Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(10):1150-1158. doi:10.7150/ijbs.10452 This issue Cite
Review
RNA Interference (RNAi) Induced Gene Silencing: A Promising Approach of Hi-Tech Plant Breeding
1. Department of Horticultural Science, Kyungpook National University, Daegu 702-701, Korea.
2. Institute of Horticultural Sciences, University of Agriculture, Faisalabad 38040, Pakistan.
3. Department of Plant Science, and Plant Genomics and Breeding Institute, Seoul National University, Seoul 151-921, Korea.
Received 2014-9-1; Accepted 2014-9-22; Published 2014-10-11
Abstract
RNA interference (RNAi) is a promising gene regulatory approach in functional genomics that has significant impact on crop improvement which permits down-regulation in gene expression with greater precise manner without affecting the expression of other genes. RNAi mechanism is expedited by small molecules of interfering RNA to suppress a gene of interest effectively. RNAi has also been exploited in plants for resistance against pathogens, insect/pest, nematodes, and virus that cause significant economic losses. Keeping beside the significance in the genome integrity maintenance as well as growth and development, RNAi induced gene syntheses are vital in plant stress management. Modifying the genes by the interference of small RNAs is one of the ways through which plants react to the environmental stresses. Hence, investigating the role of small RNAs in regulating gene expression assists the researchers to explore the potentiality of small RNAs in abiotic and biotic stress management. This novel approach opens new avenues for crop improvement by developing disease resistant, abiotic or biotic stress tolerant, and high yielding elite varieties.
Keywords: abiotic stress, biotic stress, crop improvement, functional genomics, post transcriptional gene silencing, siRNA.
Introduction
RNA interference (RNAi) is a biological mechanism which leads to post transcriptional gene silencing (PTGS) trigger by double stranded RNA (dsRNA) molecules to prevent the expression of specific genes [1, 2]. RNAi mechanism has the potential in identification and functional assessment of thousands genes within any genome that is responsible for crop improvement. This promising approach also imparts its effective and efficient role to knock down the expression of any particular gene through short interfering RNA molecules in any target cell and moreover to assess the changes that occur in signaling pathways. Recently, RNAi has become a powerful and more reliable technique to inhibit the expression of targeted genes and also determine gene loss-of-function phenotype which leads to gene functional analysis when no mutant alleles are unavailable [3]. RNAi technique was first time applied on Petunia hybrida L. plants to enhance anthocyanin pigment through introducing chalcone synthase gene (chsA) [4]. New pattern of flower color in transgenic Petunia was observed due to overexpression of chsA gene that encodes major enzymes in anthocyanin biosynthesis pathway [5].
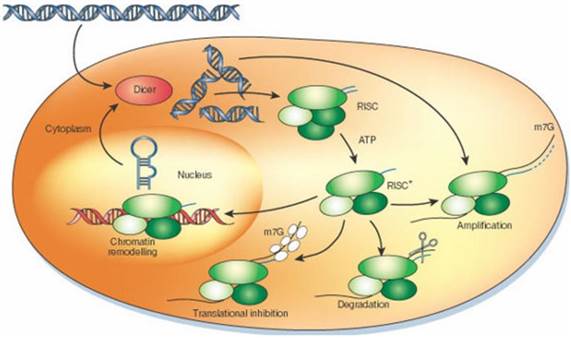
RNAi mechanism is expedited by small molecules of interfering RNA to express a gene of interest effectively. Several methods to induce RNAi, RNAi vectors, in vitro dicing and synthetic molecules are reported [6, 7]. Introduction of short pieces of double stranded RNA (dsRNA) and small interfering RNA (siRNA) into the cytosol, initiate the pathway culminating targeted degradation of the specific cellular mRNA (Figure 1). During RNAi mechanism, silencing initiate with enzyme Dicer and dsRNA is processed to convert the silencing trigger to ~22-nucleotide, small interfering RNAs (siRNAs). The antisense strand of siRNA become specific to endonuclease-protein complex, RNA-induced silencing complex (RISC), which then targets the homologous RNA and degrade it at specific site that results in the knock-down of protein expression (Figure 2) [6, 8, 9].
RNAi for plant disease resistance
Pathogens can cause huge reduction in crop yield that can have a significant negative economic impact and also they are threat to wipe-out the entire plant species. Plant pathologists and plant biotechnologists have adopted different approaches to develop pathogen resistant genotypes but in last decade RNAi-induced gene silencing emerged as an effective tool to engineer pathogen resistant plants [10]. This approach proved to be effective to create resistance against some diseases of economic importance caused by bacteria, fungi and viruses [11]. Double stranded RNA (dsRNA) act as an igniter in RNA interference and activate the homologous mRNAs to inhibit its translation and transcription to silence the susceptible genes [12]. This RNAi approach has opened new avenues in the development of eco-friendly techniques for plant improvement as specific genes are suppressed which cause stress and expression of novel genes for disease resistance.
