10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(9):983-989. doi:10.7150/ijbs.9442 This issue Cite
Review
The Role of Six1 in the Genesis of Muscle Cell and Skeletal Muscle Development
1. Department of Animal Genetics, Breeding and Reproduction, College of Animal Science and Technology, Nanjing Agricultural University, Nanjing, 210095, China;
2. Huaian Academy of Nanjing Agricultural University, Huaian, Jiangsu, 223001, China;
3. College of Life Science, Yangtze University, Jingzhou, Hubei, 434023, China.
4. Center for Basic and Applied Research, Faculty of Informatics and Management, University of Hradec Kradec Kralove, Hradec Kralove, Czech Republic.
Received 2014-4-19; Accepted 2014-6-6; Published 2014-9-5
Abstract
The sine oculis homeobox 1 (Six1) gene encodes an evolutionarily conserved transcription factor. In the past two decades, much research has indicated that Six1 is a powerful regulator participating in skeletal muscle development. In this review, we summarized the discovery and structural characteristics of Six1 gene, and discussed the functional roles and molecular mechanisms of Six1 in myogenesis and in the formation of skeletal muscle fibers. Finally, we proposed areas of future interest for understanding Six1 gene function.
Keywords: Six1, Muscle, Muscle fiber, Muscle progenitors, Regulatory network, Molecular mechanism.
Introduction
The sine oculis homeobox 1 (Six1) gene, a homologue of the Drosophila sine oculis (So) gene, belongs to the Six gene family; in vertebrates, in total of six members have been identified and which have been classified into three subgroups, Six1/Six2, Six3/Six6, and Six4/Six5 [1]. To date, Six family genes have been identified in species ranging from lower invertebrates (e.g., nematodes, planarians and cnidarians) to higher mammals (e.g., human, mouse, rat and pig) [2]. Numerous studies have demonstrated that Six family genes play important role in organogenesis [3-7] and diseases [8]. Among of the Six family genes, Six1 has been shown to play a pivotal role in skeletal muscle development [3, 9, 10].
Here, we reviewed the discovery and structural characteristics of the Six1 gene and then comprehensively summarized its functional roles and regulatory mechanisms in skeletal muscle development and the formation of skeletal muscle fibers. Finally, we proposed areas of key interest for future work.
Discovery of the Six1 gene
The chronology of Six1 discovery in different species is shown in Table 1. The first Six family gene, Drosophila sine oculis (So), was cloned from Drosophila melanogaster in 1994, and was demonstrated to play an essential role in the development of the visual system [11, 12]. One year later, three homologues of So (Six1, Six2, and Six3) were identified in mouse and were mapped to chromosome 12, 17 and 17, respectively [13, 14]. Moreover, a comprehensive analysis of the expression patterns of these homeobox genes during early mouse embryonic development was also conducted. At approximately E8.2, Six3 is restricted to the anterior neural plate with stronger expression signal, but its expression is shown to exist in developing eye, brain, and sensory regions as development proceeds [13]. By contrast, much broader and distinct expression patterns are observed for Six1 and Six2. Six1 is shown to express in mesodermally derived tissues skeletal muscle, as well as in dorsal root ganglia and Rathke's pouch, while Six2 is found to express in visceral smooth muscle, metanephros, genitalia and hindbrain [14].
The Six1 gene was first isolated in humans in 1996 and localized to human chromosome 14 [15]. Subsequently, Six1 gene has been identified and characterized in diverse species, including the vertebrates pig [16], chicken [17], Xenopus [18], zebrfish [19, 20], and the invertebrates jellyfish [21]. In addition to Six1, other homologues of the Drosophila sine oculis (So), such as Six2, Six3, Six4, Six5 and Six6 also have also been identified in a wide range of organisms throughout the animal kingdom [2].
Discovery of the Six1 gene in different species.
| Species | Tissues for function | Year | Reference |
|---|---|---|---|
| Drosophila melanogaster | Entire visual system | 1994 | (Cheyette et al., 1994; Serikaku and Otousa, 1994) |
| Mouse | Mesodermally derived tissues, e.g. skeletal muscle | 1995 | (Oliver et al., 1995a; Oliver et al., 1995b) |
| Human | Adult skeletal muscle | 1996 | (Boucher et al., 1996) |
| Chicken | Eye | 1996 | (Bovolenta et al., 1996) |
| Xenopus | Cranial ganglia, otic placodes and the eyes | 2001 | (Ghanbari et al., 2001) |
| Zebrfish | Sensory organs and muscle | 2004 | (Bessarab et al., 2004; Bessarab et al., 2008) |
| Jellyfish | Eyes | 2004 | (Stierwald et al., 2004) |
| Pig | Skeletal muscle | 2011 | (Wu et al., 2011) |
Structural characteristics of the Six1 protein
The Six protein family is a crucial developmental regulator involved in the formation of various organs, including muscle, eye, and kidney [3, 7, 10, 22-24]. As a member of the Six protein family, Six1 is characterized by two conserved domains, the homeodomain (HD) and the Six domain (SD) [1]. Based on amino acid sequence similarities of the conserved HDs and SDs, Six proteins can be subdivided into three major subgroups: Six1/2 (sine oculids), Six3/6 (optix) and Six4/5 (Dsix4) [25]. The detailed sequence similarity and phylogenetic relationships of the Six gene family members are discussed by Kumar et al [2].
