10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(1):31-37. doi:10.7150/ijbs.10615 This issue Cite
Review
Epithelial Cell Polarity Determinant CRB3 in Cancer Development
1. Center for Translational Medicine, the First Affiliated Hospital of Xi'an Jiaotong University
2. Department of Surgical Oncology, the First Affiliated Hospital of Xi'an Jiaotong University
Received 2014-9-23; Accepted 2014-10-30; Published 2015-1-1
Abstract
Cell polarity, which is defined as asymmetry in cell shape, organelle distribution and cell function, is essential in numerous biological processes, including cell growth, cell migration and invasion, molecular transport, and cell fate. Epithelial cell polarity is mainly regulated by three conserved polarity protein complexes, the Crumbs (CRB) complex, partitioning defective (PAR) complex and Scribble (SCRIB) complex. Research evidence has indicated that dysregulation of cell polarity proteins may play an important role in cancer development. Crumbs homolog 3 (CRB3), a member of the CRB complex, may act as a cancer suppressor in mouse kidney epithelium and mouse mammary epithelium. In this review, we focus on the current data available on the roles of CRB3 in cancer development.
Keywords: apical-basal polarity, CRB3, EMT, cancer
Introduction
Cell polarity, a widespread feature in unicellular and multicellular organisms, is defined as asymmetry in cell shape, organelle distribution and cell function [1]. Delicate regulation of cell polarity signaling is required for development and maintenance of tissue integrity. Cell polarity is essential in numerous biological processes, including cell growth, cell migration and invasion, and molecular transport [2]. Deregulation of cell polarity signaling may lead to the change of cell fate. There are four modes of cell polarity: planar cell polarity, apical-basal polarity, front-rear polarity, and mitotic spindle polarity [3]. In this review, we focus on apical-basal polarity, which is a characteristic of the epithelium and may be involved in cancer development.
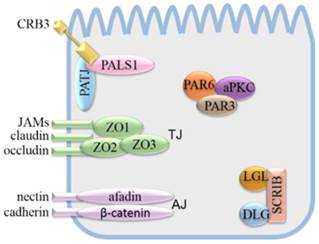
Apical-basal polarity maintenance mainly depends on cell polarity complexes and cell junction complexes. Apical-basal polarity formation starts with the establishment of various cell junction complexes [4, 5], including lateral adherens junctions (AJs), gap junctions, desmosomes, basal lamina hemidesmosomes, and apical tight junctions (TJs). AJs and TJs can interact with cell polarity complexes. The TJ is mainly composed of transmembrane proteins, including junctional adhesion molecules (JAMs), claudin and occludin, which combine with the actin cytoskeleton via the intracellular molecule ZO1, ZO2 or ZO3. The AJ isolates the apical membrane from the basolateral membrane. The AJ is mainly composed of transmembrane proteins, nectin and cadherin, which combine with the actin cytoskeleton via afadin and β-catenin, respectively.
Apical-basal polarity is established and maintained by the cooperation and antagonism of cell polarity complexes, which lead to the distribution of cellular molecules into specific cellular areas [6-8]. Apical-basal polarity is mainly regulated by three evolutionarily conserved polarity protein complexes, namely, the Crumbs (CRB) complex, partitioning defective (PAR) complex, and Scribble (SCRIB) complex. The SCRIB complex is composed of Scribble, Discs large (DLG) and Lethal giant larvae (LGL). In epithelia, the SCRIB complex is located basolaterally and is needed for maintenance of the basolateral membrane domain [9-12]. The PAR and CRB complexes have mutually antagonistic relationships with the SCRIB complex. The PAR complex consists of PAR3, PAR6 and atypical protein kinase C (aPKC). It is localized in the TJ of the epithelium and cooperates with the CRB complex in regulating the apical junction formation [13]. The CRB complex consists of the transmembrane protein CRB and the cytoplasmic proteins protein associated with Lin seven 1 (PALS1) and Pals1-associated tight junction protein (PATJ) [14]. Crb, a single-pass type membrane protein, was initially identified in Drosophila by Jürgens et al [24, 25]. To date, three vertebrate Crumbs orthologs CRB1, CRB2, and CRB3 have been identified [15]. CRB1 is related to a severe form of degenerative retinitis pigmentosa (RP12) and Leber congenital amaurosis. Diseases associated with CRB2 include Leber congenital amaurosis and cleft lip [16, 17]. CRB3 is considered a cancer suppressor [18]. Of the three CRB proteins, only CRB3 is found to be widely expressed in epithelial cells, which is consistent with the early analystic result of Drosophila crb [19].