Model of Dicer and RNA-induced silencing complex (RISC) A. silencing initiate with enzyme Dicer and dsRNA is processed to convert the silencing trigger to ~22-nucleotide, small interfering RNAs (siRNAs), B. Dicer binding and cleaving dsRNA (*Cleavage into precisely sized fragments is determined by the fact that one of the active sites in each Dicer protein is defective. Different colors show two separate molecules of Dicer). Reprinted by permission from Macmillan Publishers Ltd: Nature, 418: 244-251, (2002)
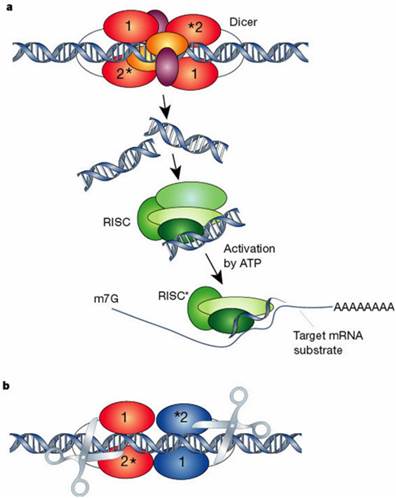
Diagrammatic representation of mechanism of RNAi. Silencing triggers in the form of double-stranded RNA presented in the cell as synthetic RNAs, replicating viruses or transcribed from nuclear genes. These are recognized and processed into small interfering RNAs by Dicer. The duplex siRNAs are passed to RISC (RNA-induced silencing complex), and the complex becomes activated by unwinding of the duplex. Activated RISC complexes can regulate gene expression at many levels. Reprinted by permission from Macmillan Publishers Ltd: Nature, 418: 244-251, (2002)
Host gene silencing -hair pin RNAi (HGS-hpRNAi) is reported as more stable gene silencing method in plants [13]. This method can be employed to increase fungal and bacterial disease resistance by changing the gene expression against pathogens through genetic engineering in the host plant. Flagellin (a bacterial component) can stimulate the expression of specific miRNA to increase disease resistance signaling pathway in Arabidopsis [14]. Over-expression of a gene AtFAAH in Arabidopsis, responsible for fatty acid (N-acylethanolamines) metabolism can alter phyto-hormone signals through intersecting with plant defense pathways to increase resistance against bacterial pathogens [15]. In rice, RNAi can knockdown OsSSI2 (OsSSI2-kd), meant for fatty acid desaturase activity that cause increase resistance against bacterial pathogen (Xanthomonas oryzae pv. Oryzae) of leaf blight and blast fungus (Magnaporthe grisea) [16]. siRNAs proved effective against the crown gall disease in Arabidopsis, Nicotiana and Lycopersicum species caused by a pathogen Agrobacterium tumefaciens by transformation of inverted repeats of this pathogen genes ipt and iaaM to encode precursors of biosynthesis for auxin and cytokinin [17]. Gene silencing can be obtained by host-induced gene (Avra10), that results in limited fungal disease attach in wheat (Triticum aestivum) and barley (Hordeum vulgare) through a transient gene expression resistant to RNAi because of silent point mutations. This suggests that the transfer of RNA from host plant to fungal pathogen Blumeria graminis, leads to RNAi-based plant protection against these pathogens [17].
RNA silencing also employs as a natural antiviral defense mechanism to cause resistance against viral diseases by virus-induced gene silencing [18]. RNA silencing hosts target protein translation and process the virus-mediated dsRNA, which results by pathogen replication into vsiRNAs (virus-mediated siRNAs). The vsiRNAs then target and suppress gene expression and protein translation in the virus genes. For stabilization of the defense system, virus encodes 'viral suppressor of RNA silencing protein', that has been identified and isolated from various plant virus [19]. Scorza et al. (2001) revealed for the first time RNAi role for virus resistance in woody perennial species and produced Plum pox virus (PPV) resistant plants containing the PPV coat protein gene [20]. Plants can also control viral diseases by RNAi and reveal resistance when having proper anti-sense or hairpin RNAi constructs [21].
RNAi for plant insects/pests resistance
Plant breeders and biotechnologists are using different approaches to develop insect/pest resistant varieties. Although classical breeders have been developed various insect/pest resistant cultivars, however, this approach is tedious and time consuming as complexity increases with some added traits. The practice of using pesticides to control pests has become a common approach around the world, but having dramatic health and environmental effects its use seems to be very limited in coming years. Among transgenic approaches to control specific insect/pest, Bt-based toxins proved effective and replaced chemical insecticides in many crops. Most of the commercially used biotechnological approaches to control insect/pests on crops are subjected to expression of Bt insecticidal proteins which help in the permeabilization of gut epithelial cell's membrane in susceptible insects [22, 23]. However, this approach is limited for some specific crops to manage some specific pests, and there is also a threat that some insects can develop resistance against Bt [24]. After the successful induction of transgene-encoded RNAi in plants [25], biotechnologists speculated about crop protection from insects through genetic-engineering to exhibit dsRNAs target insect genes and recently, application of dsRNA for knocking specific genes has been well-documented (Table 1).
RNAi offers robust and more selective pathway for battling with various destructive insect/pests that cause significant economic losses. Mao et al. (2007) reported a new strategy about plant-mediated herbivorous insects RNAi, which describes the suppression of a critical insect-gene through insect feeding on plant engineered to develop a specific dsRNA that can prompt dissection of gene functions in these insects [26]. They further revealed the herbivorous insect RNAi efficiency can be stimulated by ingestion of transgenic dsRNA producing plants that is gene-specific and proved effective against cotton bollworms (Helicoverpa armigera) damage. A gene 'CYP6AE14' was identified in Helicoverpa armigera [26]. This identified gene expressed in the insect midgut was correlated with larva growth when food contains gossypol. Therefore, after feeding on plant material exhibiting dsRNA specific to gene 'CYP6AE14', the effect of the transcript decreased in midgut and larva growth also retarded [26]. The gene silencing of 'glutathione-S-transferase' (GST1) can trigger RNAi process when herbivorous insects feed on plant material expressing dsRNA [26]. When dsRNA was injected in whitefly body cavity, RNAi was induced that knocked-down the genes expression to 70% in midgut as well as salivary glands of whitefly [27]. RNAi-mediated death of whitefly via oral dissemination of dsRNA, targets ADP/ATP, translocase, a-tubulin, ribosomal protein L9 actin ortholog and v-ATPaseA genes responsible for insects mortality [28].
