In Vitro DNA Transfection Reagent (SignaGen Laboratories, Ijamsville, MD, USA) into carp fin epithelioid cells. Transfections were performed using 12-well plates, and approximately 1x105 cells were seeded one day before transfection. Cells were lysed two days after transfection (by which time cells had grown to 90% confluency), and luciferase activity was determined using a luminometer (Promega), according to the manufacturer's protocol. Transfection efficiency was normalized by determining the activity of β-galactosidase activity in the cell lysates. All presented data are the results of three independent experiments, and were statistically analyzed by one-way ANOVA. Data represent mean ± s.d.
Total RNA isolation and reverse-transcription polymerase chain reaction (RT-PCR) analysis of zebrafish znf219L mRNA
Total RNA (from different developmental stages and tissues of adult zebrafish) was isolated using the RNAzol reagent (Tel-Test, Friendswood, TX, USA), according to the manufacturer's instructions. After treatment with RQ1 RNase-Free DNase I (Promega), 50~100 μg of total RNA were subjected to first-strand cDNA synthesis. The following primers were used to amplify zebrafish znf219L: znf219L-RT-F, 5'-GCA TGG AGC GCC TTC AGG CAG TGG CAC-3', and znf219L-RT-R, 5'-GCA GAT GCA GAA GAG CCC TCC TTC TTC-3'. Zebrafish β-actin was amplified as an internal control, using the following primers: zACT-F, 5'-GTG CTA GAC TCT GGT GAT GGT GTG-3', and zACT-R, 5'-GGT GAT GAC CTG ACC GTC AGG AAG-3'.
Morpholino oligonucleotide (MO) injection
Antisense MOs were obtained from Gene Tools (Philomath, OR, USA). The sequences of the znf219L and control morpholinos were as follows: zebrafish znf219L ATG morpholino: 5'-GTCTATGCCATGCTTCACTTCCTTG-3', znf219L splicing morpholino: 5'-GCCTCTCGCCTAAAACAAGGAATAA-3', and control morpholino: 5'-CCTCTTACCTCAGTTACAATTTATA-3'. The morpholinos were diluted to form a 24 μg/μl injection stock, which was stored at -20 °C prior to use. The MO injection stock was injected into wild-type (WT) zebrafish embryos at the 1~2-cell stage using a microinjection system consisting of an SZX9 stereomicroscope (Olympus, Tokyo, Japan) and an IM300 Microinjector (Narishige, Tokyo, Japan). The sequence of the p53 MO was as follows: 5'-AAAATGTCTGTACTATCTCCATCCG-3') [43]. The following primers were used for RT-PCR amplification of zebrafish znf219L in control (C) and znf219L splicing morpholino (MOs)-injected embryos: znf219L-F1, 5'-CCG GGA GCT TCC ATT CTG TCA TAT C -3', znf219L-F2, 5'- GCA CAC CTC AGG CGG TAT TTA ATT GG-3', and znf219L-R1, 5'-CTG GGT GCC AGC ATA ATC ACA GTG G -3'. Zebrafish β-actin was amplified as an internal control, using the following primers: zACT-F, 5'-GTG CTA GAC TCT GGT GAT GGT GTG-3', and zACT-R, 5'-GGT GAT GAC CTG ACC GTC AGG AAG-3'.
Whole-mount in situ hybridization
Digoxigenin-labeled RNA probes (Roche, Penzberg, Germany) were generated by in vitro transcription of linearized pGEM-T-easy plasmids (Promega) carrying the 5'-UTR of the following zebrafish genes (primers are indicated in parentheses): znf219L (znf219L-F, 5'- GGA AGT GAA GCA TGG CAT AGA CAC ATC-3', and znf219L-R, 5'- GAC CAG AAG AGA TCG CCT TCT CAA GAG - 3'), and col2a1a (col2a1a-F, 5'-CCT AAA ATC CCA CGC AAG AA -3', and col2a1a-R, 5'- CTT GCA GCC ATC CTC AAG TA-3'). Riboprobes for the detection of col2a1b [27], col8a1a [44], col11a1a [45], and col27a1a [46] were used as previously described. Whole-mount in situ hybridization was performed following a previously described protocol [47].
Microinjection of zebrafish embryos
In order to obtain stable transgenic fish, one-cell stage embryos were co-injected with 10 ng/μl col2a1a-2.1kb-gfp plasmid, 5 ng/μl capped Tol2 transposase mRNA, and 0.1% phenol red, as previously described [48]. The pCS-TP plasmid [49] encoding the transposase was first linearized with the restriction enzyme NotI, and then used as a template for in vitro transcription with mMessage mMachine (Ambion, Foster city, CA), according to the manufacturer's protocol.
To generate germline transgenic zebrafish, injected embryos that produced GFP signals were raised to adulthood (F0), and were crossed with wild-type fish at 3-months of age. F1 embryos were examined under a fluorescence microscope for identification of germline-transmitted F0 founders. Positive F1 embryos were raised to adulthood, and were then screened in the same way to estimate copy numbers of the transgene, and to establish stable transgenic lines.
Cell cultures
Carp fin epithelioid cells, CF [50], were maintained in Leibovitz's L-15 media supplemented with 10% fetal bovine serum at 27°C. Transfection of nucleic acid into CF cells was carried out as previously described [51]. NIH/3T3 cells (ATCC CRL-1658; Manassas, VA, USA) were cultured in high-glucose Dulbecco's modified Eagle's media (DMEM) supplemented with 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) in a humidified atmosphere of 5% CO2 at 37 ºC.
Oligonucleotide precipitation assay
NIH 3T3 cells were lysed in lysis buffer [20 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM EGTA, 1.5 mM MgCl2, 10% glycerol, 1% Triton X-100, 10 mg/ml leupeptin, 1 mM PMSF, 0.2 mM sodium orthovanadate] for use in the in vitro binding assay. The lysates were pre-incubated with streptavidin-agarose beads for 3 hours, and then incubated for 16 hours with 1 mg of poly (dI-dC) and 1 mg of a biotinylated double-stranded oligonucleotide probe (which contained one of the three zZNF219L binding element repeats present in the col2a1a gene promoter binding site 1; BS-1 sense primer: 5'-GGC AAA AAG GGG AGG GGG TGG ATG GGT TTG-3'; anti-sense primer: 5'- CAA ACC CAT CCA CCC CCT CCC CTT TTT GCC -3'; BS-2 sense primer: 5'- CAA TGG GGT GGG CTG GGG GGC TCG TAT TTC-3'; anti-sense primer: 5'- GAA ATA CGA GCC CCC CAG CCC ACC CCA TTG-3'; BS-3 sense primer: 5'- TTC AGC GCT CAT GGG GGT CGG GGT TGG ACT-3'; anti-sense primer: 5'- AGT CCA ACC CCG ACC CCC ATG AGC GCT GAA-3'). Lysates were subsequently incubated with streptavidin agarose (Streptavidin Agarose, Invitrogen) for 1 hour. The bound-proteins were then washed with lysis buffer, separated on an SDS- polyacrylamide gel, and subjected to Western blot using antibodies against the proteins of interest.