In general, the HD is composed of a 180 nucleotide motif encoding 60 amino acids, while the SD is composed of 110-150 amino acids [1, 2]. Early studies demonstrated that the HD is indispensable for DNA binding [26-30], which remains consistent with more recent research [31, 32]. Initial studies suggested that the SD might contribute to the DNA binding, because the minimal essential region of AREC3 (Six4) for the sequence-specific binding is localized at leucine 91 to aspartic acid 215, which covers part of the SD amino acid sequence [27]. However, a recent study found that the DNA binding properties of Six proteins are mainly due to the HD, and that the adjacent C-terminal region appears to increase the binding affinity of the HD to DNA; By contrast, the SD does not possess the characteristics of a DNA binding domain due to lacking the basic N-terminal arm, which is critical for canonical homeodomain-DNA binding [31].
Early studies indicated that the protein-protein interactions depend primarily upon the SD of the Six proteins [33, 34]. Further studies provided evidences that the SD is necessary to specific binding to protein partners and that the specificity in partner selection is crucial to the role of each protein [35]. Most recently, Six1 was shown to predominantly use a single helix to interact with EYA, and the mutation of a single amino acid in the helices of the HD or SD is sufficient to disrupt their interaction [36]. In addition, recent research found that the non-conserved C-terminal region of the Six proteins make important contributions to the functional specificity of the different Six proteins [37]. Thus, the conserved HD and SD are essential for Six proteins to perform their functions.
The role of Six1 in the genesis of the muscle cell lineage
The initiation of skeletal myogenesis occurs during embryogenesis and is an extremely complex and successive process, which involves the genesis, delamination and migration of muscle progenitors, followed by myoblast determination, proliferation and differentiation (Figure 1). The paraxial mesoderm of embryo segments develops into somites. The dorsal part of the somite (the dermomyotome) produces muscle progenitor cells that undergo delamination, migration, and reach to the sites of muscle formation, where they transform into myogenic regulatory factor (MRF)-expressing myoblasts, undergo proliferation and differentiation, and subsequently fuse to form myofibers and finally form the muscles of the body and limbs [38].
Six1 is a critical factor in the genesis of muscle progenitors. The myogenic progenitor cells in the limb buds of Six1-/-Six4-/- mouse embryos are lost and the expression of Pax3 in the hypaxial dermomyotome is lacking [9, 39]. Notably, Pax3 is indispensable for the genesis of muscle progenitors because hypaxial migrating muscle progenitors are lacking in the Pax3-/- mouse embryo [40]. Further studies directly demonstrated that Pax3/Pax7 is molecular mark of muscle progenitors that subsequently become myogenic and form skeletal muscle [41, 42]. These data suggest that Six1 (or Six4) is an upstream regulatory factor of Pax3 and is essential for the genesis of muscle progenitors (Figure 1A). The most study in zebrafish showed that Six1 (Six1a and Six1b) can promote the proliferation of Pax7+ muscle progenitors cells in the dermomyotome via phosphorylated Smad1/5/8 [43]. Furthermore, the Wnt signaling factors are important for the induction of myogenesis in the myotome (the first muscle mass ventral to the dermomyotome) of the embryo, and these Wnt signaling factors can activate the expression of Six1, Pax3 and Mox1 via β-catenin in vitro. These results suggesting the Wnt signaling factors lie genetically upstream of Six1, Pax3, and Mox1 [44]. In addition, Six1 frequently interacts with other factors, including Eya2/Dach2 [45] and Ski/Eya3 [46], to exert its function in embryonic skeletal myogenesis, and a reciprocal regulatory mechanism has been verified between Six1 and Pax3 [9, 47]. Although great progress has been made in understanding the mechanisms underlying the genesis of muscle progenitors, more detail about these regulatory pathways remains to be identified.
Molecular mechanisms of embryonic skeletal myogenesis. (A) Schematic representation of the underlying genetic hierarchy that controls the genesis of muscle progenitors in the dermomyotome. (B) Schematic representation of regulatory mechanisms that control the delamination and migration of muscle progenitors. (C) Schematic representation of regulatory mechanisms that control the determination, proliferation, and differentiation of myoblasts. During embryonic skeletal myogenesis, Six1 shows robust function across the entire embryonic developmental stages and may synergize with other co-factors, including Eya1[70], Eya2/Dach2 [45] and Ski/Eya3 [46], to exert its function. Of interest, a feedback regulatory loop exists among Six1, Pax3, and MyoD [9, 47, 62]. In addition, many other factors are also involved in embryonic skeletal myogenesis. Detailed information can be found in the text.
