10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(6):701-711. doi:10.7150/ijbs.11431 This issue Cite
Research Paper
HOXB5 Promotes the Proliferation and Invasion of Breast Cancer Cells
1. Department of Anatomy, Embryology Laboratory, and Brain Korea 21 plus project for Medical Science, Yonsei University College of Medicine, Seoul 120-752, Korea
2. Department of Surgery, National Health Insurance Service Ilsan Hospital, Goyang 410-719, Korea
3. Department of Surgery, Yonsei University College of Medicine, Seoul 120-752, Korea
4. Department of Pathology, National Health Insurance Service Ilsan Hospital, Goyang 410-719, Korea
*These authors contributed equally to this work.
Received 2014-12-25; Accepted 2015-4-10; Published 2015-5-1
Abstract
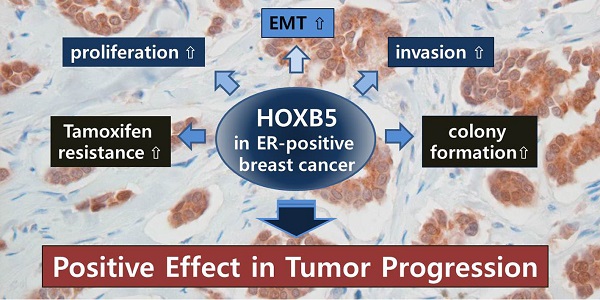
HOX transcription factors play an important role in determining body patterning and cell fate during embryogenesis. Accumulating evidence has shown that these genes act as positive and/or negative modulators in many types of cancer, including breast cancer, in a tissue-specific manner. We have previously reported that HOXB5 is aberrantly overexpressed in breast cancer tissues and cell lines. Here, we investigated the biological roles and clinical relevance of HOXB5 in breast cancer. Immunohistochemical analysis of HOXB5 on tissue microarray (TMA) including 34 normal and 67 breast cancer specimens revealed that HOXB5 was highly expressed in cancer tissues, particularly from estrogen receptor (ER)-positive breast cancer patients. An online survival analysis confirmed the correlation between HOXB5 expression and poor distant metastasis-free survival in ER-positive, but not in ER-negative, breast cancer. In vitro studies indicated that HOXB5 silencing in ER-positive cells significantly decreased cell proliferation and anchorage-independent cell growth. In contrast, overexpression of HOXB5 displayed EMT characteristics with a greater invasive ability, higher cell proliferation and colony formation in soft agar. HOXB5 knockdown or overexpression led to changes in the expression levels of RET, ERBB2, and EGFR, but not of ESR1. In conclusion, we suggest that HOXB5 acts as a positive modulator most likely by promoting cell proliferative response and invasiveness in ER-positive breast cancer. These results would help predict prognosis of breast cancer and identify a new valuable therapeutic target.
Keywords: HOXB5, aberrant expression, breast cancer, ER-positive, epithelial-mesenchymal transition
Introduction
HOX genes are a subset of homeobox genes and encode transcriptional factors which play important roles in cell proliferation, differentiation, and the determination of cell identity [1]. In addition, many HOX genes have been shown to be associated with cancer [2, 3]. In particular, several studies have shown the prognostic value of specific HOX genes, such as HOXA5, HOXA9, HOXB7, and HOXB13, as well as the molecular mechanisms underlying the regulation of gene expression and cell growth in breast cancer [4-9]. Although recent progress in this area has revealed potential targets that could increase the effectiveness of breast cancer treatments, there is still a demand to discover new oncogenes and possible treatment targets for specific types of breast cancer.
In our previous study, we have proposed 14 HOX genes, HOXA6, -A13, -B2, -B4, -B5, -B6, -B7, -B8, -B9, -C5, -C9, -C13, -D1, and -D8, as putative candidates associated with the development and progression of breast cancer based on observations of the significantly different expression pattern between non-malignant and malignant breast tissues as well as breast cancer cell lines [10]. Several HOX genes, such as HOXB7 and -B9, have already been reported to act as positive modulators in breast cancer [8, 11]. However, the other genes have not been investigated for their clinical and biological roles in breast cancer.
HOXB5 is known to play an important role in the development of the enteric nervous system (ENS) [12]. Abnormalities in HOXB5 function lead to Hirschsprung's disease via downregulation of the receptor tyrosine kinase RET (REarranged during Transfection) [12]. A recent study demonstrated that HOXB5 directly binds to the promoter of the RET gene and enhanced RET expression in a neuroblastoma cell line [13]. Interestingly, RET has been known to behave as an oncogene in several cancers including breast cancer [14]. In addition, HOXB5-regulated vascular endothelial growth factor receptor 2 (VEGFR2), angiopoietin 2 (ANG2), and alpha 2 integrin (ITGa2) have also been shown to play roles in breast cancer biology [15-20]. Therefore, we considered HOXB5 to be one of the most promising candidate associated with the development and progression of breast cancer.
To evaluate the clinical significance of HOXB5 in breast cancer and demonstrate an association between breast cancer progression and HOXB5 expression, we analyzed the expression of HOXB5 in patients with invasive breast cancer and performed in vitro studies. Our results showed that HOXB5 was highly expressed in some breast cancers, especially in estrogen receptor (ER)-positive tumors. In breast cancer cell lines, HOXB5 induced the epithelial-mesenchymal transition (EMT) and promoted tumor cell proliferation and growth as well as invasion.