Results
Cloning of znf219L cDNA from zebrafish
To clone the zebrafish cDNA sequences related to the human znf219 gene, we used the program tBLAST to search GenBank for expression sequence tags (ESTs) resembling a region of human ZNF219 that encodes the first three paired zinc fingers (amino acid residues 59 to 327). In this way, we identified one zebrafish EST clone (CR926852). We used 5'- and 3'-RACE to obtain the 5'- and 3'-untranslated regions (UTRs) of this clone, allowing us to assemble a 4101-bp cDNA with an open reading frame of 3288-bp, predicted to encode a protein of 1095 amino acid residues. The 5'- and 3'-UTRs were 237-bp and 576-bp in length, respectively. The complete sequence was deposited in GenBank with the accession number JX141438.
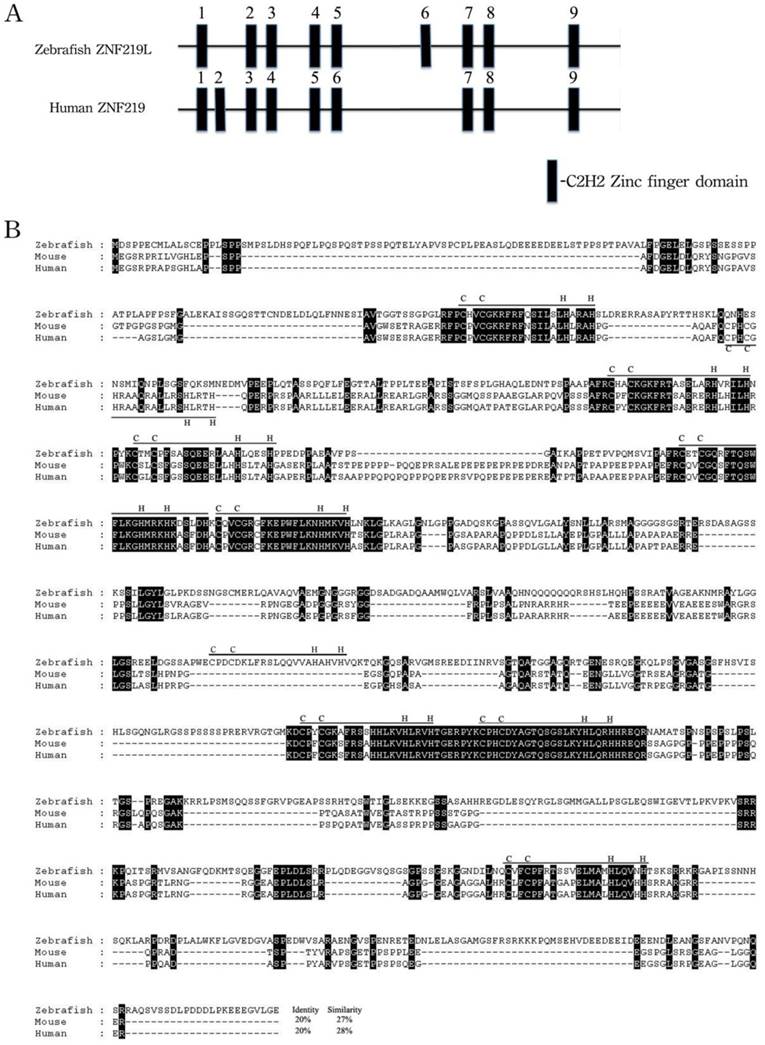
The overall predicted amino acid sequence of zebrafish ZNF219 has only 20% identity with its orthologues in human and mouse (Fig. 1B). However, the three paired zinc finger domains (aa 278-325, 358-408, and 701-749) of zebrafish ZNF219 have 58.3%, 84.3% and 95.9% identity with their equivalents in human ZNF219, respectively. In addition, zebrafish ZNF219 has two single zinc finger domains (aa 155-175 and 931-952) that have 80% identity with the corresponding domain of human ZNF219. Both human ZNF219 and zebrafish ZNF219 possess a non-homologous single zinc finger domain (the second and sixth domain, respectively (Fig. 1A). Based on its resemblance to ZNF219, we henceforth refer to the newly-identified protein as zebrafish ZNF219-like (ZNF219L).
Genomic organization of the zebrafish znf219L gene
Analysis of human, mouse, fugu, and zebrafish genomes show that znf219L are similarly present in these species. Examination of the genomic region around znf219 and a high degree of conserved genomic synteny was observed within four species, gene ARHGEF40 is flanking znf219L of four species while btr12 locates downstream of Fugu and zebrafish. (Supplementary Fig. 1). In mammals, ARHGEF40 is downstream of both human and mouse znf219L while it is upstream of fugu and zebrafish suggesting an inversion has occurred during chromosomal rearrangement after separation of the zebrafish lineage from mammals.
We then used the 4101-bp znf219L cDNA sequence to perform an online BLAST search of the GenBank database. The znf219L cDNA was found to show identity with 5 non-contiguous regions of two zebrafish BAC clones, DKEY-46I9 (GenBank accession no. BX664721, 130851-bp) and CH211-142D6 (GenBank accession no. AL929105, 178797-bp). BLAST two-sequence comparisons of each BAC clone with the zebrafish znf219L cDNA indicated that the znf219L sequence comprises 5 putative exons and 4 introns spanning at least 21.6 kb (Fig. 2).
Alignment of amino acid sequences of zebrafish ZNF219L and mouse and human ZNF219 proteins. (A) Zebrafish ZNF219L protein contains three paired zinc finger domains (aa 278-325, 358-408, and 701-749) and three single zinc finger domains. (B) The deduced amino acid sequence of zebrafish ZNF219L was aligned with those of mouse and human ZNF219 proteins. All three proteins contain eight highly-conserved C2H2-type zinc finger domains (indicated by bars labeled with C and H). Both mammalian ZNF219 and zebrafish ZNF219 possess a non-homologous single zinc finger domain (the second and sixth domain, respectively). The accession numbers for mouse and human znf219 are NM_027248.2 and NM_001102454, respectively.
Genomic organization and expression profiles of zebrafish znf219L. Genomic organization of zebrafish znf219L, and the mouse and human znf219 genes were shown in panel (A). Coding regions are shown as filled boxes numbered from 3 to 5. The 5'- and 3'-untranslated regions are shown as open boxes, while solid lines indicate introns. RT-PCR was performed with gene-specific primers. β-actin was used as an internal control to normalize the amount of cDNA prepared from different adult zebrafish tissues (B) and from zebrafish embryos at different developmental stages (C). The developmental expression profile of zebrafish znf219L mRNA was examined in embryos from 12 hpf to 144 hpf. Blank PCR was performed using gene-specific primers and β-actin primers without the addition of cDNA template. Whole-mount in situ hybridization with antisense znf219L was performed at the indicated times post-fertilization (D). The images were taken from 22 to 48 hpf in the lateral view (panels a, b) and 48 hpf in dorsal view in panel c. The boxed region is enlarged to show the signal in the notochord (nc) from 22 to 48 hpf of both lateral and dorsal view (panels a', b', and c'). Dorsal view of znf219L mRNA signals in midbrain hinfbrain boundry (mhb), and hindbrain (hb) were detected from 48 to 96 hpf (panel d, e, and f). mhb, midbrain hindbrain boundry; hb, hindbrain; ov, otic vesicle; pf, pectoral fin, nc, notochord. Scale bars=100 μm.