Interference in expression of the targeted genes results various phenotype disturbances viz. stunted growth, moulting defects, and insect mortality [29]. Baum et al. (2005) demonstrated in western corn rootworm (WCR) ingestion of dsRNAs provided in diet can trigger the RNAi which result in stunted larva growth and their death [30]. They further revealed that transgenic corn plants which are engineered for WCR dsRNAs expression exhibited a substantial reduction in insect damage that suggests RNAi pathway is effective and can be exploited further to control coleopteran insects [30]. This mechanism was also practiced in Acyrthosiphon pisum for silencing of a gene C002 expressed in insect salivary glands that proved lethal for pea aphids [31]. Later on, Pitino et al. (2011) used this approach to silence genes: Rack-1 and MpC002, expressed in salivary glands and gut tissues of Myzus persicae (green peach aphid) and reported that knock-down of these genes reduced aphid proliferation [32].
RNAi has also been exploited in plants to develop resistance against nematodes [33, 34] and this approach has appeared as a novel tool to control plant parasitic nematodes [35]. dsRNAs can be produced through engineered plants that have the ability to silence target genes in nematode body. The delivery of dsRNAs from plant to nematode occurs by the ingestion process of plant cytoplasm and after its injection into the nematode body accelerate the RNAi that results in inactivation of targeted genes through dsRNA [36]. Nevertheless, to target host genes destined for interaction, efficient regulation of dsRNA triggers expression is a prerequisite that will reduce the negative effects on plant growth and development and it also necessitate identification of nematode-responsive promoters. In plant-parasitic nematodes, it is important to understand the RNAi mechanism for loss-of-function nematode phenotypes through effective gene silencing. Transgenic plants producing RNAi triggers expression of nematode targets proved more effective against root knot nematode compared with cyst nematode [37]. It has been reported that in vitro ingestion of 16D10 dsRNA gene results the target parasitism gene silencing in root knot nematodes and reduced nematode lethality, whereas, in Arabidopsis, in vivo expression of 16D10 dsRNA gene also increase the resistance against four species of root knot nematodes. Since, there is no single natural resistant gene known against root knot nematodes, expression of dsRNA to silence target genes for disruption of the parasitism effect significantly proved a sustainable approach for robust RKN resistant cultivars [38]. RNAi permits the molecular determinants regarding parasitism and also can make it possible to identify novel specified targets vital for survival of nematodes and thus leads to the development of efficient control methods [37].
Use of RNAi technology in various plant species against different insect/pests and pathogens.
| Crop | Insect/Pathogen | Objective | Targeted genes | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | Meloidogyne species | Utilization of RNAi to silence the parasitism gene | 16D10 | [8] |
| Oryza sativa L | Magnaporthe grisea and Xanthomonas oryzae pv. oryzae | Functional analysis of a rice homolog SSI2 (OsSSI2) for disease resistance | OsSSI2 | [16] |
| Prunus domestica L. | Plum pox virus (PPV) | To exploit the role of PTGS (RNAi) for virus resistance in a woody perennial species | PPV coat protein gene | [20] |
| Gossypium hirsutum | Helicoverpa armigera | Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi | Cytochrome P450 gene (CYP6AE14) | [26] |
| Nicotiana rustica | Bemisia tabaci | Enhanced whitefly resistance via expressing double stranded RNA | v-ATPaseA | [29] |
| Zea mays | Diabrotica virgifera virgifera LeConte | Control of coleopteran insect pests through RNA interference | Genes encoding proteins | [30] |
| Medicago sativa | Acyrthosiphon pisum | RNAi knockdown of a salivary transcript | C002 | [31] |
| Nicotiana benthamiana and Arabidopsis thaliana | Myzus persicae | To develop the plant-mediated RNAi technology for aphid resistance | M. persicae Rack1, M. persicae C002 (MpC002) | [32] |
| Nicotiana rustica | Helicoverpa armigera | Improvement of pest resistance in transgenic tobacco plants expressing dsRNA | EcR | [34] |
| Citrus aurantifolia | Citrus tristeza virus (CTV) | Transformation to generate transgenic plants carrying the coat protein gene of CTV | CTV-CP | [83] |
| Juglans regia L. | Agrobacterium tumefaciens | Application of oncogene silencing technology in the generation of crown gallresistant crops. | Tryptophan mono-oxygenase (iaaM) and isopentenyl transferase (ipt) | [84] |
| Genus Malus | Agrobacterium tumefaciens | To provide effective method to produce crown gall resistant apple plants | iaaM, iaaH, and ipt | [85] |
| Oryza sativa L. | Nilaparvata lugens | Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference | NlHT1, Nlcar, Nltry | [86] |
| Genus Malus | Venturia inaequalis | To use to hairpin vector approach for resistance against V. inaequalis | GFP transgene and tri-hydroxy-naphthalene reductase gene (THN) | [87] |
Plants armed with dsRNA prevent the insect/pests damages, as the transgenic cultivars that produced dsRNAs can target certain genes in insect tissues to reveal dominance in gene expression and caused their mortality [26, 39]. A robust RNAi pathway proved to be effective against different insects and pests which have open new pathways for crop protection by developing insect resistant cultivars of commercially important plants. Nevertheless, the success of the RNAi approach to control notorious insect/pests is mainly co-related with the wise screening system for target gene selection and an appropriate delivery mechanism [40].