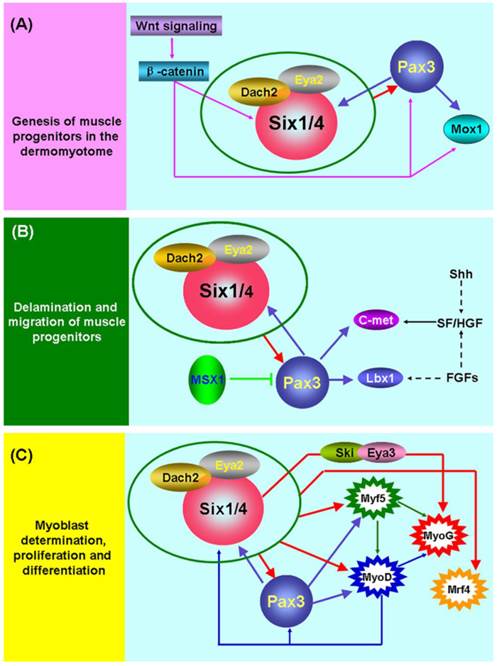
Pax3 plays a pivotal role in the delamination and migration of muscle progenitors and this biological process depends on the presence of the tyrosine kinase receptor c-Met, its ligand scatter factor/hepatocyte growth factor (SF/HGF) [38, 48-51], and the homeobox factor Lbx1 [52, 53]. The Gab1 adaptor molecule mediates the delamination, migration and survival of muscle progenitor cells, in part by transmitting c-Met signals (Sachs et al. 2000). During the delamination and migration of muscle progenitors, the homeobox factor Msx1 acts as an antagonist of Pax3 by interfering with DNA binding by Pax3 [54], and Six1 transcriptional complexes are likely to be upstream positive regulators of Pax3 (Figure 1B).
For the determination, proliferation, and differentiation of the muscle lineage (Figure 1C), MRFs are considered to be myogenic determination factors that commit muscle progenitors to the myogenic fate [38, 55]. The expression of Six1 and Pax3 factors occurs prior to MRFs in embryonic skeletal myogenesis and further studies have found that these two factors directly control MRFs expression [46, 56-58]. These data strongly suggest that the Six1 and Pax3 are indispensable for this process.
Although a great deal of research has documented that MRFs are the direct targets of Six1 and Pax3 [3, 9, 56-60], an early study found that MyoD efficiently upregulates expression of Six1 [61]. A recent study also provided strong evidence that MyoD promotes determination of the skeletal muscle lineage by directly binding and activating multiple pre-myogenic mesoderm factors, including Six1, Pax3/7, Eya2, and Meox1 [62]. These results suggest that a feedback regulatory loop may exist between MyoD and Six1, Pax3 or other pre-myogenic mesoderm factors (Figure 1C), and other MRFs may possess regulatory mechanisms similar to MyoD.
Role of Six1 in adult skeletal muscle development
An initial study did not conclude that Six1 is a critical factor for skeletal muscle development since its expression is not restricted to the muscles, even though it is weakly expressed in skeletal muscles from E13.5 to E16.0 [14]. A subsequent study showed that Six1 was exclusively expressed in human adult skeletal muscle [15], which is consistent with our recent observation that Six1 is highly expressed in primarily in skeletal muscles in pigs [16]. Activated satellite cells are able to promote the regeneration of adult skeletal muscle, and new studies have shown that Six1 is clearly expressed in activated satellite cells [43, 63, 64]. These results suggest that Six1 not only play an important role in embryonic skeletal myogenesis, but also in adult muscle regeneration.
Previous genetic experiments also demonstrated that late skeletal muscle may depend upon the Six1 gene. In 2003, Laclef et al. generated Six1-deficient mice by replacing the conserved functional domains (HD and SD) regions of the Six1 gene using a homologous recombination approach. These Six1-/- transgenic mice die at birth due to extensive muscle hypoplasia and severe rib malformations [3]. Moreover, Six4 was also shown to play an important role in myogenesis, and Six1 and Six4 have a functional redundancy since embryos of Six1-/-Six-4-/- double KO mice show more severe muscle phenotypes than the Six1-/- embryos [9]. In fact, the functional redundancy between Six1 and Six4 is strongly supported by several studies. First, Six1 and Six4 show almost identical expression patterns, and both gene products can activate the myogenin promoter [65]. Second, Six1 and Six4 regulate the same target genes; both Six1 and Six4 activate the myogenetic regulatory transcription factors myogenin and Myf5 through specific binding to the MEF3 sites in their promoters [57, 58]. However, Six1 may play a more important role in skeletal muscle development than Six4 because Six4-deficient mice do not display abnormal phenotypes in skeletal muscles development in embryos or adults [66]. In addition, Six5 has also been shown to take part in myogenesis [67] and activates myogenin through specific binding to the MEF3 site in the promoter [57, 65]. However, mice lacking Six5 show normal development and no muscle defects [68], indicating that Six5 may compensate, but not substitute Six1 or Six4 function in skeletal muscle development.