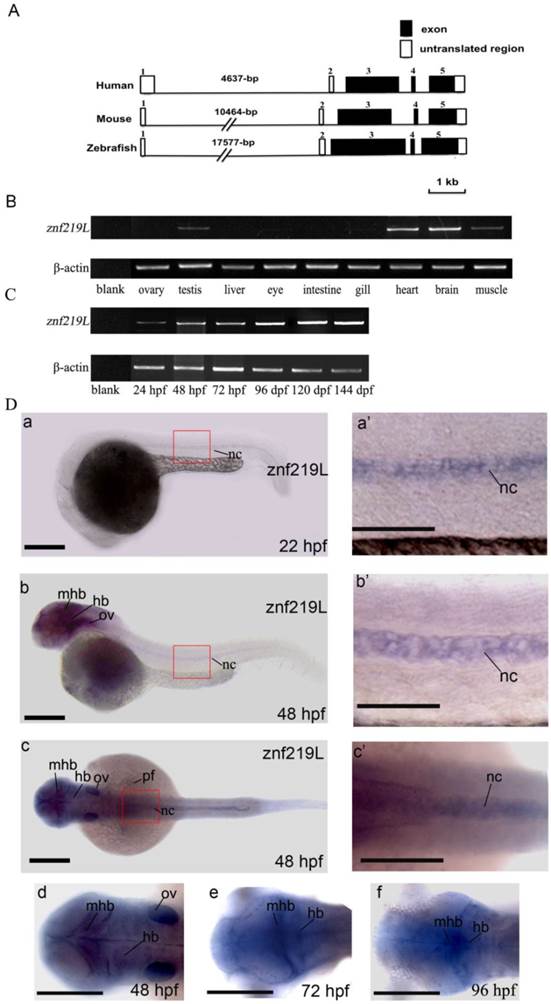
Using these putative exons as a model, a sequence alignment was produced in which each intron was defined in accordance with the GT/AG intron donor/acceptor site rule [52]. Exons 1 and 2 contain 152-bp and 81-bp of the 5'-UTR, respectively. Exon 3 contains 4-bp of the 5'-UTR and the putative translation initiation site of the first coding sequence of the znf-219L cDNA. Exon 5 contains the last 1120-bp of the coding sequence and 576-bp of the 3'-UTR. The size of the introns varied considerably, ranging from 76-bp (intron 2) to 17,577-bp (intron 1). Comparison of the exon-intron organization of zebrafish znf219L with that of the mouse and human znf219 genes indicated that their genomic structures are similar in that all genes have 5 exons and 4 introns. The mouse and human znf219 genes span approximately 14,657 and 8,595-bp, respectively. In addition, the first intron of the mouse (10,464-bp) and human znf219 genes (4,637-bp) is smaller than that of the zebrafish znf219L gene (17,577-bp) (Fig. 2A).
Expression of zebrafish znf219L in various adult tissues and at different developmental stages
Expression of zebrafish znf219L in adult tissues and embryos of different developmental stages was examined by RT-PCR analysis. Primers were used to amplify a DNA fragment spanning exons 3 to 5, to avoid interference of the PCRs by genomic DNA. The amplified product was about 1,000-bp in length. Abundant znf219L expression was detected in heart and brain, with relatively low levels in muscle and testis of adult zebrafish. Expression of znf219L was detected from 24 hours post-fertilization (hpf) onwards at early developing stages (Fig. 2B, and 2C).
Expression patterns of znf219L mRNA in zebrafish embryos
Previous studies reported that mouse znf219 mRNA was specifically expressed in the developing limb buds, suggesting the encoded protein may play an important role in regulating chondrocyte differentiation. [37]. In order to investigate whether zebrafish ZNF219L is functionally related to mammalian ZNF219, we first examined its expression pattern by performing whole mount in situ hybridization using zebrafish embryos at different developmental stages. As shown in Fig. 2D, znf219L transcripts were first detected weakly at 22 hpf in notochord only (panels a and a'). The expression level of znf219L increased in notochord, midbrain-hindbrain boundary and hindbrain from 22 to 72 hpf (panels b, b', c and c') and was nearly undetectable after 96 hpf (data not shown). The expression in pectoral fin and otic vesicle (ov) were only detected at 48 hpf (panel b).
Knockdown of znf219L caused partial notochord abnormalities
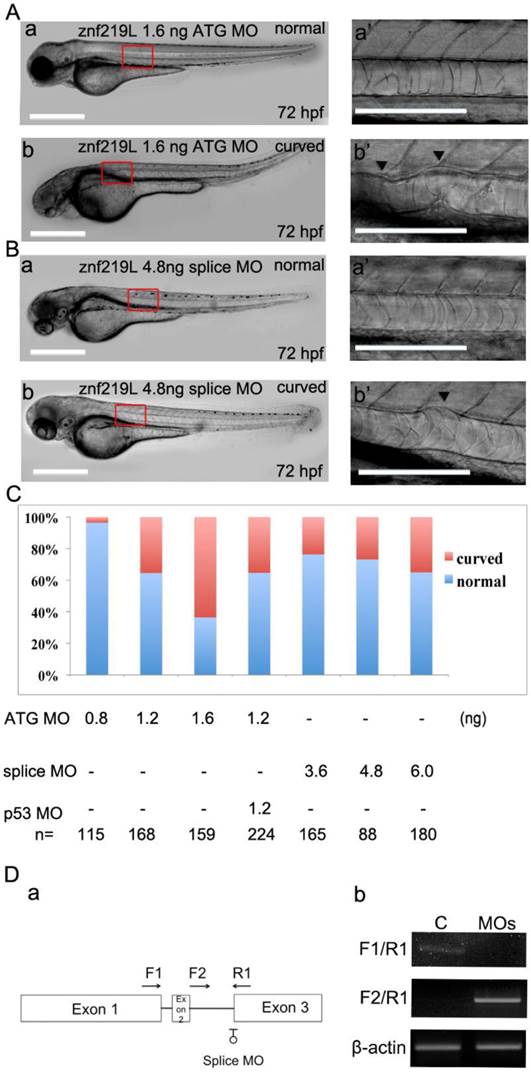
To characterize the physiological function of ZNF219L, we used ATG and splicing morpholinos to knockdown endogenous znf219L expression in zebrafish embryos. Knockdown of znf219L by ATG-MO caused a curved notochord phenotype, which showed slightly curved body axis (Fig. 3A, panels a, and a'). Similar abnormalities in the notochord were observed in embryos injected with splicing morpholinos as well (Fig. 3B, panels a and a'). Percentage of the curved notochord phenotype was correlated to the various doses of morpholino injected into the zebrafish embryos (Fig. 3C). It has been reported that concurrent knockdown of p53 can ameliorate the cell death induced by MO off-targeting [53]. Therefore, in order to rule out the possibilities that the phenotypes of the morphants were caused by the off-targeting effects of the morpholino used, coinjection of p53-MO and ATG-MO was performed. The percentage of abnormalities in the znf219L morphants coinjected with p53-MO was approximately equal to that with the various doses of morpholino used. Furthermore, co-injection of znf219L morphants with p53-MO did not result in a significant change in morphology, confirming the specificity of the znf219L morpholino (Fig. 3C).