Schematic model of cell polarity complexes and cell junction complexes.
Structure and distribution of CRB3
CRB3 is comprised of a short extracellular signal peptide and three large intracellular domains [an FERM-binding domain (FBD), a SH3-binding domain, and a carboxy-terminal PDZ-binding domain (PBD)] (Fig.1). FBD is involved in polarity formation due to its ability to bind to PALS1 and Epb4.1l5 [20, 21]. PBD interacts with the PDZ domains of PALS1 and PAR6 and is also important for polarity formation. Comparatively speaking, it is quite interesting to note that CRB3 is the only human CRB that has a PxxP motif which can bind to the SH3 domain [22-24]. SH3 domain interacts with the transcription factor ZONAB (ZO1-associated nucleic acid-binding protein), a protein that is required for normal cell proliferation [25, 26].
CRB3 includes two splice variants, CRB3A and CRB3B. CRB3A is connected with PALS1 via the ERLI motif at the C terminus of CRB3A, while CRB3B is connected with PALS1 via the CLPI motif at the C terminus of CRB3B; thus the ERLI or CLPI motif is crucial for the formation of the CRB complex [14]. The two splice variants show differential functions in human epithelial cells. CRB3A is found at the plasma membrane and is important for formation of cell junctions and repression of human cancer cell growth. In contrast, CRB3B is localized in the nucleus and plays a specific role in the ciliogenesis and mitotic spindle construction via interacting and colocalizing with importin-β [23, 24, 27-30]. Carcinogenesis is either prevented or delayed when the nuclear localization of CRB3B is disrupted [31].
Protein domain structures of Drosophila crb and human CRB.
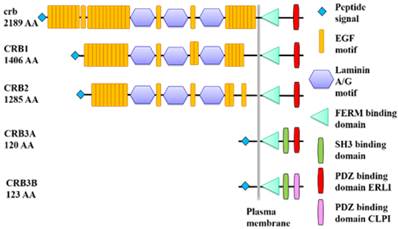
All CRB homolog proteins (crb, CRB1, CRB2, CRB3A and CRB3B) contain a signal peptide and a FERM binding domain (FBD). Extracellular parts of crb, CRB1 and CRB2 are composed of EGF and Laminin A/G domains. CRB3A and CRB3B have a SH3 binding domain. The PDZ binding domain of crb, CRB1, CRB2 or CRB3A is ended with ERLI, while the PDZ binding domain of CRB3B has a C-terminal sequence ending in CLPI.
CRB3 has been found to be expressed in humans, mice, rats, cows, and dogs, but not in Drosophila and zebrafish [27]. CRB3 is mainly localized above the TJ, a prime position at the apical membrane of the epithelium [32]. Drosophila crb has also been detected in the cytoplasm. The distribution of Drosophila crb is regulated by endocytosis [33], exocytosis [21], and the retromer. The retromer is a newly discovered retrograde complex which binds to the intracellular domain of crb via vacuolar protein sorting 35 (VPS35, a major component of the retromer) and recycles crb from endosomes to trans-Golgi network (TGN) in epithelia [34]. Unfortunately, little is known about CBR3 metabolism in mammals. The intracellular domains of crb and CRB3 are highly conserved. Therefore, it is reasonable to presume that CRB3 metabolism may be similar to that of Drosophila crb. Further studies are needed to validate this presumption.
CRB3 in apical-basal polarity maintenance
Eileen L Whiteman found that CRB3 knockout mice died shortly after birth and showed defects in epithelial morphogenesis (cystic kidneys, proteinaceous debris throughout the lungs, villus fusion and apical membrane blebs in intestines) [35]. The results suggest that CRB3 play crucial roles in growth and development of epithelial cells and the establishment and maintenance of apical-basal polarity.
