Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(1):67-74. doi:10.7150/ijbs.10468 This issue Cite
Research Paper
Silencing the HaAK Gene by Transgenic Plant-Mediated RNAi Impairs Larval Growth of Helicoverpa armigera
Key Laboratory of Oasis Eco-agriculture, College of Agriculture, Shihezi University, Shihezi 832003, China.
Received 2014-9-2; Accepted 2014-11-5; Published 2015-1-1
Abstract
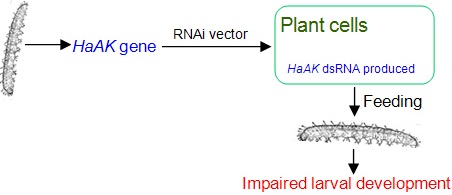
Insect pests have caused noticeable economic losses in agriculture, and the heavy use of insecticide to control pests not only brings the threats of insecticide resistance but also causes the great pollution to foods and the environment. Transgenic plants producing double-stranded RNA (dsRNA) directed against insect genes have been is currently developed for protection against insect pests. In this study, we used this technology to silence the arginine kinase (AK) gene of Helicoverpa armigera (HaAK), encoding a phosphotransferase that plays a critical role in cellular energy metabolism in invertebrate. Transgenic Arabidopsis plants producing HaAK dsRNA were generated by Agrobacterium-mediated transformation. The maximal mortality rate of 55% was reached when H. armigera first-instar larvae were fed with transgenic plant leaves for 3 days, which was dramatically higher than the 18% mortality recorded in the control group. Moreover, the ingestion of transgenic plants significantly retarded larval growth, and the transcript levels of HaAK were also knocked down by up to 52%. The feeding bioassays further indicated that the inhibition efficiency was correlated with the integrity and concentration of the produced HaAK dsRNA in transgenic plants. These results strongly show that the resistance to H. armigera was improved in transgenic Arabidopsis plants, suggesting that the RNAi targeting of AK has the potential for the control of insect pests.
Keywords: Arginine kinase, Helicoverpa armigera, RNAi, transgenic plants, double-stranded RNA, pest control.
Introduction
Insect pests significantly reduce crop yields and cause billions of dollars in economic loss in the agricultural production. Chemical pesticides are still the major approach for controlling these pests, but they are associated with significant hazards to the environment and human health. The biotechnological approach to pest control relies mainly on the expression of Bacillus thuringiensis (Bt) insecticidal proteins, the effectiveness of which is threatened by the emergence of resistance [1-3]. Alternative methods of controlling insect pests are therefore needed.
Eukaryotic organisms, including insects, possess mechanism of RNA interference (RNAi) for sequence-specific gene silencing that is triggered by the introduction of homologous double-stranded RNA (dsRNA). The transcript abundance of insect-specific target genes can be downregulated by the micro-injection of dsRNA [4] or by the uptake of exogenous dsRNA from an artificial diet [5]. Subsequently, plant-mediated RNAi technology involving the knockdown of insect genes was developed for managing pests. Baum et al. [1] engineered transgenic corn plants expressing dsRNAs that were directed against suitable insect target genes, and demonstrated that the effective RNAi responses in the western corn rootworm (WCR) resulted in high larval mortality rates. At the same time, Mao et al. [6] silenced a cytochrome P450 gene (CYP6AE14) of the cotton bollworm by transgenic plant-mediated RNAi, impairing larval tolerance on a gossypol-containing diet. Recently, further research has also confirmed the feasibility of the plant-mediated RNAi approach for pest control and showed that it dramatically decreased the levels of targeted mRNAs, resulting in larval developmental deformity and lethality [7-15]. These findings strongly suggest that plant-mediated RNAi is emerging as a powerful approach for controlling insect pests. The studies also show that genes encoding proteins with essential functions in insects are the best RNAi targets for increasing morbidity and mortality.
Arginine kinase (AK) is a phosphotransferase that plays a critical role in cellular energy metabolism in invertebrates by catalyzing the reversible transfer of phosphate from ATP to arginine, yielding phosphoarginine and ADP [16]. AK is not present in vertebrates, and phosphoarginine and its biosynthetic pathway are completely different from those in mammalian tissues, indicating that AK may be a useful target gene in the control of pests [17-19]. Most of the previous research on AK focuses on elucidating its catalytic mechanism and identifying its substrate analog inhibitors. Zhao et al. [20] produced dsRNA of Phyllotreta striolata AK by in vitro transcription, and feeding bioassays indicated that minute quantities of dsRNA greatly impaired the beetle's development, suggesting that the RNAi targeting of AK is a potentially attractive tool for controlling insect pests.