As mentioned above, the regulation of the skeletal muscle development by Six1 is the result of activating myogenin and Myf5 expression by binding to the MEF3 sites in the promoters of these genes [57, 58]. An in vitro study found that activation of the MEF3 site is required, at least in part, for the interaction of Ski with Six1 and Eya3 [46]. In fact, the interactions between the Eya and Six proteins have been observed in previous studies [65, 69]. Myogenesis is synergistically regulated by Dach2, Eya2 and Six1 in the somites of the embryo [45], while an analogous synergistic regulation relationship is also observed between Six1 and Eya1 in the transformation of adult skeletal muscle fibers [70]. Moreover, a recent report indicates that the activation of myogenin depends on a demethylated promoter which is more conducive to binding of the transcription factor Six1 and MEF2 [71].
In addition, a recent study shows that Six1 contributes to the regeneration of adult muscle by enhancing and maintaining MyoD expression in adult muscle satellite cells and that MyoD expression also depends on the MEF3 site within the core enhancer CER of MyoD [63]. Interestingly, the same feedback regulatory mechanism seems to exist between Six proteins and MRFs because MyoD is able to promote the transformation of multipotent stem cells to skeletal muscle by binding and activating the expression of a subset of pre-myogenic mesoderm genes, including Six1, Pax3/7, Eya2, and Meox1 [62] .
Pax3 is an upstream regulator of the myogenic transcription factors MyoD, Myf5 and myogenin [47, 56, 60]. Interestingly, Pax3 seems to be upstream factor of Six1 because overexpression of Pax3 in stem cells is sufficient to initiate myogenesis by promoting Six1, Mox1, and Eya2 expression [47]; Pax3 expression is not influenced in Six1-/- embryos; and delamination and migration of Pax3 positive cells is normal [3]. By contrast, the expressions of Pax3, and its downstream genes Lbx1 and Met, are lost in the hypaxial dermomyotome of the Six1-/-Six4-/- double mutant mice [9], which indicates that Six1 is the upstream regulatory factor of Pax3. One explanation for these contradictory conclusions is that a regulatory feedback loop may exist between Six1 and Pax3, and which one acts as the upstream factor may depend on different developmental stages or distinct physiological states.
Role of Six1 in the formation of skeletal muscle fibers
Initial studies concentrated on the role of Six1 in muscle development, rather than the formation of skeletal muscle fibers. Interestingly, in 2004, Grifone et al. reported that Six1 is able to drive the transformation of slow-twitch fibers to the fast-twitch phenotype in synergy with its cofactor Eya1 in adult muscles of mice [70]. Thus, the Six1/Eya1 complex was the first transcriptional complex identified to modulate the fast-twitch glycolytic phenotype of adult skeletal muscle.
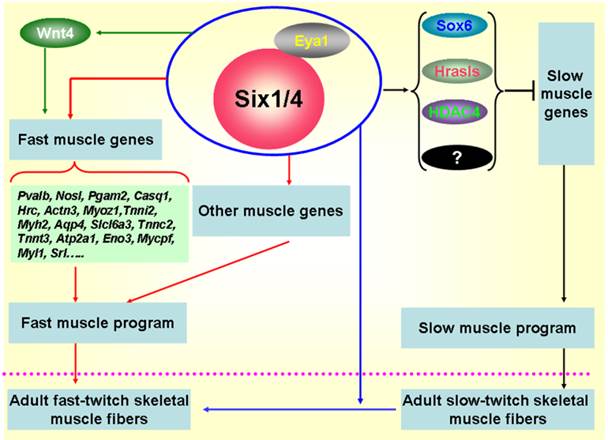
Subsequently, more investigations focused on the functional role of Six1 in the formation of skeletal muscle fibers. In zebrafish, Bessarab et al. identified two Six1 homologues, Six1a and Six1b, and found that Six1a is expressed in the fast muscle precursor region of the somite, while the loss of Six1a led to a reduction of myogenin in fast muscle precursors, suggesting that Six1a is essential for the onset of fast muscle differentiation [19]. Moreover, Maire' group conducted in-depth studies on the role of Six1 in the formation of skeletal muscle fibers using the Six1-/-/Six4-/- mice, their results indicate that the Six1 and Six4 proteins are involved in the genesis of muscle fiber-type diversity during mouse embryogenesis and are required for the activation of the fast muscle program in the mouse primary myotome by up-regulating expression of a set of fast muscle genes, including Myh2, Tnnc2, Tnnt3, Atp2a1, and Srl genes etc [10, 72] (Figure 2). In 2011, our group cloned the porcine Six1 gene and described its expression patterns in skeletal muscles with different fiber types. Six1 is expressed at higher levels in fast-twitch muscle fibers than in slow-twitch muscle fibers [16]. Furthermore, we overexpressed the Six1 gene in C2C12 cells and found that the expression of multiple fast-type muscle genes are promoted by Six1 expression, in particular, Atp2a1, Srl and Mylpf [73]. In addition, a recent study in zebrafish found that Six1a and Six1b not only regulate the proliferation and differentiation of Pax7+ muscle progenitor cells, but also are essential for fast muscle development. Six1a and Six1b are expressed in fast muscle progenitors but are absent in slow muscle progenitors and the fast muscle domain is significantly reduced in the Six1a or Six1b knock-down embryos [43]. More recently, in zebrafish, O'Brien et al. found an upstream microRNA regulatory mechanism of Six1 that miR30a directly regulate myogenesis via inhibiting Six1a/b expression [74].