RNAi for crop quality improvement
Traditionally, tremendous improvement in the crop quality has been done through conventional breeding, but this approach is time consuming and labor intensive. With the revolution in genetic engineering, biotechnologists were enthusiastic to employ this technology for improved crop quality and its nutritional status. RNAi, being a novel approach has great potential to modify the gene expression in plants for better quality traits and nutritional improvement in different crops. This approach facilitates the target gene and relative pathway identification and development of vectors for RNAi constructs for transformation and evaluation of lines for screening quality traits [41]. Seedless-ness is a desired quality trait of fruits and vegetables and RNAi can play a key role in achieving this goal. RNAi enables repression of gibberellic acid and auxin signal pathways after a reduction in the level of SlARF7 transcript responsible for pollination and fertilization in tomato plants [42]. These results by-pass the auxin signaling-fertilization pathway that leads to the development of parthenocarpic fruits having great commercial value. Carotenoid's production such as β-carotene and lutein were reported higher in potato through gene silencing of β-carotene hydroxylase [43]. The post-harvest life can enhance by knowing-out genes responsible for ethylene production in tomato [44]. This was achieved through introducing dsRNA and blocking the gene expression of ACC-oxidase which significantly reduced the ethylene formation and enhanced shelf-life in tomato. RNAi suppression of α-mannosidase and β-acetylhexosaminidase associated with fruit softening also increased the shelf-life in tomato fruits [45]. Increase in amylose contents in wheat by suppressing two genes (SBEIIa and SBEIIb) meant for starch-branching enzyme was well demonstrated by [46]. Whereas, in maize, it was used to knock-out the storage protein that had low lysine ratio (22-kD maize zein) [47]. RNAi could be exploited as a metabolic engineering tool for the production and synthesis of commercially valuable plant products such as alkaloid production (codeine, quinine, vincristine, scopolamine), biosynthesis of essential oil and flavoring agents (vanillin) [41].
RNAi for abiotic stress tolerance
Abiotic stress is a serious hazard for the life on earth, particularly plants whose growth and yield affected negatively. It is accepted as a chief source for crop devastation with respect to loss in quality and quantity as well as considered a tremendous constrains in productivity [48, 49]. It has been reckoned that nearly seventy percent of crops yield diminution is the direct consequence of abiotic stress [50]. In addition, climate alteration has aggravated the regularity and harshness of several abiotic stresses, principally elevated temperature and drought, with considerable reductions in yield of main cereals like maize, wheat and barley [51]. Years of selection and, in recent times, manipulation of the genetic architecture of crops for adaptation to abiotic stresses have been indispensable to ameliorate productivity, yield stability, and quality of product in food and fodder crop species [49]. Plants have adapted numerous physiological, bio-chemical and metabolic approaches for the purpose of encountering the abiotic stresses. Normally, it is tricky to envisage the complicated pathway of signaling that are stimulated and turned off in response to different abiotic stresses [52]. Classical techniques of breeding crop plants with greater tolerance to abiotic stresses have until now achieved inadequate success [53, 54]. It is because of a number of casual factors, including: (1) yield was the major focused of breeders rather than explicit traits; (2) the complexities in tolerance traits breeding, that include complications commenced by genotype × environment; and (3) intended traits could only be incorporated from the species that are closely related [54]. Transgenic methods are one of the numerous tools offered improvement in modern plants breeding programs. Gene detection and functional genomics programs have discovered innumerable protocols and gene families, which insure higher production and adjustment to abiotic stresses. These groups of genes can be incorporated into innovative arrangement, expressed ectopically, or delivered to the crops that are lacking these genes [49].
Molecular marker techniques are helpful to elucidate stress related traits by quantitative trait locus (QTL) mapping in order to locate the individual loci through marker-assisted selection [55]. Genomics entails genome study; transcriptome, including functional and structural examination of coding and non-coding RNA, protomics concerns with the formation of protein and post translational protein alteration together with their pathway of regulation and metabolism that offer a commanding tool in discovering the intricate network contributed in stress tolerance [56]. RNAi is an ultimate appealing and an invigorating phenomenon in which short double strand RNA (dsRNA) averts the specific gene expression by inducing degeneration in the chain sequence of particular target messenger RNA in the cytoplasm.
Current findings manifested that RNAi is playing an imperative role in abiotic stresses stimulation in different crops. The function of miRNAs (microRNA) in relation to abiotic stress like oxidative stress, cold, drought, and salinity were reported by Shaker and Zhu (2004) [57] in Arabidopsis plants under various abiotic stress and confirmed miR393 was sturdily up-regulated when exposed to higher salinity levels, dehydration, cold, and abscisic acid (ABA). Additionally, miR402, miR319c, miR397b, and miR389a were controlled by abiotic stress under varying levels in Arabidopsis [58]. RNAi technology may be a substitute of complex molecular techniques because of containing several benefits: its specificity and sequence-based gene silencing. This ability of RNAi has been efficaciously utilized for incorporating desired traits for abiotic stress tolerance in various plants species [58].