CRB3 links to actin-binding proteins
During polarization, the CRB complex forms the apical domain by recruiting actin cytoskeleton regulators Moesin and βH-spectrin to the cell membrane [36]. Partners of CRB3 (i.e., Epb4.1l5, PAR6 and PALS1) serve as adaptors linking the CRB3 to the actin cytoskeleton. Lee et al observed an apical redistribution of F-actin and the formation of ectopic F-actin foci in Epb4.1l5 knockout mice [37]. PAR6 interacts with cytoskeleton-related proteins 14.3.3 [38] and Cdc42 [39]. In PALS1-downregulated cells, the pattern of F-actin staining obtained is more diffuse [40]. Whiteman et al have observed a direct interaction between CRB3 and the membrane-cytoskeleton linker protein Ezrin, which indicates that CRB3 can link the apical membrane to the cytoskeleton [35]. In addition, recruitment and reorganization of cytoskeletal elements (such as F-actin) through these proteins may contribute to the cell junctions formation.
CRB3 induces junctions formation
CRB3 seems to be a crucial determinant of TJ and AJ formation. Immortal baby mouse kidney (iBMK) epithelial cells selected in vivo has been found to display dramatic repression of CRB3 expression, which is associated with disruption of TJ formation. CRB3 expression restores TJ formation in TDCL 5D, where CRB3 endogenous expression is undetectable. Importantly, expressed CRB3 localizes to the plasma membrane and substantially rescues TJ formation, which is accompanied by increased occluding and ZO1 at the junctions between cells [18]. Downregulation of CRB3 results in loss of TJ, while overexpression of CRB3 disrupts the cell polarity by enlarging the apical domain at the expense of the lateral domain in MCF 10A cells [20]. In the normal Drosophila embryonic tissue, crb interacts with the Baz/PAR3 and forms the AJ in epithelial cells [41]. CRB3 can also form the CRB3/PAR6/aPKC complex and regulate the actin structure, endocytosis, and AJ remodeling [12, 42, 43].
CRB3 interacts with other polarity proteins
The CRB, PAR, and SCRIB complexes maintain apical-basal polarity by interacting with each other. For example, CRB3 determines the apical identity by recruiting and activating the downstream apical PAR complex and inhibiting the basolateral SCRIB complex. Crb competes with Par3 for binding to Par6 during TJ formation in Drosophila epithelium [23, 44]; Crb can also bind to Par3 and recruit it from the subapical domain during photoreceptor remodeling [45]. aPKC can regulate CRB3 activity in vivo by phosphorylating two conserved threonine residues in the cytoplasmic tail of CRB3 [14, 46]. Phosphorylation of proteins usually plays an important role in the process of cell signal transduction. However, the function of phosphorylated CRB3 in cell growth and development is still unknown.
CRB3 associates with epithelial-mesenchymal transition (EMT)
EMT is a biologic process that allows a polarized epithelium to undergo the change in cell cytoskeleton, lose the epithelial features of junctions, and acquire the mesenchymal characteristics such as migration and invasion, anti-apoptosis, and extracellular matrix degradation. EMT is initiated by transcriptional repressors, including ZEB1, ZEB2, Snail (SNAI1), Slug (SNAI2), FOXC2, TCF/LEF, TWIST and E47. These transcriptional repressors down-regulate the expressions of E-cadherin, EVA1, MAL2, P-cadherin and cytokeratin in epithelial cells and up-regulate mesenchymal cell markers N-cadherin and vimentin [47-51]. EMT and mesenchymal-epithelial transition (MET) are rapid and reversible phenomena occurring in the natural process of cell development, a variety of chronic diseases, and human cancer development and metastasis [52-55]. Migration and invasion are the leading causes of cancer-associated mortality [56]. Recent studies have suggested that EMT is a hallmark of development, migration and invasion of epithelial cancer cells. Drosophila crb can maintain the epithelial integrity by repressing the Rac1-PI3K positive-feedback loop embryos [57]. Rac1-PI3K up-regulates N-cadherin via JNK and induce EMT in mouse mammary epithelial cells [58].