In this study, a full-length cDNA fragment of the AK gene was selected from Helicoverpa armigera, which is one of the most serious herbivorous insect pests in the Xinjiang cotton region. Transgenic Arabidopsis plants producing HaAK dsRNA were generated by Agrobacterium- mediated transformation. The feeding bioassay clearly showed that the resistance of transgenic Arabidopsis plants to H. armigera was improved, and the levels of HaAK were drastically suppressed. This is the first study to employ plant-mediated RNAi targeting of the AK gene to control an insect pest.
Materials and Methods
Vector construction and plant transformation
To construct a plasmid to express HaAK dsRNA in Arabidopsis plants, gateway cloning technology was used. The AK gene sequence was amplified using PrimeSTARTM HS DNA polymerase (Takara, China) from the pMD-HaAK vector containing the full-length HaAK cDNA (GenBank accession no. GU937510) cloned from H. armigera in our previous study. The specific primers that were used: the forward primer, 5'-CACCATGGTGGACGCCGCAACAAT-3' and the reverse primer, 5'-TTACAGCGACTTCTCAATT-3', including a 4-bp sequence CACC for directional cloning at the 5' end of the forward primer. The PCR amplification reactions contained 30 ng of plasmid DNA, 1.0 μM gene-specific primers, 10 μM of each dNTP, 0.5 U PrimeSTAR polymerase, and 1× PrimeSTAR buffer, and the final volume was adjusted to 25 μl with distilled water. The PCR cycles were performed as follows: 94°C for 5 min, followed by 30 cycles at 94°C for 1 min, 56°C for 1 min, 72°C for 1 min, and a final cycle of 72°C for 10 min. The amplified DNA fragment was cloned into pENTR/D-TOPO (Invitrogen) and was confirmed by DNA sequencing to obtain the entry vector pENTR/D-HaAK. Next, the LR recombination reaction between the entry plasmid vector pENTR/D-HaAK and gateway destination vector pANDA35HK was performed to generate the expression clone pANDA35HK-dsHaAK using the Gateway™ LR Clonase™ plus enzyme mix according to the LR Recombination Reaction manual (Invitrogen).
The dsRNA construct pANDA35HK-dsHaAK harboring the desired construct was transferred into the Agrobacterium tumefaciens strain LBA4404 by electroporation. Arabidopsis thaliana (ecotype Columbia) plants were then transformed by the floral dip method as previously described [21]. Seeds that had been bulk-harvested from each pot were sterilized using sodium hypochlorite (NaOCl, 2% active ingredient) for 20 min and then selected on 50 mg/L kanamycin MS medium. Once the kanamycin-resistant plantlets were well developed, they were transferred to pots of soil for continued growth and maturity.
Molecular analysis of transgenic plants
Total genomic DNA was isolated from the leaf tissues of Arabidopsis using a plant genomic DNA extraction kit (Takara, China). The samples were analyzed by the PCR amplification of the HaAK gene using the specific primers 5'-ATGGTGGACGCCGCAACAAT-3' and 5'-TTACAGCGACTTCTCAATT-3', while the primers 5'-CTAGTGGTACACAGAAGTCATGG-3' and 5'-GTGGGGAATCTTGGACAAT-3' that were specific for the 18S rDNA were used as a control. Total RNA was extracted from the plant leaves using the Trizol reagent (Invitrogen). The total RNA (15 µg) was separated on a 1.0% denaturing agarose gel and transferred to a Hybond-N+ filter membrane (Amersham). For the small RNA hybridization, approximately 20 µg of total RNA was separated on a denaturing 15% polyacrylamide gel containing 8 M urea. After electrophoresis, the gel was electroblotted onto a Hybond N+ membrane (Amersham). The membrane was hybridized with a DIG-labeled probe, which was obtained by PCR using the respective primers for vector construction. Hybridization and probe labeling were conducted according to the instructions that were included in the DIG Northern Starter Kit (MyLab, China).
For the RT-PCR, the RNA (1 µg) was reverse transcribed using the PrimeScriptTM 1st Strand cDNA Synthesis Kit (Takara, China). The PCR reactions were carried out with the specific primers 5'-ATGGTGGACGCCGCAACAAT-3' and 5'-TTACAGCGACTTCTCAATT-3' for HaAK and 5'- CTGCTATGTATGTGGCTATT-3' and 5'-TGGAAGGTACTGAGGGAG-3' for Actin1 of A. thaliana, which was used as a control. The PCR reactions were amplified for 35 cycles with an optimal annealing temperature of 56°C along with the appropriate control reactions. The RT-PCR reactions were performed in triplicate.