Drought stress tolerance
Drought is the most momentous ecological stress on agriculture production round the world and tremendous attempts has been made by plant scientist to increase productivity of crops in order to cope with diminishing water availability [59]. The potential of a plant to uphold enough water balance inside the tissues (turgor/turgidity) when faced a drought condition is an indication of drought tolerance. Gene expressions investigation has depicted that drought-specific allele could be classified into three major groups: (1) genes implicated in signal transduction pathways (STPs) and transcription process; (2) genes involved in protection of protein activity and membrane; and (3) genes facilitating the ion uptake and water transport [60-63].
In relation to drought responses, miR159 were reported in triggering the signaling of hormone in Arabidopsis [64]. Furthermore, miR169g and miRNA393 genes have been observed in rice crop which were stimulated under drought conditions [65]. Among genetically engineered plants the rice exhibited gene expression of RACK1 inhibition caused by RNAi, which explained the potential role of RACK1 to drought stress in rice crop. The transgenic rice was observed with a superior level of tolerance in contrast to non-transgenic rice plants [66]. In many plants such as Arabidopsis, Populus trichocarpa, and Oryza sativa the miRNA expression profiling has been performed under drought stress. miR169, miR396, miR165, miR167, miR168, miR159, miR319, miR171, miR394, miR393, miR156, and miR158 were made known to be drought-responsive [67].
Analysis of miRNAs and genome sequencing profiling were executed in drought-studied rice at a various range of growth stages, from tiller formation to inflorescence, utilizing a microarray platform. The results suggested that 16 miRNAs (miR1126, miR1050, miR1035, miR1030, miR896, miR529, miR408, miR156, miR171, miR170, miR168, miR159, miR397, miR396, miR319, miR172 and miRNA1088) were remarkably involved in down regulation in response to drought stress [67].
In contrast, 14miRNAs (miR1125, miR159, miR903, miR169, miR901, miR171, miR896, miR319, miR395, miR854, miR851, miR474, miR845, and miRNA1026) were found in up-regulation under drought stress. Few miRNAs gene families, like miR319, miR896, and miR171 were recognized as both up- and down regulated groups [68]. In Populus, miR1447, miR1445, miR171l-n, and miR1446a-e has been identified as a drought-responsive [69]. In P. vulgaris, miR2119, miR1514a, and miRS1exhibted a gentle but obvious increase in accretion upon drought treatment, on the other hand, the accumulation was higher for miR2118, miR159.2, and miR393 in reaction to the identical treatment [70]. miR169 found down-regulated in the roots only when studied in Medicago truncatula, while miR408 and miR398a,b were highly up-regulated in roots as well as shoots also under drought stress [71]. In recent studies, miRNA expressing patterns of drought tolerance wild emmer wheat in relation to drought-stress explored by utilizing a plant miRNA microarray platform [72]. At the same time, up regulation throughout drought stress in maize crop has been studied by miR474, which interact with proline dehydrogenase (PDH) [73].
Salt stress tolerance
Our planet has copious amount of salt in soil that limits the agricultural productivity, as it has been estimated that 20% of agricultural land is salt-affected, tremendously decreasing efficiency of the production potential of germplasm. Meanwhile, soil salinity is designated a serious threat due to reduction in available irrigation water quality [74]. Tolerance to salinity is a poly genic character, in many crops such as, rice, soybean, wheat, barley, tomato, and citrus its quantitative trait loci (QTL) have been identified [75].
Genetic techniques presently being used to enhance tolerance against salinity with the help of using bioinformatics, functional genomics, and genetic variations either through selection in stressed environments or via QTLs mapping followed by marker assisted selection [76]. Many regulated miRNAs have been reported in salinity stressed plants. In Arabidopsis, miR397, miR156, miR394, miR158, miR393, miR159, miR319, miR165, miR171, miR167, miR169, miR168, and miR398 were up-regulated in reaction to salinity stress, whilst the accumulation of miR398 was reduced [67]. In P. vulgaris, it was reported that increment in accumulation of miR159.2 and miRS1 with the addition of NaCl [70]. In P. trichocarpa, miR171l-n, miR530a, miR1446a-e, miR1445, and miR1447 were down regulated; on the other hand, miR1450 and miR482.2 were up-regulated in salt stress period [69]. Recently, a research investigation was carried out by using microarray to elucidate the miRNA profile salinity-tolerant and a salt-sensitive line of maize; the findings indicated that members of the miR396, miR156, miR167, and miR164 groups were down-regulated, while miR474, miR162, miR395, and miR168 groups were up-regulated in saline-stressed maize roots [77].
Cold and heat stress tolerance
In Arabidopsis, Brachypodium, and Populus species, miRNA expression has been studied in case of cold stress [67, 70, 78] miR169 and miR397 were up-regulated in all aforementioned species, and miR172 was regulated upward in Brachypodium and Arabidopsis. Additionally, many miRNAs (miR408, miR393, miR165/166, and miR396) were induced in Arabidopsis under cold stress; on the other hand, some miRNAs (miR398, miR156/157, miR394, miR159/319, and miR164) exhibited either transitory or gentle regulation when exposed to cold stress [67]. miRNA in wheat showed variant expression in heat stress response; researchers cloned the miRNAs from the leaves of wheat after treating with heat stress, with the help of Solexa high-throughput sequencing. In wheat, 32 families of miRNA distinguished, among them 9 identified miRNAs were supposed heat responsive. For instant, miR172 was distinctly decreased, while miRNAs including (miR827, miR156, miR169, miR159, miR168, miR160, miR166, and miR393) were noticed with up regulation in response to heat stress [79].