Sox6 is a transcriptional repressor of slow fiber-specific genes, but acts as an upregulator of fast fiber-specific expression [75-77]. HDAC4 also repressed MEF2-dependent slow-type muscle gene expression [78, 79]. Moreover, and in vitro myogenic cell culture experiment showed that Wnt4 promoted fast-type muscle differentiation [80]. Notably, in the remaining muscles of Six1/Six4 double mutant fetuses, Hrasls and Wnt4 expression is severely downregulated, and HDAC4 protein accumulation levels and Sox6 subcellular localization are altered, which suggests the occurrence of crosstalk between fast- and slow-type muscle gene expression mediated by Six transcriptional complexes (STC) (Figure 2). Thus, Six1 plays a critical role not only in skeletal muscle development, but also in the genesis of muscle fiber-type diversity during embryonic development and in the transformation of muscle fibers from one type to the other in the adult skeletal muscle development. The potential regulatory mechanisms are shown in Figure 2.
Regulatory mechanisms controlling the formation of fast-twitch muscle fibers. During embryogenesis, Six1 transcriptional complexes (STC) promote the fast-twitch muscle phenotype by directly controlling the expression of a set of fast muscle genes; Simultaneously, STC is also able to activate inhibitors of slow muscle genes, such as Sox6, Hrasls, and HDAC4, thereby restricting the activation of the slow muscle genes, and ultimately inhibiting the slow-twitch muscle phenotype [10, 19, 72]. In addition, the STC can drive the transformation of slow-twitch muscle fibers to fast-twitch muscle fibers in adult skeletal muscles [70].
Conclusions and prospects
As highlighted in this review, Six1 is a critical transcription factor in skeletal muscle development, participating in the genesis of muscle progenitors in embryogenesis, as well as skeletal muscle development from embryo to adulthood and in the transformation of skeletal muscle fiber types. The molecular regulatory mechanisms underlying skeletal muscle development have gradually become clear, but some unknown complex regulatory processes still need to be identified. In humans, Six1 plays an important role in skeletal muscle development, and thus, Six1 is likely to have great potential as therapeutic target for treating myopathies, despite great challenges that remain to be conquered.
In livestock, skeletal muscle is the main component of meat and the growth of skeletal muscle is a major factor affecting overall growth. Therefore, Six1 could be a candidate gene target to be investigated for the manipulation of livestock growth. Moreover, skeletal muscles are composed of different muscle fibers that display different metabolic, contractile and endurance properties, and the differences in muscle fiber type is one of the critical factors that determine meat quality. Thus, deeper understanding of the functional roles of Six1 in the formation of skeletal muscle fibers would further elucidate the molecular regulatory mechanisms influencing meat quality. Therefore, it would be of interest in future work to investigate the relationship between the Six1 gene and the production performance of livestock animals, especially in regard to meat quality traits.
Acknowledgements
We would like to thank all our coworkers, past and present, for their contributions to this manuscript. This work was supported by the Natural Science Foundation of Jiangsu Province (BK20130693 and BK20140688), the Youth Science and Technology Innovation Fund of Nanjing Agricultural College (KJ2012014), and grants from the 973 Program (2014CB138502) and the project of excellence FIM UHK.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes--structure and function as transcription factors and their roles in development. Bioessays. 2000;22(7):616-26
2. Kumar J.P. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci. 2009;66(4):565-83
3. Laclef C, Hamard G, Demignon J, Souil E, Houbron C, Maire P. Altered myogenesis in Six1-deficient mice. Development. 2003;130(10):2239-52
4. Li X, Oghi K.A, Zhang J, Krones A, Bush K.T, Glass C.K, Nigam S.K, Aggarwal A.K, Maas R, Rose D.W, Rosenfeld M.G. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426(6964):247-54
5. Fujimoto Y, Tanaka S.S, Yamaguchi Y.L, Kobayashi H, Kuroki S, Tachibana M, Shinomura M, Kanai Y, Morohashi K, Kawakami K, Nishinakamura R. Homeoproteins Six1 and Six4 regulate male sex determination and mouse gonadal development. Dev Cell. 2013;26(4):416-30