Insect culturing and feeding
Cotton bollworm (Helicoverpa armigera) eggs were obtained from Shihezi University and were incubated in an incubation chamber at 25°C and 70% relative humidity in complete darkness. The hatching larvae were then fed with a modified artificial diet as previously described [22] and reared in a growth chamber at 25°C with a 14-h light:10-h dark (14L:10D) photoperiod and 70% relative humidity.
For each feeding experiment, synchronous larvae were selected and divided into groups of 20 individuals. For the mortality bioassays, the Arabidopsis plant leaves were maintained in an 80 mm sterile plastic flask and fed to the larvae on day 1 of the first instar stage. Two to three similar leaves from the same plant were used in a duplicate feeding bioassay, and the surviving larvae were counted daily. For the insect-feeding trials with the Arabidopsis plants that were grown in soil, larvae that were at day 1 of the third instar stage were randomly released on top of mature leaves to feed on the whole plants for 5 days. The resistances of the plants to H. armigera were observed, and the larvae were weighed individually each day. Statistical analyses of the data were performed using the student's t-test.
Quantitative real-time PCR (qRT-PCR)
After feeding on the soil-grown AtdsHaAK-8 or AtdsHaAK-11 plants, the third-instar larvae were collected for total RNA extractions for the qRT-PCR analysis, respectively. Total RNA was isolated from the larvae using the Trizol reagent (Invitrogen) and then quantified using a GeneQuantTM 1300 spectrophotometer (Biochrom Ltd). One microgram of RNA was reverse transcribed to obtain first-stranded cDNA using the PrimeScriptTM 1st Strand cDNA Synthesis Kit (Takara, China). qRT-PCR reactions were performed in 20 µl volumes using the SYBR Green Master Mix (Takara, China), and reactions were performed using the LightCycler® 480 II (Roche) under the following PCR conditions: 95°C for 3 min followed by 45 cycles of 95°C for 15 s, 56°C for 15 s, and 72°C for 15 s. The reactions were carried out using the specific primers 5'-CGAAAAGTTGGAGGCTGG-3' and 5'-GGGCGTAGATTCCGACAC-3'. The Actin gene of H. armigera was amplified as an internal control using the primers 5'-GCCCATCTACGAGGGTTACGC-3' and 5'-CGTGGTGGTGAACGAGTAGCC-3'. All qRT-PCR reactions were performed in triplicate.
AK enzyme assay
The levels of AK activity of the third instar larvae that fed on the soil-grown AtdsHaAK-8 plants were analyzed. Midguts were obtained from the larvae and treated according to Mao et al. [6]. The homogenates were centrifuged (12,000 rpm) for 30 min at 4°C to obtain a crude enzyme solution. The standard AK sample was purchased from Sigma Ltd. AK activity was assayed according to Sigma's product instruction manual. One unit of the enzyme was defined as 1.0 µmole of L-arginine and ATP converted to N-phospho-L-arginine and ADP per min at a pH of 8.6 and temperature of 30°C.
Results
Generation of transgenic plants expressing HaAK dsRNA
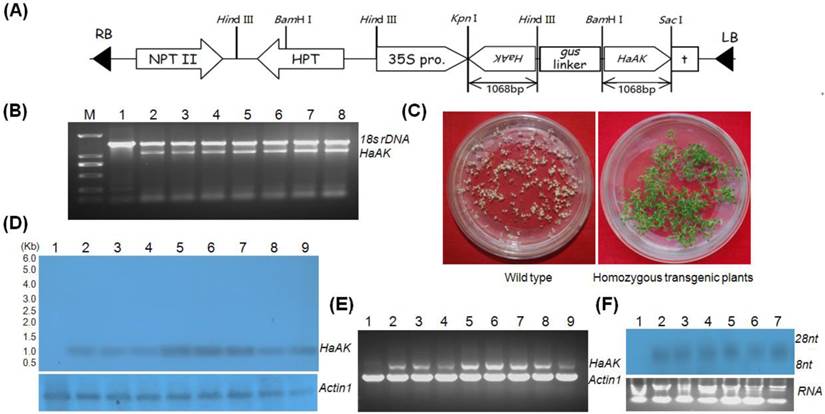
The full-length cDNA fragment of HaAK was chosen as the target for RNAi. The cDNA corresponding to the coding region of HaAK (1,068 bp), was cloned into the plasmid pANDA35HK, and the cloned fragments as inverted repeats under the control of the CaMV 35S promoter to produce the dsRNA. The resultant construct (Fig. 1A) was then transformed into Arabidopsis plants by Agrobacterium-mediated transformation, and transgenic Arabidopsis plants were obtained. The independently generated transgenic lines were analyzed by PCR amplification. The expected 1,068 bp DNA fragment of the target gene was only identified in transformants, whereas the 1,450 bp fragment of the 18s rDNA was detected in both the transformants and the untransformed control plants (Fig. 1B), showing that the target DNA fragment was successfully inserted into the Arabidopsis genomic DNA.