In addition to ATG-MO, we also synthesized splicing-MO and the positions for splicing-MO targeting sites were shown (Fig. 3D, panel a). According to the results of RT-PCR analyses of the control (C) and splicing-MO (MOs)-injected embryos (Fig. 3D, panel b), the splicing-MO was effective to block mRNA splicing. Furthermore, embryos injected with splicing-MO exhibited similar curved notochord phenotype (Fig. 3B, panels a and a') as that injected with ATG-MO. However, the ATG-MO consistently showed higher efficiency (>60% abnormal rate at 1.6 ng) than the splicing-MO (around 35% abnormal rate at 6 ng) to induce curved notochord phenotypes under our experimental conditions, we thus adopted the ATG-MO throughout our study.
Zebrafish col2a1a is down-regulated in the notochord of znf219L morphants
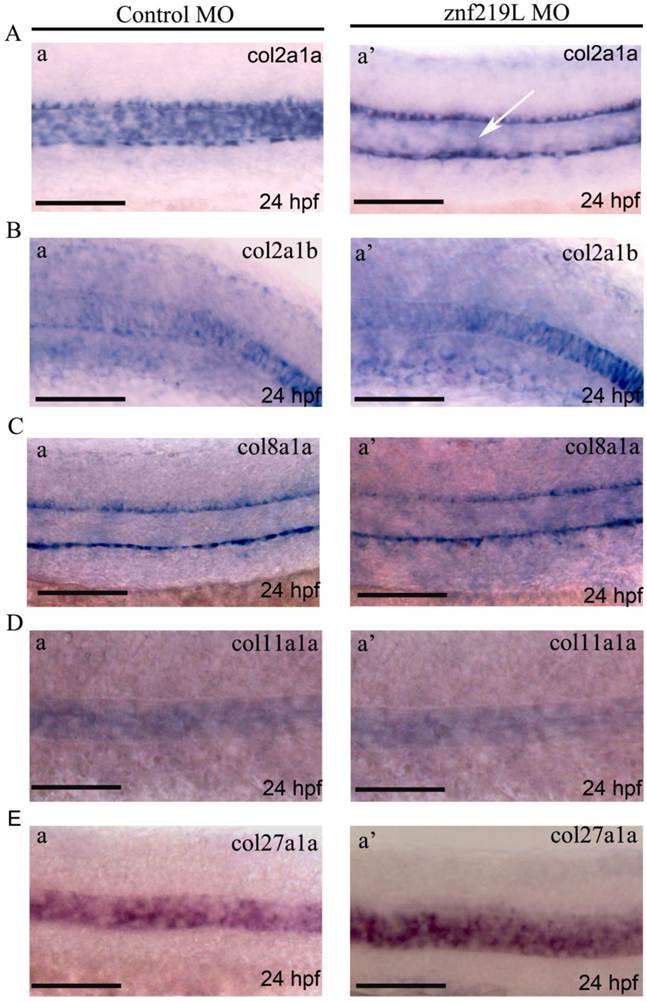
Up to date, it has been reported that at least five collagen genes; collagen type II alpha-1a (col2a1a), collagen type II alpha-1b (col2a1b), collagen type VIII alpha-1a (col8a1a), collagen type XI alpha-1a (col11a1a), and collagen type XXVII alpha-1a (col27a1a) are expressed in notochord and involved in its formation [27, 44-46, 54]. Because znf219L transcripts were first detected at 22 hpf in notochord only as shown in Fig. 2D, panels a and a', we thus wonder whether ZNF219L may regulate the expression of those five genes in notochord. Therefore, the znf219L morphants at 24 hpf were analyzed by whole-mount in situ hybridization using probes for different collagen genes. Interestingly, knockdown of znf219L by morpholino only led to the reduced expression of col2a1a specifically in zebrafish notochord (Fig. 4A, panels a and a'), while the expression of other collagen genes was not affected in notochord (Figs. 4B to 4E, panels a and a'). These data suggest that znf219L functions as a specific regulator that targets the col2a1a gene and lacking of it does not have significant effect on the formation of notochord. And, it is possible that other factors involved in regulation of these collagen genes expression in notochord other than ZNF219L.
Knockdown of znf219L caused notochord abnormalities. Knockdown of znf219L in wild-type zebrafish embryos using an ATG-MO (A) or a splicing-MO (B) resulted in abnormal notochord phenotype with partial curving of notochord (panels b, and b'). Black arrowheads in panel b' indicate partial curved notochord abnormalities. Scale bars=100 μm. (C) The percentage of abnormal notochord phenotypes in zebrafish embryos was correlated with MO dose, for both types of znf219L MO tested. Co-injection of ATG-MO with p53-MO did not decrease the ratio of abnormal notochord phenotypes. (D) Schematic depicting the binding positions of the splicing-MO and primers used for detecting znf219L-derived RNA species by RT-PCR (panel a). Representative images from RT-PCR analyses of control (C) and splicing MO (MOs)-injected embryos (panel b). RT-PCR was performed with splicing-MO F1-R1 and F2-R1 primers. β-actin was used as an internal control.
Knockdown of znf219L only caused decreased expression of col2a1a in the notochord. Expression of col2a1a (A), col2a1b (B), col8a1a (C), col11a1a (D), and col27a1a (E) was examined in znf219L ATG-MO by in situ hybridization using probes for each collagen gene. Only col2a1a expression in notochord was observed to decrease significantly, other four genes were not. White arrows indicate decreases in the expression of endogenous col2a1a in notochord. (Fig. 4A, panel a'). Scale bars=100 μm.