Materials and methods
Cell culture, plasmids, and cell line construction
MCF7, T47D, MCF10A, and MDA-MB-231 cells were kindly provided by Drs. Yong Nyun Kim and Kyung tae Kim (National Cancer Center, Korea). MCF7 and MDA-MB-231 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; WelGENE Inc., Deagu, Korea) supplemented with 10% fetal bovine serum (FBS, WelGENE Inc.) and 1x antibiotic antimycotic solution (WelGENE Inc.). T47D cells were grown in RPMI 1640 (WelGENE Inc.) with the same supplementation. MCF10A was cultured in DMEM/F12 (WelGENE Inc.) supplemented with 5% horse serum, 20 ng/ml epidermal growth factor (EGF), 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, and 100 μg/ml penicillin-streptomycin. A set of pLKO.1 lentiviral vectors containing seven different shRNA targeting HOXB5 was purchased from Thermo Fisher Scientific (Rockford, IL USA). For the control, pLKO.1 lentiviral vector harboring nonspecific shRNA (shNS-puro) were used. Lentiviral particles were produced in 293T cells by co-transfection with lentiviral packaging and envelop plasmids (pCMV Δ8.91 and pMD 1G), which were kindly provided by Dr. Seok Hyung Kim (Department of pathology, Samsung Medical Center, Seoul, Korea). The T47D cells were transduced with lentiviral particles. Through antibiotic selection using puromycin at a concentration of 0.5 μg/ml, stable cell lines were obtained and the protein levels were confirmed using Western blotting analysis. For the overexpression studies, a full-length cDNA of the HOXB5 gene was cloned into the EcoRI-XbaI site of the pcDNA3-HA-tagged expression vector. To establish stable cell lines, G418 was treated for 2~3 weeks with a concentration of 300 μg/ml.
Total RNA Isolation and RT-PCR
Total RNA was isolated from the cultured cells using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Reverse transcription was conducted with 1 μg of total RNA using ImProm-ll TM Reverse Transcriptase (Promega, Madison, WI, USA). PCR was performed using Taq polymerase (Bioneer, Seongnam, Korea). For the quantitative PCR, SYBR Green PCR Master Mix (Applied Biosystems, Calrlsbad, CA, USA) was used and then subjected to real time PCR quantification using the ABI7300 (Applied Biosystems). All reactions were done in triplicate, and the relative amounts of all mRNAs were calculated by using the comparative CT method. β-actin mRNA was used as the invariant control. All primer sequences were provided in Supplementary Table S1.
Western blot, immunocytochemistry, and antibodies
Cells were lysed in Nondet P-40 (NP-40) lysis buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1% NP-40, and Protease Inhibitor Cocktail). Protein concentrations were estimated by the BCA Protein Assay Kit (Thermo). After the immune blotting, the signals were detected using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL USA). The primary antibodies used were rabbit anti-HOXB5 (Abcam, Cambridge, MA, USA), rabbit anti-E-cadherin (Abcam), anti-β-catenin (BD, San Jose, CA, USA), anti-HA tag (Abcam), and anti- β-actin (Sigma, St. Louis, MO, USA). For the immunocytochemistry, the cells were fixed with 4% PFA and incubated in the blocking buffer (0.1% Triton X-100 containing 1% goat serum) for 30 min. Antibodies to HOXB5 (Abcam), E-cadherin (Abcam), Vimentin (Abcam), and β-catenin (BD) were used.
MTT assay
Cells were trypsinized, counted, and plated in 96-well plates at a density of 7.5x103 cells per well. On designated days, the cells were stained with 20 μl of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide, Sigma) for 3.5 hours at 37°C, followed by removal of the culture medium and incubation with 100 μl of MTT solvent (4 mM HCl, 0.1% NP40 in isopropanol). The absorbance was measured with an ELISA reader (Softmax Pro) at 560 nm. All experiments were performed in triplicate. The tamoxifen sensitivity was measured by MTT assay with the treatment of 4-hydroxytamoxifen (Sigma).
Soft agar colony-forming assay
Sterile agarose solution (1% and 0.7% agarose in sterile water) were mixed with the same volume of 2× RPMI with 20% FBS and used as bottom and top layers, respectively. The cells were adjusted to a volume of 5x103 cells in 100 μl of appropriate culture medium with serum, and then were added to the bottom layer. The plates were incubated at 37°C in a humidified incubator for 14 to 21 days. The cells were fed twice a week by adding 0.5 ml of fresh culture medium. Colonies were stained with 0.5 ml of NBT (0.5 mg/ml in PBS) and photographed with a digital camera (Kodak). The acquired images were analyzed using the ImageJ software.
Matrigel invasion assay
Matrigel™ (BD) was mixed with coating buffer (0.01 M Tris, pH 8.0 and 0.7% NaCl) to a final concentration 200-300 μg/ml. Individual inserts were coated with 100 μl of coating solution in a 24-well plate. Then, 5 × 104 cells were added to the top of this Matrigel™ layer. The culture medium with 10% FBS was added to the lower chamber as a chemoattractant. The cells were incubated at 37°C for 24-72 hours. The cells that had invaded through the Matrigel™ and reached the lower surface of the filter were stained with fluorochrome 4',6-diamidino-2-phenylindole (DAPI) and observed by fluorescent microscopy. The acquired images were analyzed using the ImageJ software.