6. Ahmed M, Xu J.S, Xu P.X. EYA1 and SIX1 drive the neuronal devel-opmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development. 2012;139(11):1965-1977
7. Anderson A.M, Weasner B.M, Weasner B.P, Kumar J.P. Dual tran-scriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development. 2012;139(5):991-1000
8. Wu W, Ren Z, Li P, Yu D, Chen J, Huang R, Liu H. Six1: A critical transcription factor in tumorigenesis. Int J Cancer. 2014 doi:10.1002/ijc.28755
9. Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller M.J, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132(9):2235-49
10. Richard A.F, Demignon J, Sakakibara I. et al. Genesis of muscle fiber-type diversity during mouse embryogenesis relies on Six1 and Six4 gene expression. Dev Biol. 2011;359(2):303-20
11. Cheyette B.N.R, Green P.J, Martin K, Garren H, Hartenstein V, Zipursky S.L. The Drosophila Sine Oculis Locus Encodes a Homeodo-main-Containing Protein Required for the Development of the Entire Visu-al-System. Neuron. 1994;12(5):977-996
12. Serikaku M.A, Otousa J.E. Sine Oculis Is a Homeobox Gene Required for Drosophila Visual-System Development. Genetics. 1994;138(4):1137-1150
13. Oliver G, Mailhos A, Wehr R. et al. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121(12):4045-55
14. Oliver G, Wehr R, Jenkins N.A, Copeland N.G, Cheyette B.N, Harten-stein V, Zipursky S.L, Gruss P. Homeobox genes and connective tissue patterning. Development. 1995;121(3):693-705
15. Boucher C.A, Carey N, Edwards Y.H, Siciliano M.J, Johnson K.J. Cloning of the human SIX1 gene and its assignment to chromosome 14. Ge-nomics. 1996;33(1):140-2
16. Wu W, Ren Z, Wang Y, Chao Z, Xu D, Xiong Y. Molecular characterization, expression patterns and polymorphism analysis of porcine Six1 gene. Mol Biol Rep. 2011;38(4):2619-32
17. Bovolenta P, Mallamaci A, Boncinelli E. Cloning and characterisation of two chick homeobox genes, members of the six/sine oculis family, expressed during eye development. Int J Dev Biol. 1996(Suppl 1):73S-74S
18. Ghanbari H, Seo H.C, Fjose A, Brandli A.W. Molecular cloning and embryonic expression of Xenopus Six homeobox genes. Mech Dev. 2001;101(1-2):271-7
19. Bessarab D.A, Chong S.W, Srinivas B.P, Korzh V. Six1a is required for the onset of fast muscle differentiation in zebrafish. Dev Biol. 2008;323(2):216-28
20. Bessarab D.A, Chong S.W, Korzh V. Expression of zebrafish six1 during sensory organ development and myogenesis. Dev Dynam. 2004;230(4):781-786
21. Stierwald M, Yanze N, Bamert R.P, Kammermeier L, Schmid V. The Sine oculis/Six class family of homeobox genes in jellyfish with and without eyes: development and eye regeneration. Dev Biol. 2004;274(1):70-81
22. Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech Dev. 2003;120(6):669-79
23. Kawakami K, Ohto H, Takizawa T, Saito T. Identification and expression of six family genes in mouse retina. FEBS Lett. 1996;393(2-3):259-63
24. Nie X.G, Xu J.S, El-Hashash A, Xu P.X. Six1 regulates Grem1 expression in the metanephric mesenchyme to initiate branching morphogenesis. Dev Biol. 2011;352(1):141-151
25. Seo H.C, Curtiss J, Mlodzik M, Fjose A. Six class homeobox genes in drosophila belong to three distinct families and are involved in head devel-opment. Mech Dev. 1999;83(1-2):127-39
26. Scott M.P. et al. The structure and function of the homeodomain. Biochim Biophys Acta. 1989;989(1):25-48
27. Kawakami K, Ohto H, Ikeda K, Roeder R.G. Structure, function and expression of a murine homeobox protein AREC3, a homologue of Drosophila sine oculis gene product, and implication in development. Nucleic Acids Res. 1996;24(2):303-10
28. Wolberger C. Homeodomain interactions. Curr Opin Struc Biol. 1996;6(1):62-68
29. Ades S.E, Sauer R.T. Specificity of Minor-Groove and Major-Groove Interactions in a Homeodomain-DNA Complex. Biochemistry. 1995;34(44):14601-14608
30. Gehring W.J, Affolter M, Burglin T. Homeodomain Proteins. Annu Rev Biochem. 1994;63:487-526
31. Hu S, Mamedova A, Hegde R.S. DNA-binding and regulation mechanisms of the SIX family of retinal determination proteins. Biochemistry. 2008;47(11):3586-94
32. Noyes M.B, Christensen R.G, Wakabayashi A, Stormo G.D, Brodsky M.H, Wolfe S.A. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133(7):1277-89
33. Kenyon K.L, Li D.J, Clouser C, Tran S, Pignoni F. Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev Dyn. 2005;234(3):497-504