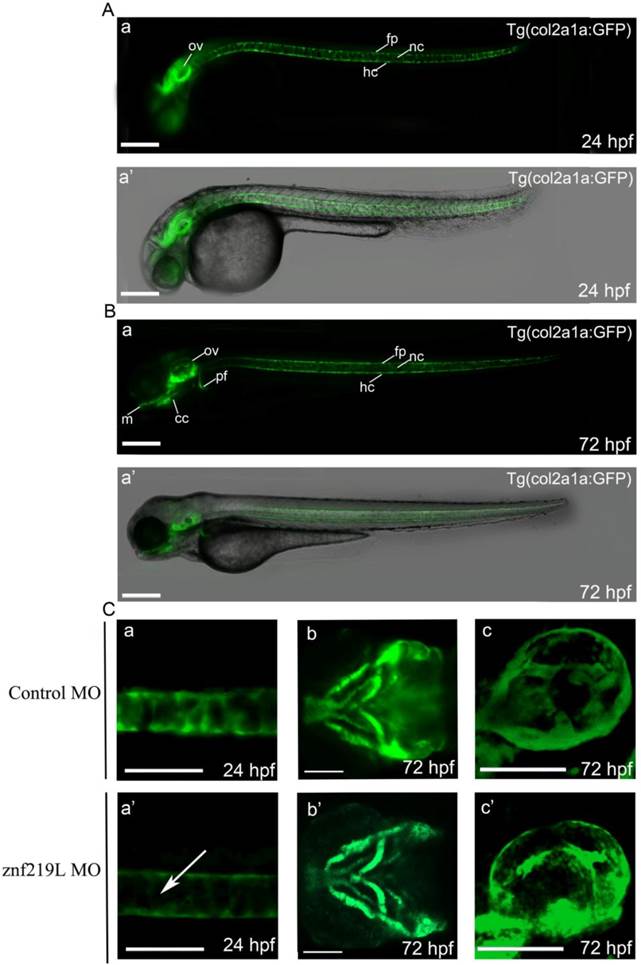
According to the previous report [27], we similarly established a transgenic Tg(col2a1a: GFP) zebrafish with GFP expression in the craniofacial cartilage, ear, notochord, floor plate, hypochord and pectoral fins (Fig. 5A, B, panels a and a'). The expression pattern was similar to that of endogenous col2a1a [27]. We then injected ATG-MO into embryos of this transgenic line at 2-4 cells stage and observed that knockdown of znf219L only decreased the GFP signal in the notochord, not in other tissues, of Tg(col2a1a: GFP) fish at 24 hpf (Fig. 5C, panels a, a' b, b' c, and c'). These data are consistent with the result mentioned above that knockdown of znf219L by morpholino led to the reduced expression of col2a1a specifically in notochord of wild-type zebrafish (Fig. 4A, panels a and a').
ZNF219L regulates col2a1a promoter activity via binding to GGGGG motifs
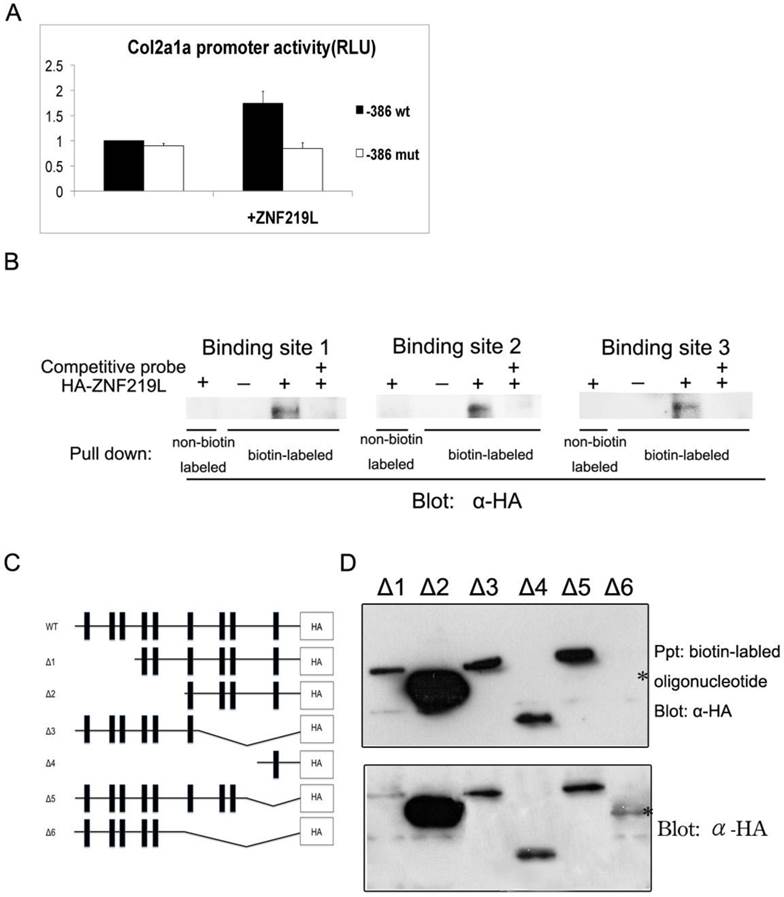
It has been previously reported that mammalian ZNF219 can localize to the nucleus, where it functions as a transcriptional regulator. In addition, ZNF219 has been reported to bind the GGGGG motif in the col2a1 promoter region [37]. According to those results mentioned above, we further examined whether ZNF219L activates the zebrafish col2a1a promoter in cultured cells. Initially, we had performed serial deletions ranged from 2.0-kb to -386 bp region of col2a1a promoter in luciferase assay. It showed no obvious difference in the activity between 2.0-kb and -386 bp region of col2a1a gene promoter. Therefore, we believe that -386 region alone could be responsible for col2a1a expression. We constructed an expression vector containing a col2a1a promoter proximal region, 386 base pairs upstream of the ATG codon (designated as the -386 region), which contains all three ZNF219L binding sites. A luciferase assay construct containing the -386 proximal region of the col2a1a promoter (henceforth referred to as the col2a1a proximal promoter luciferase construct) was used to drive luciferase expression (Fig. 6A). The col2a1a proximal promoter luciferase construct was co-transfected with a ZNF219L overexpression construct into grass carp CF cells, and luciferase activity was measured to compare promoter activity in cells with or without ZNF219L overexpression. Luciferase activity of the col2a1a proximal promoter was elevated by overexpression of ZNF219L (Fig. 6A). However, in order to confirm that binding of ZNF219L to the col2a1a proximal promoter was required for the observed increase in luciferase activity, we mutated the three ZNF219L binding sites. It was previously reported that mutation of the ZNF219 recognition sequence GGGGG to GGCGG reduced its binding affinity in mammals [37]. Therefore, we mutated all three ZNF219L binding sites from GGGGG to GAAAG (thereby mutating two extra base pairs as compared to the earlier study) in the col2a1a proximal promoter. As shown in panel b, mutation of these motifs reduced promoter activity in cells with and without ZNF219L overexpression. These results suggest that zebrafish ZNF219L can specifically up-regulate the transcriptional activity of the col2a1a promoter via binding to the GGGGG motifs. To confirm this hypothesis, we performed a pull-down assay with biotin-labeled oligonucleotide probes, each containing one of the three GGGGG motifs. We found that ZNF219L could directly bind to all three GGGGG motifs in an independent manner. The association between ZNF219L and each motif was specific, because the addition of a competitive non-biotin-labeled probe effectively inhibited binding (Fig. 6B).