UV-B radiation stress tolerance
A computer based technique was used to identify miRNAs in Arabidopsis stimulated by UV-B radiation. Among the 21 miRNAs associating to eleven miRNAs groups sorted out in the study, the under mentioned were expected to be involved in up regulation in response to UV-B stress: miR401, miR156/157, miR393, miR159/319, miR160, miR172, miR165/166, miR170/171, miR167, and miR398 [80]. Few of the similar families that were identified to be up-regulated in Arabidopsis by UV-B radiation (miR168, miR156, miR167, miR160, miR398, and miR165/166) were discovered to be involved in up-regulation in Populus termula by UV-B radiation. Moreover, 3 families (miR393, miR159, and miR169) that were found as a down regulating in P. termula were up-regulated in Arabidopsis implying that some UB-V radiation stress responses could be species-specific [81].
Mechanical stress tolerance
Plants face mechanical stress that is also attributed to a dynamical and static stress when stems or branches are twisted by external forces, as wind or gravity. In an investigation of P. trichcarpa, Pt-miRNA levels of transcript were compared in compression stressed or tension stressed xylem with the non-stressed xylem. miR408 showed up-regulation while miR156, miR48, miR162, miR475, miR164, and miR480 were down-regulated by compressing and tension. miR168 was regulated upward only under tension stressed tissue, but miR172 and miR160 were down regulated in compressed tissue. These findings revealed that miRNAs may be regulated in mechanical stress and could play a role in defense system for mechanical and structural fitness [82].
Conclusion
RNA interference (RNAi) has recently become a highly effective and powerful tool of functional genomics for silencing the gene expression for crop improvement. Nevertheless, RNAi stability in plants is critical, but RNAi-mediated gene suppression approach open new avenues in the development of eco-friendly biotech approaches for crop improvement by knocking-out the specific genes for better stress tolerance and integrating novel traits in various plant species including insect/pest/pathogen resistance and enhanced nutritional status. The RNAi is a sophisticated technology having revolutionary capabilities could be further exploited for functional analysis of target genes and regulation of gene expression for crop protection and improvement.
Acknowledgements
This work was carried out with the support of "Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ00929609)" Rural Development Administration, Republic of Korea.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bosher JM, Labouesse M. RNA interference: genetic wand and genetic watchdog. Nat Cell Bio. 2000;2:31-36
2. Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Gen. 2007;8:173-84
3. Tierney MB, Lamour KH. An introduction to reverse genetic tools for investigating gene function. The Plant Health Instructor. 2005 doi: 10.1094/PHI-A-2005-1025-01
4. Napoli C, Lemieux C, Jorgensen R. Introduction of chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279-289
5. van der Krol AR, Mur LA, Beld M, Mol JNM, Stuitje AR. Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2:291-299
6. Agrawal N, Dasaradhi PVN, Mohommed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA Interference: Biology, Mechanism, and Applications. Microbiol Mol Biol Rev. 2003;67:657-685
7. Arenz C, Schepers U. RNA interference: from an ancient mechanism to a state of the art therapeutic application? Naturwissenschaften. 2003;90:345-359
8. Hannon GJ. RNA interference. Nature. 2002;418:244-251
9. Watson JM, Fusaro AF, Wang M, Waterhouse PM. RNA silencing platforms in plants. FEBS Lett. 2005;579:5982-5987
10. Duan CG, Wang CH, Guo HS. Application of RNA silencing to plant disease resistance. Silence. 2012;3:5
11. Wani SH, Sanghera GS, Singh NB. Biotechnology and plant disease control-role of RNA interference. Am J Plant Sci. 2010;1:55-68
12. Almeida R, Allshire RC. RNA Silencing and genome regulation. Trends Cell Bio. 2005;15:251-258
13. Senthil-Kumar M, Mysore KS. RNAi in Plants: recent developments and applications in agriculture. Gene Silencing: Theory, Techniques and Applications. 2010 183-199
14. Fritz JH, Girardin SE, Philpott DJ. Innate immune defense through RNA interference. Science STKE. 2006;2006:pe27 doi: 10.1126/stke.3392006pe27
15. Kang L, Yuh-Shuh W, Srinivasa RU, Keri W, Yuhong T, Vatsala V, Barney JV, Kent DC, Elison BB, Kirankumar SM. Overexpression of a fatty acid amide hydrolase compromises innate immunity in Arabidopsis. Plant J. 2008;56:336-349
16. Jiang CJ, Shimono M, Maeda S, Inoue H, Mori M, Hasegawa M, Sugano S, Takatsuji H. Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol Plant-Microbe In. 2009;22:820-829
17. Escobar MA, Civerolo EL, Summerfelt KR, Dandekar AM. RNAi-mediated oncogene silencing confers resistance to crown gall tumorigenesis. Proc Nat Acad Sci. USA. 2001;98:13437-42
18. Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10:632-644
19. Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413-426
20. Scorza R, Callahn A, Levy L, Damsteegt V, Webb K, Ravelonandro M. Post-transcriptional gene silencing in plum pox virus resistant European plum containing the plum pox potyvirus coat protein gene. Transgenic Res. 2001;10:201-209
21. Sudarshana MR, Roy G, Falk BW. Methods for engineering resistance to plant viruses. Methods Mol Bio. 2007;354:183-195
22. Rajamohan F, Lee MK, Dean DH. Bacillus thuringiensis insecticidal proteins: Molecular mode of action. In: Progress in nucleic acid research and molecular biology. 1998;60:1-27
23. Vaughn T, Cavato T, Brar G, Coombe T, DeGooyer T, Ford S, Groth M, Howe A, Johnson S, Kolacz K, Pilcher C, Purcell J, Romano C, English L, Pershing J. A method of controlling corn rootworm feeding using a Bacillus thuringiensis protein expressed in transgenic maize. Crop Sci. 2005;45:931-93
24. Gordon KHJ, Waterhouse PM. RNAi for insect-proof plants. Nat Biotechnol. 2007;25:1231-1232
25. Waterhouse PM, Graham MW, Wang MB. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Nat Acad Sci USA. 1998;95:13959-13964
26. Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY. Silencing a cotton bollworm P450 monooxygenase gene by plant mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25:1307-1313
27. Ghanim M, Kontsedalov S, Czosnek H. Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius). Insect Biochem Mol Biol. 2007;37:732-738
28. Upadhyay SK, Chandrashekar K, Thakur N, Singh PK, Tuli R. et al. RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J Biosci. 2011;36:153-161
29. Thakur N, Upadhyay SK, Verma PC, Chandrashekar K, Tuli R, Singh PK. Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase a gene. PLoS ONE. 2014;9:e87235. doi:10.1371/journal.pone.0087235
30. Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, Vaughn T, Roberts J. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322-1326
31. Mutti NS, Park Y, Reese JC, Reeck GR. RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J Insect Sci. 2006;6:1-7
32. Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA. Silencing of aphid genes by dsRNA feeding from plants. PLoS ONE. 2011;6:e25709. doi:10.1371/journal.pone.0025709
33. Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811
34. Graham LD, Kotze AC, Fernley RT, Hill RJ. An ortholog of the ecdysone receptor protein (EcR) from the parasitic nematode Haemonchu contortus. Mol Biochem Parasitol. 2010;171:104-107
35. Lilley CJ, Davies LJ, Urwin PE. RNA interference in plant parasitic nematodes: a summary of the current status. Parasitol. 2012;139:630-40
36. Gheysen G, Vanholme B. RNAi from plants to nematodes. Trends in Biotechnol. 2007;25:89-92
37. Rosso MN, Jones JT, Abad P. RNAi and functional genomics in plant parasitic nematodes. Ann Rev Phytopathol. 2009;47:207-32
38. Huang G, Allen R, Davis EL, Baum TJ, Hussey RS. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Nat Acad Sci. USA. 2006;103:14302-14306
39. Zhu JQ, Liu S, Ma Y, Zhang JQ, Qi HS, Wei ZJ, Yao Q, Zhang WQ, Li S. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS ONE. 2012;7:e38572. doi:10.1371/journal.pone.0038572
40. Wang Y, Zhang H, Li H, Miao X. Second-generation sequencing supply an effective way to screen RNAi targets in large scale for potential application in pest insect control. PLoS ONE. 2011;6:e18644
41. Saurabh S, Vidyarthi AS, Prasad D. RNA interference: concept to reality in crop improvement. Planta. 2014;239:543-564
42. DeJong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 2009;5:160-170
43. Eck JV, Conlin B, Garvin DF, Mason H, Navarre DA, Brown CR. Enhanced beta-carotene content in potato via RNAi silencing of the beta-carotene hydroxylase gene. Am J Potato Res. 2007;84:l331-l1342
44. Xiong A, Yao Q, Peng R, Li X, Han P, Fan H. Different effects on ACC oxidase gene silencing triggered by RNA interference in transgenic tomato. Plant Cell Rep. 2005;23:639-646
45. Meli VS, Ghosh S, Prabha TN, Chakraborty N, Chakraborty S, Datta A. Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc Nat Acad Sci. USA. 2010;107:2413-2418
46. Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li Z, Rahman S, Morell M. High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc Nat Acad Sci. USA. 2006;103:3546-3551
47. Segal G, Song R, Messing J. A new opaque variant of maize by a single dominant RNA-interference-inducing transgene. Genetics. 2003;165:387-97
48. Tardieu F, Tuberosa R. Dissection and modelling of abiotic stress tolerance in plants. Curr Opin Plant Biol. 2010;13:206-212
49. Jewell MC, Campbell BC, Godwin ID. Transgenic plants for abiotic stress resistance. Transgenic Crop Plants. 2010 doi 10.1007/978-3-642-04812-8-2
50. Acquaah G. Principles of plant genetics and breeding. Oxford, UK: Blackwell. 2007
51. Lobell DB, Field CB. Global scale climate - crop yield relationships and the impacts of recent warming. Environ Res Lett. 2007;2:014002
52. Chawla K, Barah P, Kuiper M, Bones AM. Systems biology: a promising tool to study abiotic stress responses. Omics and Plant Abiotic Stress Tolerance. 2011:163-172