Tissue microarray and immunohistochemistry (TMA-IHC)
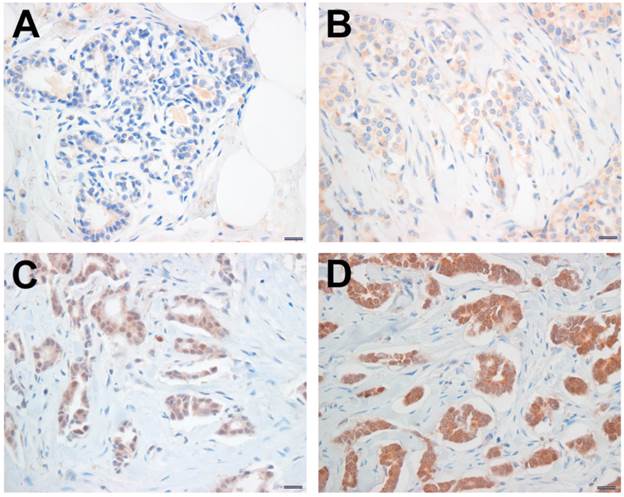
Among patients who had undergone surgical resection for breast cancer between January 2000 and December 2004 at National Health Insurance Service Ilsan Hospital, Gyeonggi-do, Korea, 67patients were selected because their paraffin blocks had been well preserved and tumor in a single tissue block was sufficient for the construction of TMA. Data regarding patient demographics and the histopathology of the primary tumor were retrospectively obtained by reviewing medical records. In this particular case, no consent from patients was needed according to the South Korea's Bioethics and Safety Act. The Ethics Committee for the Clinical Research of the Institutional Review Board of the National Health Insurance Service Ilsan Hospital, Gyeonggi-do, Korea, approved this study protocol (SU-YON 2013-129). Formalin-fixed, paraffin-embedded tissue blocks were arrayed using a tissue-arraying instrument (AccuMax_Array; Petagen Inc., Seoul, Korea). Briefly, representative areas of each tumor were selected and marked on the H&E slide by breast pathologists. In 34 cases, normal tissues adjacent to each tumor were also circled for scoring. The designated zone of each donor block was punched with a tissue cylinder 2 mm in diameter and the sample was transferred to a recipient block in a grid pattern. Immunohistochemical staining was carried out in the tissue microarray blocks. Thick sections of 4 μm were obtained with a microtome, transferred into adhesive slides, and dried at 59°C for an hour. After deparaffinization and rehydration, the sections were treated with a 3% hydrogen peroxide solution for 10 min to block endogenous peroxidase and then pretreated for antigen retrieval in 10 mM citrate buffer (pH 6.0) in a microwave oven for 20 min. After incubation with primary antibodies against HOXB5 (Abcam), immunodetection was carried out with biotinylated anti-mouse immunoglobulin antibody (DAKO, Kyoto, Japan), followed by peroxidase-labeled streptavidin using a labeled streptavidin biotin kit with 3,3'-diaminobenzidine chromogen as the substrate. Slides were counterstained with Harris hematoxylin. Brown nuclear or cytoplasmic staining for HOXB5 was considered positive. For semi-quantitative analysis, HOXB5 immunostaining signals were scored using the Allred scoring system. The proportion score was rated as 0 = no cells stained positive, 1 = less than 1/100 positive, 2 = between 1/100 and 1/10, 3 = between 1/10 and 1/3, 4 = between 1/3 and 2/3, and 5 = more than 2/3. The intensity score was made on the basis of the average intensity of staining: 0 = negative, 1 = weak, 2 = moderate and 3 = strong (Fig. 1). The final score was the sum of the intensity score and the proportion score. A final staining score ≥ 3 was considered to be positive. For the Kaplan-Meier analysis, an online database [21] was used with gene expression data and survival information on 4,142 patients downloaded from the NCBI Gene Expression Omnibus (GEO; Affymetrix Microarrays only), the European Genome-phenome Archive (EGA), and the Cancer Genome Atlas (TCGA).
Immunohistochemical staining for HOXB5. The HOXB5 expression levels were categorized as negative (A), weak (B), moderate (C), and strong positive (D), according to the intensity score of HOXB5 immunostaining signal. Scale bar; 100 μm.
Statistical analysis
The clinicopathologic variables were compared by the Chi-square test or Fisher's exact test. The independent t-test was used for comparison of age. Distant metastasis-free survival (DMFS) time was measured from the date of operation to the date of the first distant metastasis. Survival curves based on the Kaplan-Meier method were compared using a log-rank test. All statistical tests were two-sided and p-values of less than 0.05 were considered statistically significant. SPSS for Windows version 21.0 (SPSS Inc., Chicago, IL, USA) was used for statistical analyses. Results from the in vitro assay are expressed as mean ± SEM and were analyzed using the paired t-test. Differences were considered significant at p < 0.05.