We next used an in vitro binding assay to identify which zinc finger domain is required for association of ZNF219L with the col2a1a gene promoter. A series of deletion mutants of ZNF219L were generated for use in the assay, as shown in Fig. 6C. A deletion mutant without the sixth, seventh, eighth, and ninth zinc finger domains (upper panel) was not pulled down by biotin-labeled oligonucleotides containing the three GGGGG motifs, indicating that it could not bind to the col2a1a gene promoter (△6, Fig. 6D). However, the deletion mutant without the seventh, eighth, and ninth zinc finger domains (△3) and another deletion mutant with only the ninth zinc finger domain (△4) interacted with biotin-labeled oligonucleotides. Thus, both sixth and ninth zinc finger domains are involved in binding to the GGGGG motifs Lysates of cells expressing different ZNF219L mutants were analyzed by immunoblotting with anti-HA antibody as control in the lower panel.
Altogether, our data indicate that ZNF219L is required for the regulation of col2a1a promoter activity via binding to GGGGG motifs in cultured cells.
Discussion
In this study, we described the cloning of zebrafish ZNF219-related protein based on mammalian ZNF219; this protein consists of 1095 amino acids and 9 classical C2H2 zinc finger domains. The overall amino acid sequence of this protein has low identity (20%) with that of human ZNF219, but higher identity was observed in each C2H2 zinc finger domain (Fig. 1). We further demonstrated that (1) its expression in the midbrain hindbrain boundary, hindbrain and notochord (Fig. 2); (2) its morpholino knockdown caused partial notochord phenotype and led to down-regulation of col2a1a mRNA expression in notochord (Figs. 3-5); (3) its ability to bind to the GGGGG motifs in the col2a1a gene promoter (Fig. 6) and (4) the sixth and/or ninth zinc finger domains of ZNF219L are required for binding to the col2a1a gene promoter (Fig. 6D). Taken together, these data indicate that zebrafish ZNF219L functions in regulating col2a1a expression in the notochord.
Knockdown of znf219L resulted in decreasing GFP expression in notochord of a transgenic zebrafish line, Tg(col2a1a:GFP). According to the previous report [27], we similarly established a transgenic Tg(col2a1a: GFP) zebrafish using the upstream 2.1-kb promoter region. The GFP signals in this zebrafish line were observed in ear, notochord, floor plate, and hypochord at 24 hpf (A), while GFP signals in other regions, such as craniofacial cartilage, and pectoral fins were observed at 72 hpf (B). Merged bright-field and fluorescence images are shown in panel a', while fluorescence images are shown in panel a. Injection of this transgenic zebrafish line with znf219L ATG-MO at 2-4 cells stage (C) resulted in a decrease in GFP expression specifically in the notochord (panels a, and a', lateral view), but not in craniofacial cartilage (panels b and b', ventral view) and ear (panels c, and c', lateral view). White arrows indicate the decrease in GFP expression in notochord. m, Meckel's cartilage; cc, craniofacial cartilage; ov, optic vesicle; nc, notochord; fp, floor plate; hc, hypochord. Scale bars=100 μm.
ZNF219L regulates the col2a1a proximal promoter in cultured cells. (A) The effect of ZNF219L on col2a1a promoter activity was studied in grass carp CF cells. The col2a1 gene -386 proximal promoter luciferase constructs, containing either wild-type (GGGGG) or mutant (GAAAG) potential ZNF219L binding sites, were co-transfected with a ZNF219L expression vector into grass carp CF cells. The luciferase activity of co-transfected cell lysates was measured at 48 hours after transfection. Data represent mean ± s.d. (n=3) (panel b). (B) ZNF219L binds to the three GGGGG motifs in the col2a1 proximal promoter. NIH3T3 cell lysates were transfected with empty or HA-ZNF219L vector, and then incubated with biotinylated oligonucleotide containing one of the three GGGGG motifs of the col2a1 gene promoter. Association of the ZNF219L protein with each oligonucleotide was determined by immunoblotting with anti-HA antibody after precipitation with streptavidin agarose beads. (C) Schematic diagram of HA-tagged deletion mutants of ZNF219L. The black bars represent zinc finger domains. (D) Binding of ZNF219L deletion mutants to the zebrafish col2a1a gene promoter in vitro (upper panel). Lysates of cells expressing each ZNF219L mutant were precipitated with biotinylated oligonucleotide containing the col2a1a gene promoter GGGGG motif. Bound protein was subsequently detected by immunoblotting with anti-HA antibody. The star (*) indicates the failure of the ZNF219L Δ6 mutant to interact with the probe. Lysates of cells expressing each ZNF219L mutant were detected by immunoblotting with anti-HA antibody as control in lower panel.
Zinc finger proteins contain one to forty zinc finger domains [38]. Most of these proteins can be classified into three groups, based on the number and pattern of the zinc fingers: triple-C2H2, multiple-adjacent-C2H2, and separated-paired-C2H2 finger proteins [55]. Human ZNF219 is a separated-paired-C2H2 finger protein, which contains nine zinc finger domains. Proteins with nine zinc finger domains may require two to four of these domains for DNA binding, while the others act as protein-protein interaction domains. For example, the friend of GATA (FOG) family-FOG-1 protein uses the first, fifth, sixth, and ninth zinc finger domains for protein-protein interaction, while the FOG-2 protein uses the first, fifth, sixth, and eighth zinc finger domains to interact with GATA-1 [56]. The first three zinc finger domains of TFIIIA (also a nine zinc finger domain protein) are used for DNA binding, and the fourth to seventh zinc finger domains are used for RNA binding [57, 58]. Human ZNF219 was reported to recognize and bind to specific DNA sequences through two regions: the first three zinc finger domains, and a paired zinc finger domain (fifth and sixth fingers) [35]. The latter was also demonstrated to interact with Sox9 [37]. Thus, the fifth and sixth zinc finger domains of human ZNF219 are required for DNA binding. In this study, we demonstrated that zebrafish ZNF219L is a protein with nine zinc finger domains, and its sixth and ninth zinc finger domains are required for binding to the promoter of col2a1a (Fig. 6D). However, the sixth zinc finger domain of zebrafish ZNF219L is not present in mammalian ZNF219 (Fig. 1), and the ninth zinc finger domain of mammalian ZNF219 is not involved in DNA binding [35, 37]. Therefore, zebrafish ZNF219L may bind DNA through specific and unique zinc finger domains.
Human ZNF219 was determined to be either a transcriptional repressor or enhancer, depending on the target gene and the cell type. Human ZNF219 represses expression of high mobility group nucleosomal binding protein 1 (HMGN1) in 293 cells (a human embryonic kidney cell line) [35], but enhances expression of aggrecan, Col2a1, and Col11a2 in C3H10T1/2 mesenchymal cells through associating with Sox9 [37]. In this study, we demonstrated that knockdown of zebrafish znf219L resulted in curved notochord phenotype with only down-regulation of col2a1a without affecting other four collagen genes-col2a1b, col8a1a, col11a1a, and col27a1a expression the notochord (Figs. 3-5). Also, zebrafish ZNF219L enhances transcription of col2a1a in carp CF-cells through recognized the same GGGGG motifs as in mammals (Fig. 6). However, zebrafish sox9a regulates expression of col2a1a in craniofacial cartilage, but sox9a knockdown did not affect col2a1a expression in the notochord [27]. Interestingly, the GFP signal in the Tg(col2a1a:GFP) transgenic zebrafish line was detected in many tissues, including craniofacial cartilage, ear, notochord, floor plate, hypochord and pectoral fins, in a pattern similar to that of endogenous col2a1a [27]. Knockdown of znf219L and sox9a only affected the GFP signal in the notochord and craniofacial cartilage, respectively. This suggests that other factors may be required for col2a1a expression in other tissues.