53. Richards RA. Defining selection criteria to improve yield under drought. Plant Growth Regul. 1996;20:57-166
54. Tester M, Bacic A. Abiotic stress tolerance in grasses from model plants to crop plants. Plant Physiol. 2005;137:791-793
55. Witcombe JR, Hollington PA, Howarth CJ, Reader S, Steele KA. Breeding for abiotic stresses for sustainable agriculture. Phil Trans R Soc Lond B Biol Sci. 2008;363:703-716
56. Gupta B, Sengupta A, Saha J, Gupta K. Plant abiotic stress: 'Omics' approach. Plant Biochem Physiol. 2013;1:e108. doi:10.4172/2329-9029.1000e108
57. Sunkar R, Zhu JK. Novel and stress-regulated micro RNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16:2001-2019
58. Jagtap UB, Gurav RG, Bapat VA. Role of RNA interference in plant improvement. Naturwissenschaften. 2011;98:473-492
59. Cattivelli L, Rizza F, Badeck FW, Mazzucotelli E, Mastrangelo AM, Francia E, Mare C, Tondelli A, Stanca AM. Drought tolerance improvement in crop plants: an integrated view from breeding to genomics. Field Crops Res. 2008;105:1-14
60. Vijn I, Smeekens S. Fructan: more than a reserve carbohydrate? Plant Physiol. 1999;120:351-359
61. Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377-403
62. Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross talk between two stress signalling pathways. Curr Opin Plant Biol. 2000;3:217-233
63. Smirnoff N. Plant resistance to environmental stress. Curr Opin Biotechnol. 1998;9:214-219
64. Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by gibberellin-regulated microRNA. Development. 2004;131:3357-3365
65. Zhao BT, Liang RQ, Ge LF, Li W, Xiao HS, Lin HX, Ruan KC, Jin YX. Identification of drought-induced microRNAs in rice. Biochem Biophys Res Commun. 2007;354:585-590
66. Jian X, Zhang L, Li G, Zhang L, Wang X, Cao X, Fang X, Zha FC. Identification of novel stress-regulated microRNAs from Oryza sativa L. Genomics. 2010;95:47-50
67. Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836-843
68. Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot. 2010;61:4157-4168
69. Lu SF, Sun YH, Chiang VL. Stress-responsive microRNAs in Populus. Plant J. 2008;55:131-151
70. Arenas-Huertero C, Pérez B, Rabanal F. et al. Conserved and novel miRNAs in the legume Phaseolus vulgaris in response to stress. Plant Mol Biol. 2009;70:385-401
71. Trindade I, Capitão C, Dalmay T, Fevereiro M, Santos D. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta. 2010;231:705-716
72. Kantar M, Lucas S, Budak H. miRNA expression patterns of Triticum dicoccoides in response to shock drought stress. Planta. 2010;233:471-84
73. Wei L, Zhang D, Xiang F, Zhang Z. Differentially expressed miRNAs potentially involved in the regulation of defense mechanism to drought stress in maize seedlings. Int J Plant Sci. 2009;170:979-989
74. Flowers TJ. Improving crop salt tolerance. J Exp Bot. 2004;55:307-319
75. Flowers TJ, Flowers SA. Why does salinity pose such a difficult problem for plant breeders? Agr Water Manag. 2005;78:15-24
76. Yamaguchi T, Blumwald E. Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci. 2005;10:615-620
77. Ding D, Zhang L, Wang H, Liu Z, Zhang Z, Zheng Y. Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot. 2009;103:29-38
78. Zhang J, Xu Y, Huan Q, Chong K. Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics. 2009;10:449 doi:10.1186/1471-2164-10-449
79. Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 2010;10:123
80. Zhou X, Wang G, Zhang W. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol Syst Biol. 2007;3:103
81. Jia X, Ren L, Chen QJ, Li R, Tang G. UV-B-responsive microRNAs in Populus tremula. J Plant Physiol. 2009;166:2046-2057
82. Lu SF, Sun YH, Shi R, Clark C, Li LG, Chiang VL. Novel and mechanical stress responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186-2203
83. Domínguez A, De Mendoza AH, Guerri J, Cambra M, Navarro L, Moreno P, Peña L. Efficient production of transgenic citrus plants expressing the coat protein gene of Citrus tristeza virus. Plant Cell Rep. 2000;19:427-433
84. Escobar MA, Leslie CA, Mcgranahan GH, Dandekar AM. Silencing crown gall disease in walnut (Julgans regia L.). Plant Sci. 2002;163:591-597
85. Viss WJ, Pitrak J, Humann J, Cook M, Driver J, Ream W. Crown-gall-resistant transgenic apple trees that silence Agrobacterium tumefaciens oncogenes. Mol Breeding. 2003;12:283-295
86. Zha W, Peng X, Chen R, Du B, Zhu L, He G. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the Hemipteran insect Nilaparvata lugens. PLos ONE. 2011;6:e20504. doi:10.1371/journal.pone.0020504
87. Fitxgerald A, Van Kha JA, Plummer KM. Simultaneous silencing of multiple genes in the apple scab fungus Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet Biol. 2004;41:963-971
Author contact
![]() Corresponding author: kblimac.kr, phone: +82-53-950-5726; fax: +82-53-950-5722.
Corresponding author: kblimac.kr, phone: +82-53-950-5726; fax: +82-53-950-5722.

 Global reach, higher impact
Global reach, higher impact