Recent reports have provided evidence that members of the CRB complex are direct targets of the EMT transcriptional repressors, and CRB3 in turn affects on the expression of EMT transcriptional repressors. It has been found that CRB3, PATJ and PALS1 are up-regulated after the E-cadherin repressor ZEB1 is silenced in colorectal carcinoma cells SW480 and undifferentiated breast tumor cells MDA-MB-231 [59, 60]. The transcriptional repressor Snail alters epithelial cell polarity by repressing the transcription of CRB3 and abolishing the localization of Par and CRB complexes at cell-cell junctions in MDCK cells [61]. ZEB1 or Snail/Slug represses the mRNA expressions of CRB3 by directly binding to their promoter elements in normal and transformed epithelial cell lines [62]. CRB3 knockdown increases the expression of Snail in TGF-β-treated Eph4 cells grown at high density [63]. These findings suggest that CRB3 may inhibit the process of EMT via transcriptional repressors such as ZEB1 or Snail/Slug.
CRB3 in cancer
Cancer development is a multistep process caused by initial benign overproliferation and inhibition of apoptosis. The process by which benign cells are transformed to malignant cancer cells includes altered cell-matrix junctions and migration and metastasis to distal parts. Mammalian organs mainly consist of epithelial cells and epithelial cells are the source of approximately 90% of human cancers. Changes in multiple polarity proteins have been found in cancer tissues. For example, PAR6 is up-regulated in breast cancer cells and it induces cell proliferation by interacting with aPKC and cdc42 [64, 65]. Depletion of SCRIB in Drosophila cells results in activation of downstream oncogenes such as c-myc, destroying cell polarity, blocking 3D morphogenesis of cells, inhibiting apoptosis, enhancing proliferation, and promoting carcinogenesis [66].
Loss of CRB3 occurs concomitantly with increased expression of vimentin and reduced expression of E-cadherin, two important hallmarks of EMT. In addition, the expression level of CRB3 negatively correlates with migration and invasion of the mouse kidney epithelial cells. Reduced expression of the murine CRB3 gene also can promote carcinogenesis of kidney epithelia [18]. Loss of CRB3 enhances TGF-β signaling and predisposes cells to TGF-β-mediated EMT in mouse mammary epithelia [63].The above studies show that loss of CRB3 may favor EMT and migration and invasion of cancer cells. Presently, studies have been mainly focused on the regulatory role of CRB3 in Hippo and Notch signaling.
CRB3 regulates Hippo signaling
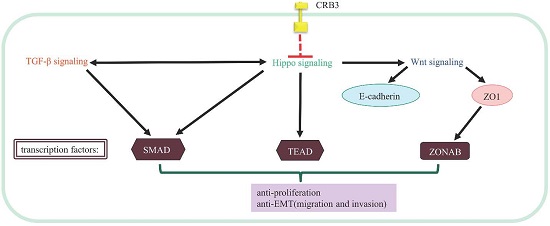
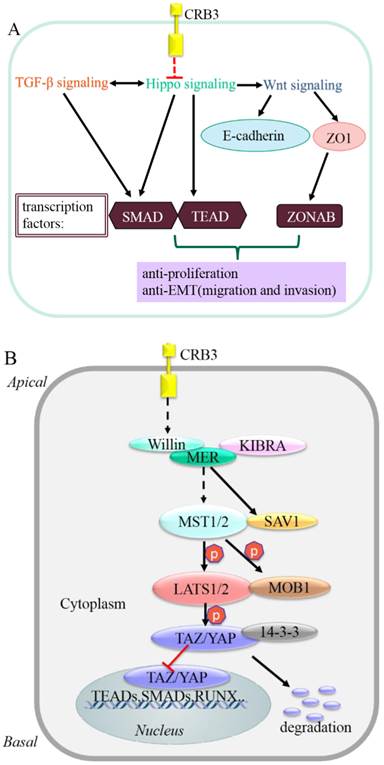
The molecular mechanism by which CRB3 exerts its cancer suppressive function may be related to its interaction with the Hippo signaling [9, 10, 67]. The Hippo signaling is a critical regulator of cell cycle, proliferation, apoptosis, differentiation and contact inhibition [68]. CRB3 regulates proliferation and apoptosis via mediation of the Hippo signaling. Crb suppresses cancer development in Drosophila by regulating the Hippo signaling via the FERM-domain protein Expanded [9, 10, 67]. However, the mechanism underlying the interaction between CRB3 and Hippo signaling in human is unclear. It has been suggested that FRMD6/Willin is a homologue of Expanded [69], and it also has an FERM domain. Whether CRB3 regulates the Hippo signaling in human needs to be further verified [70]. FRMD6/Willin may be regulated by CRB3 and forms a complex with KIBRA and MER. The complex ultimately phosphorylates the transcription co-activator Yes-associated protein (YAP)/the transcriptional co-activator with PDZ-binding domain (TAZ, also known as WWTR1). The phosphorylated YAP/TAZ binds to the 14-3-3 protein and is retained in cytoplasm. Cytoplasmic YAP/TAZ suppresses TGF-β signaling, which in turn prevents the nuclear accumulation of the SMAD complex and inhibits the Wnt/β-catenin signaling [71-74]. Inhibition of the Wnt/β-catenin signaling downregulates TJ markers ZO1 and E-cadherin, induces EMT [63], and ultimately triggers cancer metastasis [70, 75, 76].