Results
Clinical significance of HOXB5 in breast cancer
To assess whether there is a difference in HOXB5 protein expression between normal breast tissues and breast cancer tissues, 34 breast cancer tissues and paired 34 normal breast tissues which were obtained from the same specimens were analyzed by TMA-IHC. Among 34 pairs, 41.2% of breast cancer tissues and 29.4% of normal breast tissues were HOXB5 positive (Table 1). Especially, among 17 pairs which were ER positive cases, HOXB5 protein expression was significantly higher in breast cancer tissues than in normal breast tissues (64.7% in cancers and 11.8% in normal tissues, p = 0.001; Table 1). When correlation analysis was performed between the expression of HOXB5 and various clinicopathological features, ER-positive patients had higher HOXB5 expression (p < 0.001; Table 2). Other clinical characteristics such as tumor size, lymphatic metastasis, tumor stage, and histologic grade were not positively related to the expression of HOXB5.
Analysis of HOXB5 expression in breast specimens
| HOXB5 positivity | Normal | Cancer | p-value | |
|---|---|---|---|---|
| All cases | negative | 24 (70.6%) | 20 (58.8%) | 0.310 |
| positive | 10 (29.4%) | 14 (41.2%) | ||
| ER positive cases | negative | 15 (88.2%) | 6 (35.3%) | 0.001 |
| positive | 2 (11.8%) | 11 (64.7%) |
ER, estrogen receptor
Correlation analysis between HOXB5 expression and various clinicopathological features
| Parameter | HOXB5 negative (n=35) | HOXB5 positive (n=32) | p-value |
|---|---|---|---|
| Age | 48.0 ± 12.5 | 50.2 ± 12.3 | 0.468 |
| Tumor size | |||
| ≤ 2 cm | 14 (40.0%) | 15 (46.9%) | 0.570 |
| > 2 cm | 21 (60.0%) | 17 (53.1%) | |
| Node status | |||
| No metastasis | 17 (48.6%) | 16 (50.0%) | 0.907 |
| Metastasis | 18 (51.4%) | 16 (50.0%) | |
| Stage | |||
| I, II | 25 (71.4%) | 25 (78.1%) | 0.529 |
| III, IV | 10 (28.6%) | 7 (21.9%) | |
| Histologic grade | |||
| I | 6 (18.8%) | 10 (32.3%) | 0.218 |
| II, III | 26 (81.2%) | 21 (67.7%) | |
| ER status | |||
| Negative | 23 (65.7%) | 5 (15.6%) | < 0.001 |
| Positive | 12 (34.3%) | 27 (84.4%) | |
| PR status | |||
| Negative | 9 (25.7%) | 5 (15.6%) | 0.310 |
| Positive | 26 (74.3%) | 27 (84.4%) | |
| HER2 status | |||
| Negative | 26 (74.3%) | 26 (81.3%) | 0.495 |
| Positive | 9 (25.7%) | 6 (18.8%) |
HOXB5 positive was defined by Allred score ≥3.
ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
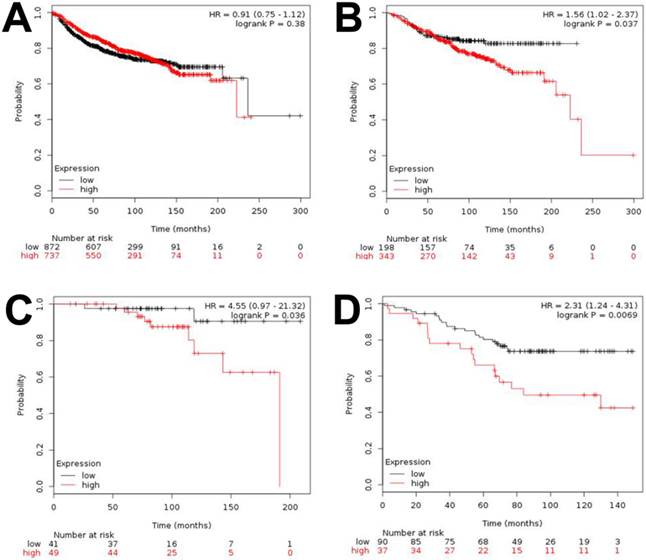
To assess the survival of breast cancer patients in the context of HOXB5 expression levels, DMFS curves were drawn and compared using the Kaplan-Meier method and the log-rank test, respectively. However, we found no significant difference between the HOXB5-positive and HOXB5-negative groups in terms of DMFS (data not shown). The reliability of the subgroup analysis was poor due to the limited number of cases in this set. Therefore, we further analyzed a publicly available database [21] and confirmed that HOXB5 had no impact on DMFS when all patients were analyzed together without bias, which was consistent with the result based on our sample set (Fig. 2A). However, a higher expression of HOXB5 was related to poor DMFS when only the ER-positive breast cancer patients were analyzed (p = 0.037; Fig. 2B). The higher expression of HOXB5 was also linked to poor DMFS for patients with ER-positive, lymph node-negative, and histologic grade (HG) I breast cancer (p = 0.036), who usually had good prognosis and were occasionally treated with endocrine therapy alone (Fig. 2C). Furthermore, a higher expression of HOXB5 was associated with poor endocrine responsiveness (p = 0.0069; Fig. 2D).
Kaplan-Meier analysis of distant metastasis-free survival (DMFS) and overall survival (OS) based on HOXB5 expression in the online dataset. Survival was compared between the HOXB5-high and -low expression groups. (A) DMFS for all of the breast cancer patients in the dataset. (B) DMFS for ER-positive patients. (C) DMFS for the patients with ER-positive, lymph node-negative, histologic grade (HG) I. (D) OS for the patients who received endocrine therapy only.