34. Kenyon K.L, Yang-Zhou D, Cai C.Q, Tran S, Clouser C, Decene G, Ranade S, Pignoni F. Partner specificity is essential for proper function of the SIX-type homeodomain proteins Sine oculis and Optix during fly eye development. Dev Biol. 2005;286(1):158-168
35. Weasner B, Salzer C, Kumar J.P. Sine oculis, a member of the SIX family of transcription factors, directs eye formation. Dev Biol. 2007;303(2):756-71
36. Patrick A.N, Cabrera J.H, Smith A.L, Chen X.J.S, Ford H.L, Zhao R. Structure-function analyses of the human SIX1-EYA2 complex reveal insights into metastasis and syndrome BOR. Nat Struct Mol Biol. 2013;20(4):447
37. Weasner B.P, Kumar J.P. The non-conserved C-terminal segments of Sine Oculis Homeobox (SIX) proteins confer functional specificity. Genesis. 2009;47(8):514-23
38. Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: from somite to limb. J Anat. 2003;202(1):59-68
39. Grifone R, Demignon J, Giordani J, Niro C, Souil E, Bertin F, Laclef C, Xu P.X, Maire P. Eya1 and Eya2 proteins are required for hypaxial somitic myogenesis in the mouse embryo. Dev Biol. 2007;302(2):602-16
40. Tremblay P, Dietrich S, Mericskay M, Schubert F.R, Li Z, Paulin D. A crucial role for Pax3 in the development of the hypaxial musculature and the long-range migration of muscle precursors. Dev Biol. 1998;203(1):49-61
41. Relaix F, Rocancourt D, Mansouri A, Buckingham M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435(7044):948-953
42. Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, Jory A, Gomes D, Tajbakhsh S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Gene Dev. 2005;19(12):1426-1431
43. Nord H, Skalman L.N, von Hofsten J. Six1 regulates proliferation of Pax7-positive muscle progenitors in zebrafish. J Cell Sci. 2013;126(8):1868-1880
44. Petropoulos H, Skerjanc I.S. Beta-catenin is essential and sufficient for skeletal myogenesis in P19 cells. J Biol Chem. 2002;277(18):15393-9
45. Heanue T.A, Reshef R, Davis R.J, Mardon G, Oliver G, Tomarev S, Lassar A.B, Tabin C.J. Synergistic regulation of vertebrate muscle develop-ment by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 1999;13(24):3231-43
46. Zhang H, Stavnezer E. Ski Regulates Muscle Terminal Differentiation by Transcriptional Activation of Myog in a Complex with Six1 and Eya3. J Biol Chem. 2009;284(5):2867-2879
47. Ridgeway A.G, Skerjanc I.S. Pax3 is essential for skeletal myogenesis and the expression of Six1 and Eya2. J Biol Chem. 2001;276(22):19033-9
48. Epstein J.A, Shapiro D.N, Cheng J, Lam P.Y, Maas R.L. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc Natl Acad Sci U S A. 1996;93(9):4213-8
49. Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376(6543):768-71
50. Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnen-berg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C. The role of SF/HGF and c-Met in the development of skeletal muscle. Development. 1999;126(8):1621-9
51. Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Scatter Factor/Hepatocyte Growth-Factor Is Essential for Liver Development. Nature. 1995;373(6516):699-702
52. Schafer K, Braun T. Early specification of limb muscle precursor cells by the homeobox gene Lbx1h. Nat Genet. 1999;23(2):213-6
53. Mennerich D, Schafer K, Braun T. Pax-3 is necessary but not sufficient for lbx1 expression in myogenic precursor cells of the limb. Mech Dev. 1998;73(2):147-58
54. Bendall A.J, Ding J, Hu G, Shen M.M, Abate-Shen C. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development. 1999;126(22):4965-76
55. Sabourin L.A, Rudnicki M.A. The molecular regulation of myogenesis. Clin Genet. 2000;57(1):16-25
56. Tajbakhsh S, Rocancourt D, Cossu G, Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act up-stream of MyoD. Cell. 1997;89(1):127-138
57. Spitz F, Demignon J, Porteu A, Kahn A, Concordet J.P, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci U S A. 1998;95(24):14220-5
58. Giordani J, Bajard L, Demignon J, Daubas P, Buckingham M, Maire P. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proc Natl Acad Sci U S A. 2007;104(27):11310-5
59. Fougerousse F, Durand M, Lopez S, Suel L, Demignon J, Thornton C, Ozaki H, Kawakami K, Barbet P, Beckmann J.S, Maire P. Six and Eya expression during human somitogenesis and MyoD gene family activation. J Muscle Res Cell Motil. 2002;23(3):255-64
60. Bajard L, Relaix F, Lagha M, Rocancourt D, Daubas P, Buckingham M.E. A novel genetic hierarchy functions during hypaxial myogenesis: Pax3 directly activates Myf5 in muscle progenitor cells in the limb. Genes Dev. 2006;20(17):2450-64