Direct biochemical interactions between proteins are indicated by solid lines. Dashed lines indicate genetic interactions for which no direct interactions have been reported. Arrowed and blunted ends indicate activation and inhibition, respectively.
CRB3 regulates Notch signaling
The Notch signaling is involved in regulating many processes (proliferation, differentiation, cell death, and cell fate) during development and renewal of adult tissues [77]. The Notch signaling mediates TGF-β-induced EMT through the induction of Snail [78]. The Notch function changes during the development of cell differentiation and can be either oncogenic or tumour-suppressive, depending on cellular context [79]. Crb has been shown to prevent endocytosis of the Notch receptor and/or its ligand Delta in Drosophila and zebrafish by directly interacting with these proteins via its lager extracellular structure; this may also negatively affect the epithelial tissue growth [11, 80, 81]. However, overexpression of human CRB3 does not have this effect on Notch signaling in mammalian cells [80]. Drosophila crb and human CRB3 have different roles in the Notch signaling because crb interacts with the Notch signaling mainly via the extracellular section of crb, whereas the extracellular CRB3 contains only signal peptide.
Schematic model of CRB3 in carcinogenesis and Hippo signaling cascade in mammals.
Conclusion
CRB3 is an essential determinant of epithelial apical identity in epithelia. CRB3 maintains apical-basal polarity by ensuring the delivery and stabilization of junctional proteins and interacting with other polarity proteins such as ZO1 and PAR6. CRB3 is mainly deposited in the plasma membrane; therefore, it may mediate the extracellular signal transduction in cancer development via intracellular signal, such as Hippo signal. However, little is known about its mechanisms. Currently, most CRB studies are done on Drosophila. Drosophila crb greatly differs in the structure from mammalian CRB3; therefore, studies on mammalian CRB3 are needed. Based on the above review, new questions arise: Does CRB3 inhibit the Hippo signal by directly affecting Willin? Does CRB3 regulate activation of the Notch and other signals and how? In-depth research is needed to reveal the role of CRB3 in tumorigenesis.
Together, CRB3 plays important roles in epithelial polarity maintenance and may relate to cancer development. Understanding of CRB3-related molecular signaling pathways and their interactions with CRB3 will be of great help in exploitation of possible therapeutic interventions for cancers.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 81272342) and Program for New Century Excellent Talents, Ministry of Education of China (No.NCET-10-0677).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Nelson WJ. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422:766-74
2. Ellenbroek SI, Iden S, Collard JG. Cell polarity proteins and cancer. Seminars in cancer biology. 2012;22:208-15
3. Muthuswamy SK, Xue B. Cell polarity as a regulator of cancer cell behavior plasticity. Annual review of cell and developmental biology. 2012;28:599-625
4. Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335-44
5. Miyoshi J, Takai Y. Structural and functional associations of apical junctions with cytoskeleton. Biochimica et biophysica acta. 2008;1778:670-91
6. Suzuki A, Ohno S. The PAR-aPKC system: lessons in polarity. Journal of cell science. 2006;119:979-87
7. Matter K, Balda MS. Signalling to and from tight junctions. Nature reviews Molecular cell biology. 2003;4:225-36
8. Bilder D. Epithelial polarity and proliferation control: links from the Drosophila neoplastic tumor suppressors. Genes Dev. 2004;18:1909-25
9. Ling C, Zheng Y, Yin F, Yu J, Huang J, Hong Y. et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10532-7
10. Parsons LM, Grzeschik NA, Allott ML, Richardson HE. Lgl/aPKC and Crb regulate the Salvador/Warts/Hippo pathway. Fly (Austin). 2010;4:288-93