HOXB5 promotes breast cancer cell growth and invasion in vitro
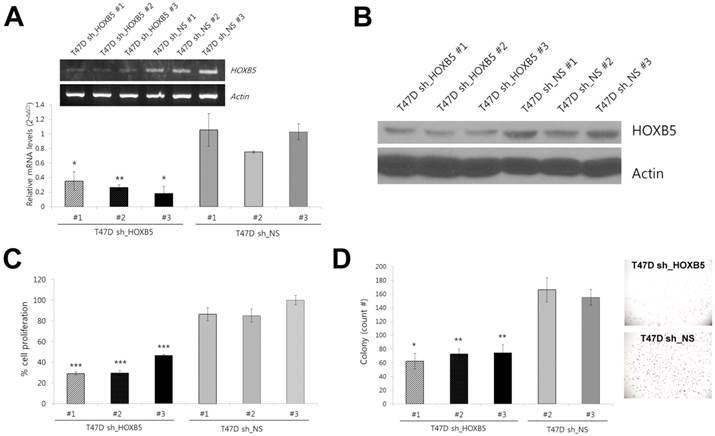
To explore the functional relevance of HOXB5 to ER-positive breast cancer, we examined the effect of HOXB5 knockdown and ectopic expression on tumor cell behavior. Lentiviral infection of shRNA constructs in ER-positive T47D cells resulted in a reduction of HOXB5 expression at the mRNA and protein level (Fig. 3A, B). Three stable cell lines (sh_HOXB5#1, -#2, and -#3) with low HOXB5 expression exhibited reduced cell proliferation and lower levels of anchorage-independent growth (Fig. 3C, D).
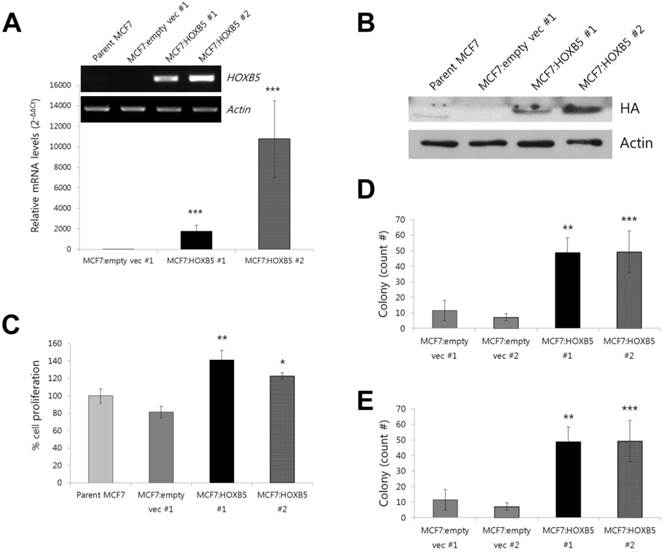
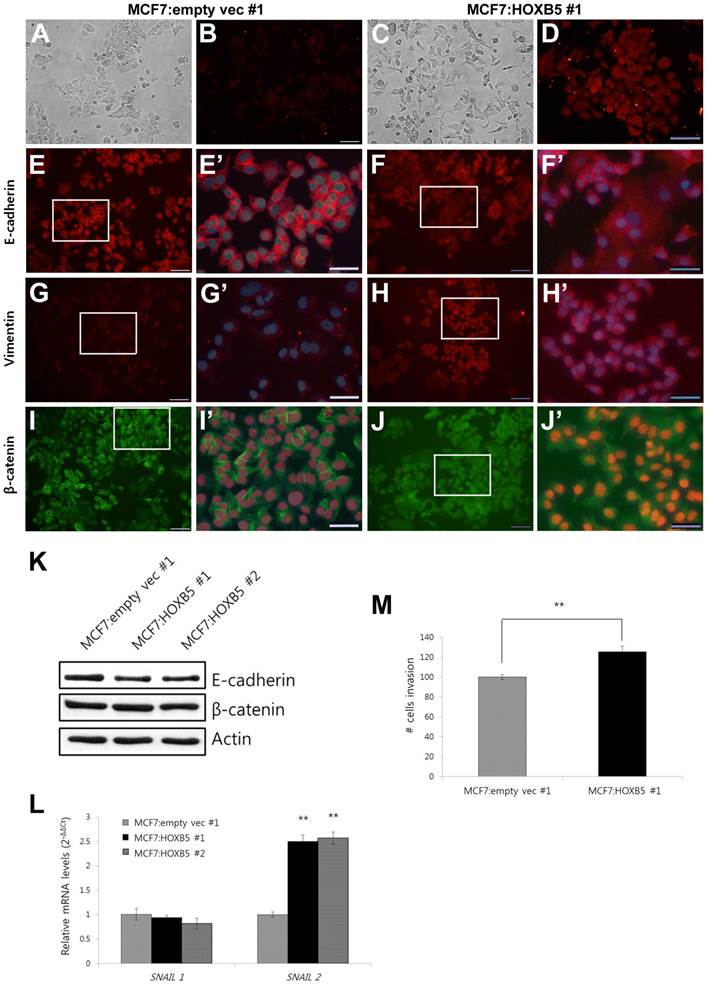
For the overexpression studies, the MCF7 cell line was chosen as a model system since it expresses very low levels of endogenous HOXB5 [10]. Compared to the clonal cells containing empty vector only (MCF7:empty vec), the cells with HOXB5 expression vector highly expressed HOXB5, which was confirmed by PCR (Fig. 4A) and western blotting (Fig. 4B), as well as by immunocytochemistry (Fig. 5B, D). Out of two independent clonal cell lines, MCF7:HOXB5 #2 cells had a 5-fold greater HOXB5 expression than MCF7:HOXB5 #1 cells, based on qPCR data, however, these two clonal cells behaved similarly. High expression of HOXB5 promoted cell proliferation compared with the cells harboring empty vector as well as parent MCF7 cells (Fig. 4C). In addition, the cells with high HOXB5 expression facilitated anchorage-independent cell growth and showed significant increases in both number and size of colonies in soft agar (Fig. 4D, E). More interestingly, the strong ectopic expression of HOXB5 in MCF7 cells showed typical features of EMT, with a cellular morphological change from cobblestone-like to a spindle and fibroblast-like shape (Fig. 5A, C). In addition, E-cadherin expression was weaker and dispersed into the cytoplasm in HOXB5-overexpressing MCF7 cells (Fig. 5E-F'). Western blotting results confirmed that the protein levels of E-cadherin were reduced in HOXB5 overexpressing cells (Fig. 5K). Together with up-regulated expression of Vimentin at the cell membrane borders (Fig. 5G-H'), the β-catenin expression in the cell membrane was shifted to the cytoplasm or in the nucleus, without changes in total protein levels (Fig. 5G-J', K). At the transcriptional level, induction of Snail2 (also known as Slug), but not Snail1, was prominent in HOXB5 overexpressing cells (Fig. 5L). Consistent with these functional changes, HOXB5 overexpression increased ability to invade through the Matrigel (Fig. 5M). These findings indicate that HOXB5 can induce EMT and positively regulate tumor cell proliferation and invasion.