61. Ishibashi J, Perry R.L, Asakura A, Rudnicki M.A. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J Cell Biol. 2005;171(3):471-82
62. Gianakopoulos P.J, Mehta V, Voronova A, Cao Y, Yao Z, Coutu J, Wang X, Waddington M.S, Tapscott S.J, Skerjanc I.S. MyoD directly up-regulates premyogenic mesoderm factors during induction of skeletal myogenesis in stem cells. J Biol Chem. 2011;286(4):2517-25
63. Liu Y, Chakroun I, Yang D. et al. Six1 Regulates MyoD Expression in Adult Muscle Progenitor Cells. PLoS One. 2013;8(6):e67762
64. Le Grand F, Grifone R, Mourikis P, Houbron C, Gigaud C, Pujol J, Maillet M, Pages G, Rudnicki M, Tajbakhsh S, Maire P. Six1 regulates stem cell repair potential and self-renewal during skeletal muscle regeneration. J Cell Biol. 2012;198(5):815-832
65. Ohto H, Kamada S, Tago K, Tominaga S.I, Ozaki H, Sato S, Kawakami K. Cooperation of six and eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol. 1999;19(10):6815-24
66. Ozaki H, Watanabe Y, Takahashi K, Kitamura K, Tanaka A, Urase K, Momoi T, Sudo K, Sakagami J, Asano M, Iwakura Y, Kawakami K. Six4, a putative myogenin gene regulator, is not essential for mouse embryonal development. Mol Cell Biol. 2001;21(10):3343-50
67. Kirby R.J, Hamilton G.M, Finnegan D.J, Johnson K.J, Jarman A.P. Drosophila homolog of the myotonic dystrophy-associated gene, SIX5, is required for muscle and gonad development. Curr Biol. 2001;11(13):1044-1049
68. Klesert T.R, Cho D.H, Clark J.I, Maylie J, Adelman J, Snider L, Yuen E.C, Soriano P, Tapscott S.J. Mice deficient in Six5 develop cataracts: implications for myotonic dystrophy. Nat Genet. 2000;25(1):105-9
69. Ikeda K, Watanabe Y, Ohto H, Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol. 2002;22(19):6759-66
70. Grifone R, Laclef C, Spitz F, Lopez S, Demignon J, Guidotti J.E, Kawakami K, Xu P.X, Kelly R, Petrof B.J, Daegelen D, Concordet J.P, Maire P. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Mol Cell Biol. 2004;24(14):6253-67
71. Palacios D, Summerbell D, Rigby P.W, Boyes J. Interplay between DNA methylation and transcription factor availability: implications for developmental activation of the mouse Myogenin gene. Mol Cell Biol. 2010;30(15):3805-15
72. Niro C, Demignon J, Vincent S, Liu Y.B, Giordani J, Sgarioto N, Favier M, Guillet-Deniau I, Blais A, Maire P. Six1 and Six4 gene expression is necessary to activate the fast-type muscle gene program in the mouse primary myotome. Dev Biol. 2010;338(2):168-182
73. Wu W.J, Ren Z.Q, Zhang L, Liu Y, Li H.G, Xiong Y.Z. Overexpression of Six1 gene suppresses proliferation and enhances expression of fast-type muscle genes in C2C12 myoblasts. Mol Cell Biochem. 2013;380(1-2):23-32
74. O'Brien J.H, Hernandez-Lagunas L, Artinger K.B, Ford H.L. Mi-croRNA-30a regulates zebrafish myogenesis via targeting the transcription factor Six1. J Cell Sci. 2014;127:2291-2301
75. Hagiwara N, Ma B, Ly A. Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p(100H). Dev Dynam. 2005;234(2):301-311
76. Hagiwara N, Yeh M, Liu A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Dev Dynam. 2007;236(8):2062-2076
77. von Hofsten J, Elworthy S, Gilchrist M.J, Smith J.C, Wardle F.C, Ingham P.W. Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. Embo Reports. 2008;9(7):683-689
78. Potthoff M.J, Wu H, Arnold M.A, Shelton J.M, Backs J, McAnally J, Richardson J.A, Bassel-Duby R, Olson E.N. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117(9):2459-2467
79. Cohen T.J, Waddell D.S, Barrientos T, Lu Z, Feng G, Cox G.A, Bodine S.C, Yao T.P. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J Biol Chem. 2007;282(46):33752-33759
80. Takata H, Terada K, Oka H, Sunada Y, Moriguchi T, Nohno T. Involvement of Wnt4 signaling during myogenic proliferation and differentiation of skeletal muscle. Dev Dynam. 2007;236(10):2800-2807
Author contact
![]() Corresponding authors: Department of Animal Genetics, Breeding and Reproduction, College of Animal Science and Technology, Nanjing Agricultural University, Nanjing, 210095, China; E-mail: wuwangjun2009com; Tel: +86 25 84399762; Fax: +86 25 84399762.
Corresponding authors: Department of Animal Genetics, Breeding and Reproduction, College of Animal Science and Technology, Nanjing Agricultural University, Nanjing, 210095, China; E-mail: wuwangjun2009com; Tel: +86 25 84399762; Fax: +86 25 84399762.

 Global reach, higher impact
Global reach, higher impact