Recent studies have indicated that other zinc finger proteins are also involved in chondrogenesis. For example, zinc finger protein 521 (ZNF521) acts as a transcriptional regulator during bone formation, and is also a target gene of parathyroid hormone-related peptide, which regulates growth plate chondrocyte proliferation and differentiation [59-61]. Zinc finger protein 145 (ZNF145) functions in chondrogenesis by enhancing the expression of sox9 in human mesenchymal stem cells (MSCs) [62]. This suggests that zebrafish ZNF219L may regulate col2a1a expression in the notochord directly, as a single factor. It is likely that ZNF219L and Sox9a act separately, to regulate col2a1a expression in the notochord and craniofacial cartilage, respectively.
In summary, our results demonstrate that the reduction of zebrafish ZNF219L causes down-regulation of col2a1a gene expression in notochord and partial notochord abnormalities. Our data indicates that zebrafish ZNF219L acts as an important transcriptional factor in regulating the col2a1a gene expression in notochord.
Supplementary Material
Supplementary Figure 1Acknowledgements
This study was supported by a grant (NSC97-2313-B-001-002-MY3) from the National Science Council, Taipei, Taiwan. The AB wild type zebrafish were kindly provided by the Taiwan Zebrafish Core Facility at Academia Sinica (ZCAS), which is supported by the NSC (National Science Council, NSC 100-2321-B-001-030), Taiwan.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Shimeld SM, Holland PW. Vertebrate innovations. Proc Natl Acad Sci U S A. 2000;97:4449-4452
2. Keller R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 2002;298:1950-1954
3. Monsoro-Burq AH, Bontoux M, Vincent C, Le Douarin NM. The developmental relationships of the neural tube and the notochord: short and long term effects of the notochord on the dorsal spinal cord. Mech Dev. 1995;53:157-170
4. Melby AE, Warga RM, Kimmel CB. Specification of cell fates at the dorsal margin of the zebrafish gastrula. Development. 1996;122:2225-2237
5. Stemple DL. Structure and function of the notochord: an essential organ for chordate development. Development. 2005;132:2503-2512
6. Haga Y, Dominique VJ 3rd, Du SJ. Analyzing notochord segmentation and intervertebral disc formation using the twhh:gfp transgenic zebrafish model. Transgenic Res. 2009;18:669-683
7. Trout JJ, Buckwalter JA, Moore KC. Ultrastructure of the human intervertebral disc: II. Cells of the nucleus pulposus. Anat Rec. 1982;204:307-314
8. Erwin WM, Islam D, Inman RD, Fehlings MG, Tsui FW. Notochordal cells protect nucleus pulposus cells from degradation and apoptosis: implications for the mechanisms of intervertebral disc degeneration. Arthritis Res Ther. 2011;13:R215
9. McCann MR, Tamplin OJ, Rossant J, Seguin CA. Tracing notochord-derived cells using a Noto-cre mouse: implications for intervertebral disc development. Dis Model Mech. 2012;5:73-82
10. Fratzl P. Collagen: Structure and Mechanics. 2008.
11. Ricard-Blum S, Ruggiero F. The collagen superfamily: from the extracellular matrix to the cell membrane. Pathol Biol (Paris). 2005;53:430-442
12. Asanbaeva A, Tam J, Schumacher BL, Klisch SM, Masuda K, Sah RL. Articular cartilage tensile integrity: modulation by matrix depletion is maturation-dependent. Arch Biochem Biophys. 2008;474:175-182
13. Cheah KS, Stoker NG, Griffin JR, Grosveld FG, Solomon E. Identification and characterization of the human type II collagen gene (COL2A1). Proc Natl Acad Sci U S A. 1985;82:2555-2559
14. Cheah KS, Lau ET, Au PK, Tam PP. Expression of the mouse alpha 1(II) collagen gene is not restricted to cartilage during development. Development. 1991;111:945-953
15. Matsumoto T, Deguchi T, Kawasaki T, Yuba S, Sato J. Molecular cloning and expression of the col2a1a and col2a1b genes in the medaka, Oryzias latipes. Gene Expr Patterns. 2012;12:46-52
16. Miller EJ, Matukas VJ. Chick cartilage collagen: a new type of alpha 1 chain not present in bone or skin of the species. Proc Natl Acad Sci U S A. 1969;64:1264-1268
17. Warman ML, Cormier-Daire V, Hall C, Krakow D, Lachman R, LeMerrer M. et al. Nosology and classification of genetic skeletal disorders: 2010 revision. Am J Med Genet A. 2011;155A:943-968
18. Parke DW. Stickler syndrome: clinical care and molecular genetics. Am J Ophthalmol. 2002;134:746-748
19. Snead MP, Yates JR. Clinical and Molecular genetics of Stickler syndrome. J Med Genet. 1999;36:353-359
20. Stickler GB, Belau PG, Farrell FJ, Jones JD, Pugh DG, Steinberg AG. et al. Hereditary Progressive Arthro-Ophthalmopathy. Mayo Clin Proc. 1965;40:433-455
21. Hoornaert KP, Vereecke I, Dewinter C, Rosenberg T, Beemer FA, Leroy JG. et al. Stickler syndrome caused by COL2A1 mutations: genotype-phenotype correlation in a series of 100 patients. Eur J Hum Genet. 2010;18:872-880
22. Van Camp G, Snoeckx RL, Hilgert N, van den Ende J, Fukuoka H, Wagatsuma M. et al. A new autosomal recessive form of Stickler syndrome is caused by a mutation in the COL9A1 gene. Am J Hum Genet. 2006;79:449-457
23. Richards AJ, Laidlaw M, Meredith SP, Shankar P, Poulson AV, Scott JD. et al. Missense and silent mutations in COL2A1 result in Stickler syndrome but via different molecular mechanisms. Hum Mutat. 2007;28:639
24. Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A. et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711-1714
25. Huang KY, Chen GD, Cheng CH, Liao KY, Hung CC, Chang GD. et al. Phosphorylation of the zebrafish M6Ab at serine 263 contributes to filopodium formation in PC12 cells and neurite outgrowth in zebrafish embryos. PLoS One. 2011;6:e26461
26. Yan YL, Hatta K, Riggleman B, Postlethwait JH. Expression of a type II collagen gene in the zebrafish embryonic axis. Dev Dyn. 1995;203:363-376
27. Dale RM, Topczewski J. Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev Biol. 2011;357:518-531
28. Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T. et al. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 2004;50:3561-3573
29. Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T. et al. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9:15-20
30. Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW. et al. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174-178
31. Zhao Q, Eberspaecher H, Lefebvre V, De Crombrugghe B. Parallel expression of Sox9 and Col2a1 in cells undergoing chondrogenesis. Dev Dyn. 1997;209:377-386
32. Yan YL, Willoughby J, Liu D, Crump JG, Wilson C, Miller CT. et al. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069-1083
33. Delous M, Yin C, Shin D, Ninov N, Debrito Carten J, Pan L. et al. Sox9b is a key regulator of pancreaticobiliary ductal system development. PLoS Genet. 2012;8:e1002754
34. Crotwell PL, Mabee PM. Gene expression patterns underlying proximal-distal skeletal segmentation in late-stage zebrafish, Danio rerio. Dev Dyn. 2007;236:3111-3128
35. Sakai T, Hino K, Wada S, Maeda H. Identification of the DNA binding specificity of the human ZNF219 protein and its function as a transcriptional repressor. DNA Res. 2003;10:155-165
36. Sakai T, Toyoda A, Hashimoto K, Maeda H. Isolation and characterization of a novel zinc finger gene, ZNF219, and mapping to the human chromosome 14q11 region. DNA Res. 2000;7:137-141
37. Takigawa Y, Hata K, Muramatsu S, Amano K, Ono K, Wakabayashi M. et al. The transcription factor Znf219 regulates chondrocyte differentiation by assembling a transcription factory with Sox9. J Cell Sci. 2010;123:3780-3788
38. Brayer KJ, Kulshreshtha S, Segal DJ. The protein-binding potential of C2H2 zinc finger domains. Cell Biochem Biophys. 2008;51:9-19