Effect of HOXB5 knockdown in T47D breast cancer cell lines. (A) Comparison of mRNA expression levels of HOXB5 in stable cell lines (sh_HOXB5 #1, -#2, and -#3 vs. sh_NS #1, -#2, and -#3) acquired after knockdown with HOXB5 shRNA (sh_HOXB5) or non-specific (NS) shRNA (sh_NS). The fold change shown in the graph was quantitated by realtime PCR. *p < 0.05, **p < 0.01 vs. sh_NS #1. (B) Immunoblot analysis of HOXB5 expression in three independent T47D stable cells expressing shRNA against HOXB5 (T47D-sh_HOXB5 #1, -#2, and -#3) and three control cells (T47D-sh_NS #1, -#2, and -#3). (C) Cell proliferation was measured by colorimetric MTT assay. ***p < 0.001 vs. sh_NS #2. (D) Soft agar colony formation by T47D-sh_HOXB5 and sh_NS cells. The photographs on the right show a representative image of each HOXB5 KD and control group. *p < 0.05, **p < 0.01 vs. sh_NS #2.
Effect of HOXB5 overexpression in a MCF7 breast cancer cell line. (A) Comparison of mRNA expression levels of HOXB5 in HOXB5 overexpressing cells (MCF7:HOXB5 #1 and -#2) with control cells (MCF7:empty vec #1 and parent MCF7). The fold change shown in the graph was quantitated by realtime PCR. (B) Immunoblot analysis of HOXB5 expression in two clones with high HOXB5 and control cells (MCF7:empty vec #1 and parent MCF7). Antibody against HA tag was used to confirm the protein expression. (C) Effect of HOXB5 overexpression in cell proliferation was measured by colorimetric MTT assay. (D-E) Soft agar colony forming assay. The total count number (D) and the average size of colonies (E) are shown in the graph. *p < 0.05, **p < 0.01, ***p < 0.001 vs. MCF7:empty vec #1 for all (A) to (E).
Effect of HOXB5 on the expression of breast cancer-related genes and tamoxifen resistance
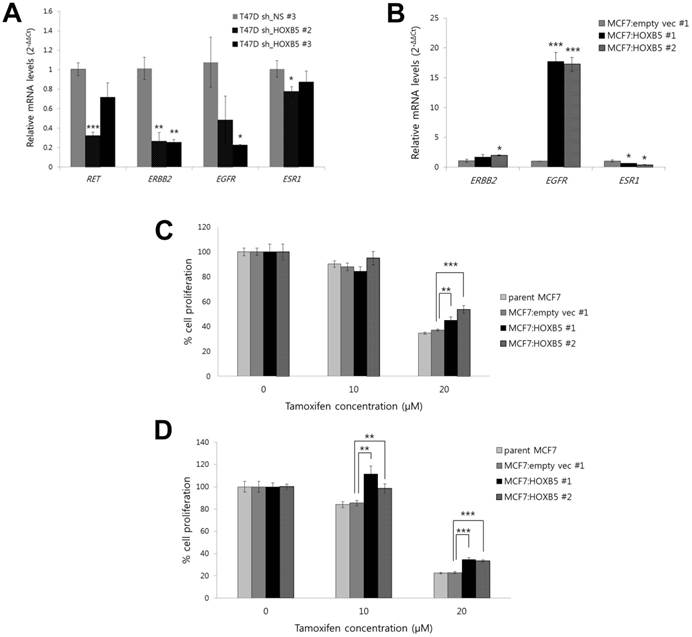
In breast cancer cells, overexpression of the receptor tyrosine kinase gene RET, as well as the ErbB/HER family members and other growth factor receptors, has been linked to the tamoxifen resistance and the development of more aggressive, estrogen-independent tumors. The results from our in vitro models demonstrate that knockdown of HOXB5 led to a reduction in transcript levels of RET, ERBB2 and EGFR (Fig. 6A). In contrast, a high ectopic expression of HOXB5 in MCF7 cells showed an opposite effect on the expression of EGFR (Fig. 6B). In both knockdown and overexpression systems, ESR1, which encodes estrogen receptor 1, had small changes in transcriptional levels (Fig. 6A, B). Interestingly, MTT proliferation assays revealed that MCF7 cells overexpressing HOXB5 were more resistant to tamoxifen (Fig. 6C, D). Taken together, these data suggest a possible role of HOXB5 on endocrine resistance in breast cancer.