11. Richardson EC, Pichaud F. Crumbs is required to achieve proper organ size control during Drosophila head development. Development. 2010;137:641-50
12. Lee M, Vasioukhin V. Cell polarity and cancer--cell and tissue polarity as a non-canonical tumor suppressor. Journal of cell science. 2008;121:1141-50
13. Hirose T, Izumi Y, Nagashima Y, Tamai-Nagai Y, Kurihara H, Sakai T. et al. Involvement of ASIP/PAR-3 in the promotion of epithelial tight junction formation. Journal of cell science. 2002;115:2485-95
14. Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A, Massey-Harroche D. Polarity complex proteins. Biochimica et biophysica acta. 2008;1778:614-30
15. Omori Y, Malicki J. oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Current biology: CB. 2006;16:945-57
16. Alves CH, Sanz AS, Park B, Pellissier LP, Tanimoto N, Beck SC. et al. Loss of CRB2 in the mouse retina mimics human retinitis pigmentosa due to mutations in the CRB1 gene. Human molecular genetics. 2013;22:35-50
17. Bulgakova NA, Knust E. The Crumbs complex: from epithelial-cell polarity to retinal degeneration. Journal of cell science. 2009;122:2587-96
18. Karp CM, Tan TT, Mathew R, Nelson D, Mukherjee C, Degenhardt K. et al. Role of the polarity determinant crumbs in suppressing mammalian epithelial tumor progression. Cancer research. 2008;68:4105-15
19. Elsum I, Yates L, Humbert PO, Richardson HE. The Scribble-Dlg-Lgl polarity module in development and cancer: from flies to man. Essays in biochemistry. 2012;53:141-68
20. Fogg VC, Liu CJ, Margolis B. Multiple regions of Crumbs3 are required for tight junction formation in MCF10A cells. Journal of cell science. 2005;118:2859-69
21. Gosens I, Sessa A, den Hollander AI, Letteboer SJ, Belloni V, Arends ML. et al. FERM protein EPB41L5 is a novel member of the mammalian CRB-MPP5 polarity complex. Experimental cell research. 2007;313:3959-70
22. Lemmers C, Medina E, Delgrossi MH, Michel D, Arsanto JP, Le Bivic A. hINADl/PATJ, a homolog of discs lost, interacts with crumbs and localizes to tight junctions in human epithelial cells. The Journal of biological chemistry. 2002;277:25408-15
23. Lemmers C, Michel D, Lane-Guermonprez L, Delgrossi MH, Medina E, Arsanto JP. et al. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Molecular biology of the cell. 2004;15:1324-33
24. Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933-45
25. Balda MS, Matter K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. The EMBO journal. 2000;19:2024-33
26. Balda MS, Garrett MD, Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. The Journal of cell biology. 2003;160:423-32
27. Fan S, Fogg V, Wang Q, Chen XW, Liu CJ, Margolis B. A novel Crumbs3 isoform regulates cell division and ciliogenesis via importin beta interactions. The Journal of cell biology. 2007;178:387-98
28. Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct interaction of two polarity complexes implicated in epithelial tight junction assembly. Nature cell biology. 2003;5:137-42
29. Roh MH, Fan S, Liu CJ, Margolis B. The Crumbs3-Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. Journal of cell science. 2003;116:2895-906
30. Schluter MA, Pfarr CS, Pieczynski J, Whiteman EL, Hurd TW, Fan S. et al. Trafficking of Crumbs3 during cytokinesis is crucial for lumen formation. Molecular biology of the cell. 2009;20:4652-63
31. A S. Investigation of Role of CRB3a in Tumorigenesis and Further Characterization of Its Roles Through Binding Partner Analysis. Université Laval. 2012
32. Martin-Belmonte F, Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nature reviews Cancer. 2012;12:23-38
33. Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nature cell biology. 2005;7:1232-9
34. Zhou B, Wu Y, Lin X. Retromer regulates apical-basal polarity through recycling Crumbs. Developmental biology. 2011;360:87-95
35. Whiteman EL, Fan S, Harder JL, Walton KD, Liu CJ, Soofi A. et al. Crumbs3 is essential for proper epithelial development and viability. Molecular and cellular biology. 2014;34:43-56