The single-copy transgenic plants were selected through Mendelian segregation and their progeny were screened for homozygous plants used for subsequent experiments (Fig. 1C). Northern blot was performed to detect HaAK dsRNA expression in transgenic plants. The transgenic Arabidopsis plants producing HaAK dsRNA (AtdsHaAK) were identified, and the expression levels of hairpin dsRNA varied among several independent transformant lines (Fig. 1D). The plants of the AtdsHaAK-8 line accumulated higher levels of the long dsRNA of HaAK. RT-PCR analysis also revealed the presence of large amounts of undegraded, full-length HaAK dsRNA in different transgenic plant lines (Fig. 1E). Several independent transgenic lines produced the small interfering RNAs (siRNAs, ~21-23 nt) (Fig. 1F). This indicated that a portion of HaAK dsRNA had been processed into small RNA fragments in the Arabidopsis leaves. The transgenic plants did not display any morphologcal changes compared with the wild-type plants.
Transgenic plants expressing HaAK dsRNA show enhanced resistance to H. armigera
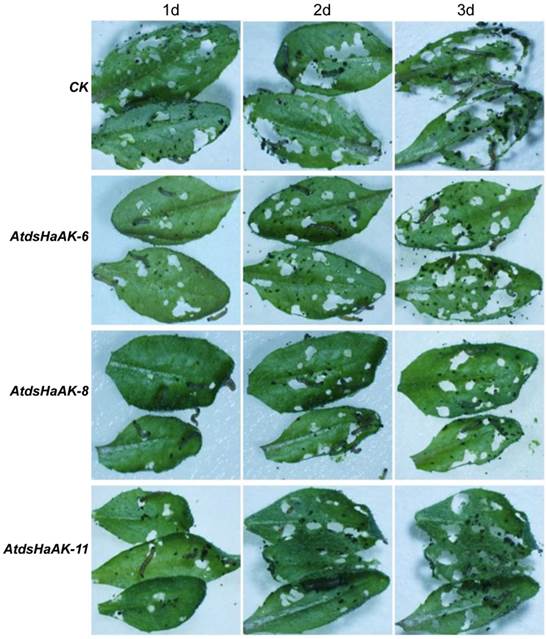
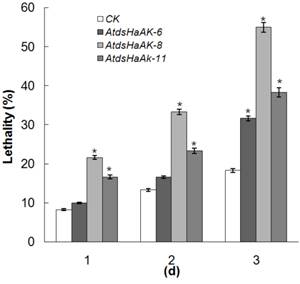
To evaluate whether the transgenic Arabidopsis plants expressing HaAK dsRNA possessed improved resistance to H. armigera, the first-instar larvae were transferred to detached leaves of the Arabidopsis plant lines. After 1 day of feeding, it was evident that the transgenic Arabidopsis plants, and especially the AtdsHaAK-8 line, exhibited much higher resistances to H. armigera than the wild-type plants. The AtdsHaAK-8 leaves were ingested significantly less by H. armigera larvae compared with the untransformed leaves (Fig. 2). Compared with the untransformed plants, the transgenic plants expressing HaAK dsRNA were much less damaged after 3 days of feeding. Moreover, feeding on the leaves of the transgenic plants caused significantly higher rates of lethality compared with feeding on the control leaves. In the control group, approximately 80% of the adults survived for 3 days, which was lower than that was recorded in the field. However, there were significant differences between the control and experimental groups. The mortality of larvae in the experimental groups ranged from 32% to 55% when they were fed with transgenic plants containing dsRNA after three days (Fig. 3), which was dramatically higher than the 18% that was recorded for the control group (P < 0.05). Among the larvae that were placed on the A. thaliana leaves, those on the AtdsHaAK-8 leaves had higher mortality rates than those on the AtdsHaAK-6 and AtdsHaAK-11 leaves. Plants of AtdsHaAK8 line had higher levels of undegraded, full-length HaAK dsRNA compared with those of AtdsHaAK6 or AtdsHaAK11 line (Fig. 1D), suggesting that lethality was related to HaAK dsRNA expression levels in the transgenic plants.