Discussion
In this study, we demonstrated that the expression level of the HOXB5 protein was higher in breast cancer tissues compared to normal tissues. In particular, HOXB5-positive patients showed a higher rate of ER-positive tumors. Although more cases are necessary to validate our data, this result suggests that it is worth investigating the function of HOXB5 in breast cancer biology. Furthermore, survival analysis using online resources have shown an association between high HOXB5 and poor DMFS in patient groups with ER-positive breast cancer, implying a clinical significance of HOXB5 in ER-positive breast cancer. To correlate and validate the results of this human study, we performed an in vitro assay for HOXB5 in breast cancer cell lines. HOXB5 knockdown in T47D led to reduced cell proliferation and a lower rate of anchorage-independent growth. MCF7 cells overexpressing HOXB5 had an opposite effect on cell proliferation as well as on tumor cell growth in soft agar. Moreover, an ectopic expression of HOXB5 enhanced invasion and induced an EMT.
Morphological changes with EMT features in the cells with high HOXB5 expression. (A-D) Cell morphology detected under bright field microscope (A and C) and the immunofluorescence analysis of HOXB5 expression (B and D). (E-J') E-cadherin (E-F'), Vimentin (G-H'), and β-catenin (I-J') expression in MCF7:empty vec #1 (E, E', G, G', I, and I') and MCF7:HOXB5 #1 (F, F', H, H', J, J') cells. The boxed regions in E, F, G, H, I, and J (200x, Scale bar; 100 μm) were magnified and overlayed with a DAPI nuclear counterstain (E', F', G', H', I', and J'; 600x, Scale bar; 50 μm). (K) Immunoblot analysis of E-cadherin and β-catenin in two clones with high HOXB5 (MCF7:HOXB5 #1 and -#2) and control MCF7:empty vec #1 cells. (L) Relative mRNA expression levels of Snail1 and Snail2 in the cells with high HOXB5 (MCF7:HOXB5 #1 and -#2) and control cells. (M) Matrigel invasion assay with the same cells used for immunofluorescence assay shown in A to J'. The invasive cells were stained with DAPI. **p < 0.01 vs. MCF7:empty vec #1 in (L) and (M).
Effect of HOXB5 on the expression of breast cancer-related genes and tamoxifen resistance. (A) Realtime PCR analysis of RET, ERBB2, EGFR, and ESR1 in HOXB5 KD cells (T47D sh_HOXB5 #2 and -#3) and control cells. *p < 0.05, **p < 0.01, ***p < 0.001 vs. sh_NS control. (B) Realtime PCR analysis of ERBB2, EGFR, and ESR1 in the cells with high HOXB5 (MCF7:HOXB5 #1 and -#2) and control cells. **p < 0.01, ***p < 0.001 vs. MCF7:empty vec #1. (C-D) Cell proliferation was measured by the MTT assay at day 1 (C) and day 2 (D) with treatment of 10 μM and 20 μM tamoxifen in the cells with high HOXB5 (MCF7:HOXB5 #1 and -#2) and control cells (parent MCF7 and MCF7: empty vec #1). **p < 0.01, ***p < 0.001 vs. MCF7: empty vec #1.
In order to assess whether the function of HOXB5 is specific to ER-positive cells or not, we also analyzed the effect of HOXB5 overexpression in several different breast cancer cell lines. Similar to its effect in ER-positive MCF7 cells, HOXB5 overexpression promoted cell proliferation in ER-negative MDA-MB-231 cells, and moreover, HOXB5 slightly induced cell proliferation even in MCF10A normal human mammary epithelial cells (Supplementary Fig. S1). The proliferative response of mammary epithelial cells to HOXB5 does not seem to be unique to ER-positive cells. However, the HOXB5-induced EMT and invasive potential seem to be phenotypic features of MCF7 cells, as EMT-like properties were not prominent in T47D cells. No phenotypic changes by HOXB5 overexpression in MDA-MB-231 cells were detectable, probably due to their intrinsically high invasive and aggressive properties with fibroblastic morphology. Although the overexpression of HOXB5 seems to induce EMT and cell proliferation in a cell context-dependent manner similar to those previously reported [22, 23], there is still a possibility that HOXB5 plays a role as a modulator of EMT predominantly in ER-positive breast cancer cells because Snail2, an essential mediator of Twist1-induced EMT, was induced by HOXB5 overexpression in ER-positive MCF7 and T47D cells, but was not significantly induced in ER-negative MDA-MB-231 cells (Supplementary Fig. S2). Further analysis in ER-negative breast cells will be necessary to define the role of HOXB5 in ER-negative breast tumors.