36. Medina E, Williams J, Klipfell E, Zarnescu D, Thomas G, Le Bivic A. Crumbs interacts with moesin and beta(Heavy)-spectrin in the apical membrane skeleton of Drosophila. The Journal of cell biology. 2002;158:941-51
37. Jensen AM, Westerfield M. Zebrafish mosaic eyes is a novel FERM protein required for retinal lamination and retinal pigmented epithelial tight junction formation. Current biology: CB. 2004;14:711-7
38. Etienne-Manneville S, Manneville J-B, Nicholls S, Ferenczi MA, Hall A. Cdc42 and Par6-PKCζ regulate the spatially localized association of Dlg1 and APC to control cell polarization. The Journal of cell biology. 2005;170:895-901
39. Grindstaff KK, Yeaman C, Anandasabapathy N, Hsu S-C, Rodriguez-Boulan E, Scheller RH. et al. Sec6/8 complex is recruited to cell-cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epithelial cells. Cell. 1998;93:731-40
40. van Rossum AG, Aartsen WM, Meuleman J, Klooster J, Malysheva A, Versteeg I. et al. Pals1/Mpp5 is required for correct localization of Crb1 at the subapical region in polarized Müller glia cells. Human molecular genetics. 2006;15:2659-72
41. Campbell K, Knust E, Skaer H. Crumbs stabilises epithelial polarity during tissue remodelling. Journal of cell science. 2009;122:2604-12
42. Georgiou M, Marinari E, Burden J, Baum B. Cdc42, Par6, and aPKC regulate Arp2/3-mediated endocytosis to control local adherens junction stability. Current biology: CB. 2008;18:1631-8
43. Leibfried A, Fricke R, Morgan MJ, Bogdan S, Bellaiche Y. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-cadherin endocytosis. Current biology: CB. 2008;18:1639-48
44. Morais-de-Sa E, Mirouse V, St Johnston D. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 2010;141:509-23
45. Walther RF, Pichaud F. Crumbs/DaPKC-dependent apical exclusion of Bazooka promotes photoreceptor polarity remodeling. Current biology: CB. 2010;20:1065-74
46. Sotillos S, Diaz-Meco MT, Caminero E, Moscat J, Campuzano S. DaPKC-dependent phosphorylation of Crumbs is required for epithelial cell polarity in Drosophila. The Journal of cell biology. 2004;166:549-57
47. Wirtz D, Konstantopoulos K, Searson PC. The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nature reviews Cancer. 2011;11:512-22
48. Chua KN, Poon KL, Lim J, Sim WJ, Huang RY, Thiery JP. Target cell movement in tumor and cardiovascular diseases based on the epithelial-mesenchymal transition concept. Advanced drug delivery reviews. 2011;63:558-67
49. Berx G, Raspe E, Christofori G, Thiery JP, Sleeman JP. Pre-EMTing metastasis? Recapitulation of morphogenetic processes in cancer. Clinical & experimental metastasis. 2007;24:587-97
50. Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Developmental cell. 2008;14:818-29
51. Castro Alves C, Rosivatz E, Schott C, Hollweck R, Becker I, Sarbia M. et al. Slug is overexpressed in gastric carcinomas and may act synergistically with SIP1 and Snail in the down-regulation of E-cadherin. The Journal of pathology. 2007;211:507-15
52. Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH. et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS genetics. 2011;7:e1002218
53. Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Annals of oncology: official journal of the European Society for Medical Oncology / ESMO. 2010;21(Suppl 7):vii89-92
54. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-90
55. Savagner P, Kusewitt DF, Carver EA, Magnino F, Choi C, Gridley T. et al. Developmental transcription factor slug is required for effective re-epithelialization by adult keratinocytes. Journal of cellular physiology. 2005;202:858-66
56. Sanaa Al Saleh LHS, Yunus A. Luqmani. Signalling pathways involved in endocrine resistance in breast cancer and associations with epithelial to mesenchymal transition (Review). International Journal of Oncology. 2011;38:1197-217
57. Chartier FJ, Hardy EJ, Laprise P. Crumbs controls epithelial integrity by inhibiting Rac1 and PI3K. Journal of cell science. 2011;124:3393-8
58. Shintani Y, Wheelock MJ, Johnson KR. Phosphoinositide-3 kinase-Rac1-c-Jun NH2-terminal kinase signaling mediates collagen I-induced cell scattering and up-regulation of N-cadherin expression in mouse mammary epithelial cells. Molecular biology of the cell. 2006;17:2963-75
59. Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M. et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26:6979-88
60. Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A. et al. The transcriptional repressor ZEB1 promotes metastasis and loss of cell polarity in cancer. Cancer research. 2008;68:537-44
61. Whiteman EL, Liu CJ, Fearon ER, Margolis B. The transcription factor snail represses Crumbs3 expression and disrupts apico-basal polarity complexes. Oncogene. 2008;27:3875-9
62. Godde NJ, Galea RC, Elsum IA, Humbert PO. Cell polarity in motion: redefining mammary tissue organization through EMT and cell polarity transitions. J Mammary Gland Biol Neoplasia. 2010;15:149-68
63. Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG. et al. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Developmental cell. 2010;19:831-44
64. Warner SJ, Yashiro H, Longmore GD. The Cdc42/Par6/aPKC polarity complex regulates apoptosis-induced compensatory proliferation in epithelia. Current biology: CB. 2010;20:677-86
65. Nolan ME, Aranda V, Lee S, Lakshmi B, Basu S, Allred DC. et al. The polarity protein Par6 induces cell proliferation and is overexpressed in breast cancer. Cancer research. 2008;68:8201-9
66. Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB. et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865-78
67. Robinson BS, Huang J, Hong Y, Moberg KH. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Current biology: CB. 2010;20:582-90
68. McCaffrey LM, Macara IG. Epithelial organization, cell polarity and tumorigenesis. Trends in cell biology. 2011;21:727-35
69. Visser-Grieve S, Hao Y, Yang X. Human homolog of Drosophila expanded, hEx, functions as a putative tumor suppressor in human cancer cell lines independently of the Hippo pathway. Oncogene. 2011;31:1189-95
70. Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nature cell biology. 2011;13:877-83
71. Mauviel A, Nallet-Staub F, Varelas X. Integrating developmental signals: a Hippo in the (path)way. Oncogene. 2012;31:1743-56
72. Harvey KF, Hariharan IK. The hippo pathway. Cold Spring Harbor perspectives in biology. 2012;4:a011288
73. Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA. et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Developmental cell. 2010;18:579-91
74. Pan D. The hippo signaling pathway in development and cancer. Developmental cell. 2010;19:491-505
75. Varelas X, Wrana JL. Coordinating developmental signaling: novel roles for the Hippo pathway. Trends in cell biology. 2012;22:88-96
76. Chan SW, Lim CJ, Guo K, Ng CP, Lee I, Hunziker W. et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer research. 2008;68:2592-8
77. Ilagan MX, Kopan R. SnapShot: notch signaling pathway. Cell. 2007;128:1246
78. Matsuno Y, Coelho AL, Jarai G, Westwick J, Hogaboam CM. Notch signaling mediates TGF-beta1-induced epithelial-mesenchymal transition through the induction of Snai1. The international journal of biochemistry & cell biology. 2012;44:776-89
79. Aster JC. In brief: Notch signalling in health and disease. The Journal of pathology. 2014;232:1-3
80. Herranz H, Stamataki E, Feiguin F, Milán M. Self-refinement of Notch activity through the transmembrane protein Crumbs: modulation of γ-Secretase activity. EMBO reports. 2006;7:297-302
81. Ohata S, Aoki R, Kinoshita S, Yamaguchi M, Tsuruoka-Kinoshita S, Tanaka H. et al. Dual roles of Notch in regulation of apically restricted mitosis and apicobasal polarity of neuroepithelial cells. Neuron. 2011;69:215-30
Author contact
![]() Corresponding author: Peijun Liu, Ph.D.&M.D., Professor, Email: liupeijunxjtu.edu.cn, Tel: 086-18991232306, Address: 277 Yanta Western Rd., Xi'an, Shaan Xi Province, China, 710061.
Corresponding author: Peijun Liu, Ph.D.&M.D., Professor, Email: liupeijunxjtu.edu.cn, Tel: 086-18991232306, Address: 277 Yanta Western Rd., Xi'an, Shaan Xi Province, China, 710061.

 Global reach, higher impact
Global reach, higher impact