Generation and molecular analysis of transgenic plants. (A) Schematic representation of the pANDA35HK-dsHaAK expression cassettes used for Arabidopsis transformation. 35S pro, CaMV 35S promoter; HPT, hygromycin phosphotralsferase gene; NPT II, neomycin phosphotransferase II gene; HaAK, cDNA sequence of AK gene from H. armigera; RB, right border; LB, left border; (B) Detection of HaAK in non-transformed control and transgenic plants by PCR. A 1,068 bp fragment of HaAK was amplified and 18s rDNA was served as a control. Lane M, DNA marker DL2,000; Lane 1, untransformed control; Lane 2-8, the transformants; (C) The wild and transgenic plants were grown on kanamycin-containing medium. The homozygous single-copy transgenic plants were selected through Mendelian segregation; (D) Northern blot detection of HaAK dsRNA in control and different transgenic lines. Lane 1, the non-transformed control; Lane 2-9, the transformants (line AtdsHaAK-2, AtdsHaAK-3, AtdsHaAK-6, AtdsHaAK-7, AtdsHaAK-8, AtdsHaAK-9, AtdsHaAK-11 and AtdsHaAK-12); Actin1 was used as a reference and the DIG labeled probe for Actin1 was obtained by PCR using primers as described for RT-PCR. (E) RT-PCR amplification of HaAK in control and transgenic plants. A 1,068 bp fragment of HaAK was amplified and Arabidopsis Actin1 gene was used as a control. Lane 1, untransformed plant; Lane 2-9, the transformants (line AtdsHaAK-2, AtdsHaAK-3, AtdsHaAK-6, AtdsHaAK-7, AtdsHaAK-8, AtdsHaAK-9, AtdsHaAK-11, AtdsHaAK-12); (F) Northern blot detection of small RNA fragments of HaAK in transgenic plants leaves. Lane 1, non-transformed plant; Lane 2-7, the transformants.
Arabidopsis plants leaves were used for feeding larvae. Twenty H. armigera (day 1 of the first instar larvae) were released on the detached leaves maintained in a sterile plastic flask. The images were taken using a dissecting microscope (Olympus SZX7, Japan). Only some larvae have been shown because they were active in plastic flask and the images of leaves were magnified for ease of observation. The non-transformants (CK) were used as a control, and three different transgenic lines, AtdsHaAK-6, AtdsHaAK-8 and AtdsHaAK-11, were used as experimental groups. Leaves from transgenic plants expressing HaAK dsRNA were ingested relative less by larvae compared with the untransformed leaves after 3 days of feeding. Especially, the mortality rate of larvae feeding on leaves of AtdsHaAK-8 line was significantly high and larval body sizes were distinctly reduced.
The mortality of the first instar larvae fed on the leaves of different transgenic lines. Feeding on transgenic leaves expressing HaAK dsRNA caused significantly higher lethality than in the control (*P < 0.05). The error bar means standard error of three independent experiments.
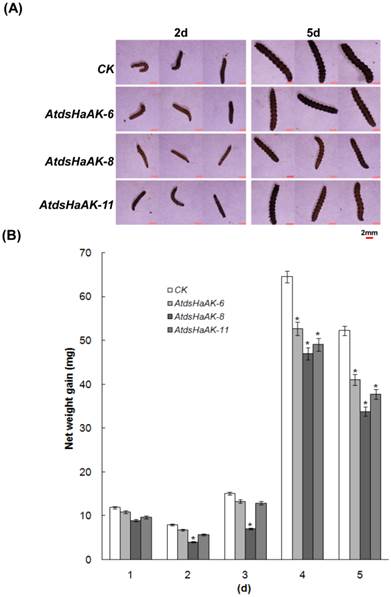
Additionally, the third-instar larvae that were previously reared on artificial diets were randomly released onto mature Arabidopsis leaves to feed on the whole plants that were growing in soil. It was evident that transgenic Arabidopsis plants expressing HaAK dsRNA exhibited much higher resistances to H. armigera compared with the control after 5 days (Fig. 4). Likewise, the average body weight of the larvae that were reared on transgenic lines was similar to the weight of those that were reared on the wild-type plants on day 1. However, growth retardation became evident by day 4. Accordingly, the increase in body weight became significantly different on day 4 (Fig. 5). The growth of the larvae that fed on the AtdsHaAK8 line was significantly impaired, and they had smaller body sizes and exhibited lower net weight gains compared with those that fed on the control plants. This was in accordance with the aforementioned analysis of the larvae that fed on the detached leaves of the Arabidopsis plants. The insect feeding trials demonstrated that transgenic plants expressing dsRNA targeting AK could adversely affect cotton bollworm growth and survival.
Resistance to H. armigera was improved in transgenic plants expressing HaAK dsRNA. Plants were grown in nutrient-rich Metro-Mix in individual 12×25-inch pots. Relative humidity was maintained at 60 to 70%. Similar sizes of homozygous transgenic plants (25 days after sowing) were utilized in the bioassay. Twenty H. armigera (day 1 of the third instar larvae) were randomly released on the whole plants to evaluate RNAi effects. After 5d of feeding, transgenic plants with less damage exhibited higher resistance to H. armigera than the control. The non-transformants (CK) were used as a control, and three different transgenic lines, AtdsHaAK-6, AtdsHaAK-8 and AtdsHaAK-11, were used as experimental groups.