39. Brayer KJ, Segal DJ. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys. 2008;50:111-131
40. Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39-46
41. Westerfield M. THE ZEBRAFISH BOOK, 5th Edition; A guide for the laboratory use of zebrafish (Danio rerio). 2007.
42. Docchio F, Sacchi CA. Nd:YAG laser irradiation of an eye model: experimental analysis. Lasers Surg Med. 1987;6:520-529
43. Chen J, Ruan H, Ng SM, Gao C, Soo HM, Wu W. et al. Loss of function of def selectively up-regulates Delta113p53 expression to arrest expansion growth of digestive organs in zebrafish. Genes Dev. 2005;19:2900-2911
44. Gansner JM, Gitlin JD. Essential role for the alpha 1 chain of type VIII collagen in zebrafish notochord formation. Dev Dyn. 2008;237:3715-3726
45. Fang M, Adams JS, McMahan BL, Brown RJ, Oxford JT. The expression patterns of minor fibrillar collagens during development in zebrafish. Gene Expr Patterns. 2010;10:315-322
46. Christiansen HE, Lang MR, Pace JM, Parichy DM. Critical early roles for col27a1a and col27a1b in zebrafish notochord morphogenesis, vertebral mineralization and post-embryonic axial growth. PLoS One. 2009;4:e8481
47. Chen YC, Cheng CH, Chen GD, Hung CC, Yang CH, Hwang SP. et al. Recapitulation of zebrafish sncga expression pattern and labeling the habenular complex in transgenic zebrafish using green fluorescent protein reporter gene. Dev Dyn. 2009;238:746-754
48. Kawakami K. Transposon tools and methods in zebrafish. Dev Dyn. 2005;234:244-254
49. Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133-144
50. Leu JH, Yan SJ, Lee TF, Chou CM, Chen ST, Hwang PP. et al. Complete genomic organization and promoter analysis of the round-spotted pufferfish JAK1, JAK2, JAK3, and TYK2 genes. DNA Cell Biol. 2000;19:431-446
51. Chang MS, Chang GD, Leu JH, Huang FL, Chou CK, Huang CJ. et al. Expression, characterization, and genomic structure of carp JAK1 kinase gene. DNA Cell Biol. 1996;15:827-844
52. Breathnach R, Benoist C, O'Hare K, Gannon F, Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978;75:4853-4857
53. Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA. et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78
54. Duran I, Mari-Beffa M, Santamaria JA, Becerra J, Santos-Ruiz L. Actinotrichia collagens and their role in fin formation. Dev Biol. 2011;354:160-172
55. Iuchi S. Three classes of C2H2 zinc finger proteins. Cell Mol Life Sci. 2001;58:625-635
56. Fox AH, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999;18:2812-2822
57. Del Rio S, Setzer DR. The role of zinc fingers in transcriptional activation by transcription factor IIIA. Proc Natl Acad Sci U S A. 1993;90:168-172
58. Friesen WJ, Darby MK. Phage display of RNA binding zinc fingers from transcription factor IIIA. J Biol Chem. 1997;272:10994-10997
59. Hesse E, Kiviranta R, Wu M, Saito H, Yamana K, Correa D. et al. Zinc finger protein 521, a new player in bone formation. Ann N Y Acad Sci. 2010;1192:32-37
60. Correa D, Hesse E, Seriwatanachai D, Kiviranta R, Saito H, Yamana K. et al. Zfp521 is a target gene and key effector of parathyroid hormone-related peptide signaling in growth plate chondrocytes. Dev Cell. 2010;19:533-546
61. Hesse E, Saito H, Kiviranta R, Correa D, Yamana K, Neff L. et al. Zfp521 controls bone mass by HDAC3-dependent attenuation of Runx2 activity. J Cell Biol. 2010;191:1271-1283
62. Liu TM, Guo XM, Tan HS, Hui JH, Lim B, Lee EH. Zinc-finger protein 145, acting as an upstream regulator of SOX9, improves the differentiation potential of human mesenchymal stem cells for cartilage regeneration and repair. Arthritis Rheum. 2011;63:2711-2720
Author contact
![]() Corresponding author: Dr. Chang-Jen Huang, Institute of Biological Chemistry, Academia Sinica, 128 Academia Rd., Sec. 2, Taipei 115, Taiwan. Tel: 886-2-2785-5696; Fax: 886-2-2788-9759; E-mail: cjibcsinica.edu.tw or Dr. Yu-San Han, Institute of Fisheries Sciences, National Taiwan University, Taipei 106, Taiwan. Tel: 886-2-3366-3726; Fax: 886-2-3366-5864; E-mail: yshanedu.tw.
Corresponding author: Dr. Chang-Jen Huang, Institute of Biological Chemistry, Academia Sinica, 128 Academia Rd., Sec. 2, Taipei 115, Taiwan. Tel: 886-2-2785-5696; Fax: 886-2-2788-9759; E-mail: cjibcsinica.edu.tw or Dr. Yu-San Han, Institute of Fisheries Sciences, National Taiwan University, Taipei 106, Taiwan. Tel: 886-2-3366-3726; Fax: 886-2-3366-5864; E-mail: yshanedu.tw.

 Global reach, higher impact
Global reach, higher impact