The reduction in body size and net weight gain of H. armigera larvae fed on transgenic plants. (A) The larvae fed on different plant lines were very variable in both sizes. The non-transformants (CK) were used as a control, and three different transgenic lines, AtdsHaAK-6, AtdsHaAK-8 and AtdsHaAK-11, were used as experimental groups. Twenty H. armigera (day 1 of the third instar larvae) were randomly released on the whole plants to evaluate RNAi effects. Bar = 2mm. (B) The growth of larvae feeding with transgenic plants expressing HaAK dsRNA was significantly impaired. The error bar means standard error of three independent experiments. The increase in body weight became significantly different by day 5(*P < 0.05).
HaAK expression was suppressed by ingesting transgenic plants
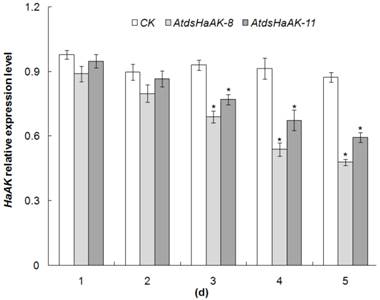
To confirm whether the ingestion of transgenic plants triggered gene-specific silencing, the transcript levels of the target gene in H. armigera larvae were detected using qRT-PCR. According to the above mortality bioassays, the larvae that fed on the AtdsHaAK-8 and AtdsHaAK-11 lines were used for qRT-PCR analysis, and time-course analyses of HaAK mRNA was performed (Fig. 6). In the larvae that were reared on the AtdsHaAK-8 and AtdsHaAK-11 plants, HaAK transcript levels remained largely unchanged on day 1 and began to decrease on day 3. Five days after the transfer, the larvae that were fed with AtdsHaAK-8 and AtdsHaAK-11 plants had fewer HaAK transcripts compared with those that were fed with wild-type plants (Fig. 6). The larvae that fed on the leaves of the AtdsHaAK-8 plants exhibited significantly decreased levels of endogenous AK mRNA, and their relative transcripts levels were reduced by 52% by day 5 (Fig. 6).
AK activity analysis
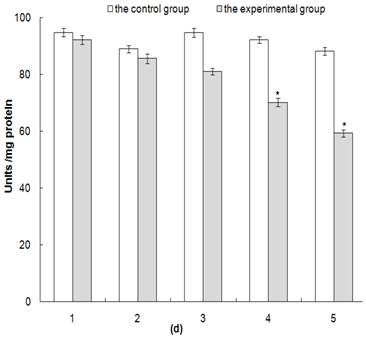
Our data also suggest that AK knockdown efficiency in H. armigera is correlated with dsHaAK expression levels in transgenic plants. Because the negative effect on AK enzyme activity is likely to be magnified if AK transcripts levels are downregulated by RNAi, AK activity was analyzed using an NADH-linked enzyme assay. The level of AK activity in the experimental group that fed on AtdsHaAK-8 plants grown in the field was significantly reduced compared with that in the controls over 5 days (Fig. 7). On the first day, the inhibition of AK activity was very weak, and enzyme activity levels remained at >90%. However, the midguts of the larvae that were reared on the AtdsHaAK-8 plants showed losses of almost 40% in the catalytic activity of AK (P < 0.05) after 5 days, while there was no obvious change in the control group (Fig. 7).
HaAK expression in midguts of H. armigera larvae. Suppression of HaAK transcript levels in midguts of third-instar larve fed on AtdsHaAK-8 and AtdsHaAK-11 plants, respectively. The mRNA abundance was determined by qRT-PCR analysis and Actin gene was amplified as an internal control. The error bar means standard error of three biological replicates (*P < 0.05).
Detection of enzyme activity of AK in midguts of the third instar larvae feed on transgenic plants expressing HaAK dsRNA. Enzyme activity of AK in midguts reflecting RNAi effects at different times after the third instar larvae were fed with control and AtdsHaAK-8 plants, respectively. One unit of the enzyme was defined as 1.0 µmole of L-arginine and ATP converted to N-phospho-L-arginine and ADP per minute at a pH 8.6 and 30°C. The error bar means standard error of three biological replicates (*P < 0.05).
Discussion
Using dsRNA to knock down specific genes has been well documented in recent years. The application of RNAi technology to suppress critical insect gene(s) by allowing insects to feed on transgenic plant tissues has shown that insect development can be delayed and the damage to plants can be reduced [1,6,8]. AK plays a critical role in cell energy metabolism in invertebrates [23] and therefore seems to be an excellent target for RNAi in the control of pests.
In this study, the AK gene from H. armigera was selected for a dsRNA construct, which was then transferred into Arabidopsis plants. The exact dsRNA size that is needed to trigger RNAi in plants and other eukaryotes is still not entirely clear. The targeting sequence for gene silencing in plants using RNAi was approximately 300 to 700 nucleotides in length [24]. In feeding experiments, most sequences have ranged from 300 to 520 bp [25]. In our study, the full-length cDNA sequence (1,068 bp) of HaAK was used as the target fragment. The RNAi AK expression in H. armigera using several transgenic lines suggests that the use of a longer target fragment was successful in generating RNAi in this experiment. Dicer-like enzymes are ubiquitously generated in plants and may degrade intact dsRNA [2,26,27]. In our study, some of the intact dsRNA was processed into small dsRNAs. Both intact dsRNA and small dsRNAs were present in the transgenic plants. Mao et al. [6] reported that the intact, long dsRNA that is produced in plants could effectively suppress insect gene expression. Our study also showed that the inhibition efficiency of the targeted gene was related to the integrity of the dsRNA produced in transgenic plants.
Among the transgenic Arabidopsis lines, plants of the AtdsHaAK8 line produced the highest levels of long, undegraded HaAK dsRNA. When first-instar larvae were fed AtdsHaAK-8 leaves for 3 days, a maximal mortality rate of 55% was reached. Likewise, the adverse effects on third-instar larvae growth and improved resistance of plants to H. armigera in the AtdsHaAK-8 experimental group were more apparent than in other transgenic lines. The reduced expression levels of long dsRNA in transgenic plants weakened their efficacies in controlling the insects. Our results also suggest that the long intact dsRNA that is produced in the transgenic plants allow for the effective plant-mediated RNAi control of pest damage.
It had been reported that delivering dsRNA to herbivorous insects by feeding them transgenic plants results in the reduction of target mRNA expression by 40% to 70% [8]. In this study, the ingestion of the AtdsHaAK-8 line suppressed HaAK expression by up to 52% over 5 days. This was consistent with the observation that dsRNA-mediated gene silencing did not completely eliminate the gene products. Insect genes can be knocked down, rather than knocked out, by the microinjection of dsRNA [4,28,29], the uptake of exogenous dsRNA from an artificial diet [5], or through plant-mediated strategies [1,2,6]. Although the transcript levels of HaAK were distinctly suppressed in the third-instar larvae, we did not observe any significant lethal phenotypes. It seemed that the third-instar larvae had developed resistance to the transgenic plants. There are several possible reasons for this observation. First, the transgenic plants may not have produced sufficient dsRNA due to the expression of heterologous genes that were derived from different plant backgrounds [30,31]. Dicer-like enzymes in plants could have decreased the expression levels of the intact dsRNA in the transgenic lines. We found that the higher the concentrations of intact dsRNA that were produced in transgenic plants, the greater the RNAi inhibitory effects. Second, third-instar larvae of this species are large in size. Arabidopsis plants are short-lived, which is adverse for the long-term observational experiments. In our feeding experiments, the first-instar larvae had higher mortality rates. This feeding regime did not seem to meet the need of insects considering their growth rates. There may be a time-lag between gene silencing and larval death. Moreover, we do not know whether there are energy metabolic pathways that may compensate for the RNAi targeting HaAK.
The nucleotide sequences that were used determine the possible RNAi effects in the target organism but also in other insects. Baum et al. [1] reported that the larvae of WCR were killed when fed with an artificial diet containing specific dsRNAs that were designed to target WCR genes. When these dsRNAs were tested on other insect pests, the RNAi effects remained relatively high due to the sequence identities between the WCR genes and their orthologs in other insect species [1]. Because the AK gene is highly conserved in insects and has a similar sequence identity in other insect species, dsRNA targeting HaAK could potentially adversely affect other insects and could be used as a general biocontrol agent for pests. This study demonstrated that dsRNA targeting HaAK was effective at suppressing the growth and development of H. armigera. Further experiments are necessary to determine the effectiveness of dsRNA targeting AK in controlling other insect species. The persistence of the silencing effect also requires further investigation due to the low mortality rate of the third-instar larvae.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (31201521, 31101094) and the Doctor Funds of Xinjiang Bingtuan (2012BB004) and the foundation of Shihezi University (2011gxjs-yz06).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Baum JA, Bogaert T, Clinton W. et al. Control of coleopteran insect pests through RNA interference. Nat Biotechnol. 2007;25:1322-1326
2. Gordon KH, Waterhouse PM. RNAi for insect-proof plants. Nat Biotechol. 2007;25:1231-1232
3. Tabashnik BE, Gassmann AJ, Crowder DW. et al. Insect resistance to Bt crops: evidence versus theory. Nat Biotechnol. 2008;26:199-202
4. Bettencourt R, Terenius O, Faye I. Hemolin gene silencing by dsRNA injected into Cecropia pupae is lethal to next generation embryos. Insect Mol Biol. 2002;11:267-271
5. Turner CT, Davy MW, MacDiarmid RM. et al. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol Biol. 2006;15:383-391
6. Mao YB, Cai WJ, Wang JW. et al. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechol. 2007;25:1307-1313
7. Mao YB, Tao XY, Xue XY. et al. Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res. 2011;20:665-673
8. Zha W, Peng X, Chen R. et al. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PLoS One. 2011;6:e20504
9. Pitino M, Coleman AD, Maffei ME. et al. Silencing of aphid genes by dsRNA feeding from plants. PLoS One. 2011;6:e25709
10. Zhu JQ, Liu S, Ma Y. et al. Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS One. 2012;7:e38572
11. Kumar P, Pandit SS, Baldwin IT. Tobacco rattle virus vector: A rapid and transient means of silencing manduca sexta genes by plant mediated RNA interference. PLoS One. 2012;7:e31347
12. Xiong Y, Zeng H, Zhang Y. et al. Silencing the HaHR3 gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. Int J Biol Sci. 2013;9:370-381
13. Wuriyanghan H, Falk BW. RNA Interference towards the Potato Psyllid, Bactericera cockerelli, Is Induced in Plants Infected with Recombinant Tobacco mosaic virus (TMV). PLoS One. 2013;8:e66050
14. Wang Z, Dong Y, Desneux N. et al. RNAi silencing of the HaHMG-CoA reductase gene inhibits oviposition in the Helicoverpa armigera cotton bollworm. PLoS One. 2013;8:e67732
15. El-Shesheny I, Hajeri S, El-Hawary I. et al. Silencing abnormal wing disc gene of the Asian citrus psyllid, Diaphorina citri disrupts adult wing development and increases nymph mortality. PLoS One. 2013;8:e65392
16. Newsholme EA, Beis I, Leech AR. et al. The role of creatine kinase and arginine kinase in muscle. Biochem J. 1978;172:533-537
17. Pereira CA, Alonso GD, Paveto MC. et al. Trypanosoma cruzi arginine kinase characterization and cloning. J Biol Chem. 2000;275:1495-1501
18. Brown AE, Grossman SH. The mechanism and modes of inhibition of arginine kinase from the cockroach (Periplaneta americana). Arch Insect Biochem. 2004;57:166-177
19. Wu QY, Li F, Zhu WJ. et al. Cloning, expression, purification, and characterization of arginine kinase from Locusta migratoria manilensis. Comp Biochem Physiol Part B. 2007;148:355-362
20. Zhao YY, Yang G, Wang-Pruski GF. et al. Phyllotreta striolata (Coleoptera: Chrysomelidae): Arginine kinase cloning and RNAi-based pest control. Eur J Entomol. 2008;105:815-822
21. Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735-743
22. Abbasi BH, Ahmed K Khalique F. et al. Rearing the cotton bollworm, Helicoverpa armigera, on a tapioca-based artificial diet. J Insect Sci. 2007;7:35
23. Ellington WR. Evolution and physiological roles of phosphagen systems. Ann Rev Physiol. 2001;63:289-325
24. Preuss S, Pikaard CS. Targeted gene silencing in plants using RNA interference. In: (ed.) Engelke D. RNA Interference (RNAi)~Nuts& Bolts of siRNA Technology. DNA Press, LLC. 2003:23-36
25. Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. J Insect Physiol. 2010;56:227-235
26. Bouche N, Lauressergues D, Gasciolli V. et al. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347-3356
27. Xie Z, Johansen LK, Gustafson AM. et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104
28. Tomoyasu Y, Denell RE. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol. 2004;214:575-578
29. Ghanima M, Kontsedalov S, Czosnek H. Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius). Insect Biochem Mol Biol. 2007;37:732-738
30. Kusnadi AR, Nikolov ZL, Howard JA. Production of recombinant protein in transgenic plants: practical considerations. Biotechnol Bioeng. 1997;56:473-483
31. Teli NP, Timko MP. Recent developments in the use of transgenic plants for the production of human therapeutics and biopharmaceuticals. Plant Cell Tissue Organ Cult. 2004;79:125-145
Author contact
![]() Corresponding author: 86-993-2067-366, Fax: 86-993-2057-990, e-mail address: sunjieedu.cn.
Corresponding author: 86-993-2067-366, Fax: 86-993-2057-990, e-mail address: sunjieedu.cn.

 Global reach, higher impact
Global reach, higher impact