One of the interesting findings in this study was the dysregulation of RET, ERBB2/HER2, and EGFR as a consequence of HOXB5 silencing or overexpression. Trastuzumab, an inhibitor of ERBB2, is very well known as a therapeutic agent for HER2-positive breast cancer. An agent targeting EGFR is also currently under development for triple negative breast cancer [24]. Taken together with our HOXB5 knockdown experiments, in which cell proliferation and colony formation were suppressed, HOXB5 is expected to be a putative therapeutic target for breast cancer. In addition, cross-talk between ER signaling and the ERBB2 or EGFR pathway is a well-known mechanism of endocrine therapy resistance [25]. RET has also been reported to be associated with endocrine resistance [26]. Although there has been no strong evidence to link between HOXB5 and the key genes involved in endocrine resistance, we showed here a drastic up-regulation of EGFR transcripts when the cells over-expressed the HOXB5, which may contribute to endocrine-resistant phenotype. Together with the current in vitro studies showing tamoxifen resistance in HOXB5 overexpressing MCF7 cells, the survival analysis showed a correlation between high HOXB5 expression and a poor response to endocrine therapy. Since it strongly suggests a possible role of HOXB5 in endocrine therapy resistance, it is worthwhile to investigate further whether HOXB5 could be a therapeutic target to overcome this resistance.
Several HOXB cluster genes, such as HOXB2, HOXB7, HOXB9, and HOX13, have been reported to play a role in breast cancer. HOXB2 has been proposed to be a negative growth regulator, whereas HOXB7, HOXB9, and HOXB13 confer more aggressive phenotypes and are responsible for poorer prognoses [8, 9, 11]. HOXB7 and HOXB9 promoted EMT phenotypes [11, 21], and HOXB7 and HOXB13 have recently been identified as mediating tamoxifen resistance [8, 9]. Recent advances in elucidating different functions of HOX genes shed valuable light on the molecular links between the dysregulation of each HOX gene and the altered cellular functions that contribute to the development of cancer. Findings such as those in one study reporting the cluster-based regulation of HOXB genes in Barrett's esophagus (BE) [27] might open a way to target several HOX genes together for cancer therapy, for example, by controlling common enhancers or epigenetic factors affecting cluster genes.
In summary, our work provides new insight into the physiological significance and potential clinical applications of HOXB5, which modulates cell proliferation and tumor progression in ER-positive breast cancer. Because a single gene may have different function depending on the tissue or cell type, further studies to identify the molecular basis of HOXB5 regulation and its impacts on particular types of breast cancer are needed to validate HOXB5 as a potential therapeutic target and biomarker in breast cancer.
Supplementary Materials
Supplementary Table S1 and Figures S1-S2.
Abbreviations
TMA: tissue microarray; IDC: invasive ductal carcinoma; ER: estrogen receptor; EMT: epithelial-mesenchymal transition; ENS: enteric nervous system; RET: REarranged during Transfection; VEGFR2: vascular endothelial growth factor receptor 2; ANG2: angiopoietin 2; ITGa2: alpha 2 integrin; DMFS: distal metastasis-free survival; OS: overall survival; HG: histologic grade; BE: Barrett's esophagus.
Acknowledgements
This research was supported by funds from the Basic Science Research Program through the National Research Foundation (NRF) funded by the Ministry of Education, Science and Technology (NRF-2013R1A1A2008399, NRF-2010-0025149, NRF-2014R1A1A2056986), and partly from the Faculty Research Grant (6-2014-0147) of Yonsei University College of Medicine, Seoul, Korea.
Conflict of Interests
The authors have no conflicts of interest to declare.
References
1. Wang KC, Helms JA, Chang HY. Regeneration, repair and remembering identity: the three Rs of Hox gene expression. Trends Cell Biol. 2009;19:268-275
2. Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361-371
3. Grier D, Thompson A, Kwasniewska A. et al. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205:154-171
4. Raman V, Tamori A, Vali M. et al. HOXA5 regulates expression of the progesterone receptor. J Biol Chem. 2000;275:26551-26555
5. Chen HX, Chung S, Sukumar S. HOXA5-induced apoptosis in breast cancer cells is mediated by caspases 2 and 8. Mol Cell Biol. 2004;24:924-935
6. Gilbert P, Mouw J, Unger M. et al. HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. J Clin Invest. 2010;120:1535-1550
7. Sun M, Song C-X, Huang H. et al. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci U S A. 2013;110:9920-9925
8. Jin K, Kong X, Shah T. et al. The HOXB7 protein renders breast cancer cells resistant to tamoxifen through activation of the EGFR pathway. Proc Natl Acad Sci U S A. 2012;109:2736-2741
9. Shah N, Jin K, Cruz L-A. et al. HOXB13 mediates tamoxifen resistance and invasiveness in human breast cancer by suppressing ERα and inducing IL-6 expression. Cancer Res. 2013;73:5449-5458
10. Hur H, Lee J-Y, Yun H. et al. Analysis of HOX gene expression patterns in human breast cancer. Mol Biotechnol. 2014;56:64-71
11. Hayashida T, Takahashi F, Chiba N. et al. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci U S A. 2010;107:1100-1105
12. Lui VC, Cheng WW, Leon TY. et al. Perturbation of hoxb5 signaling in vagal neural crests down-regulates ret leading to intestinal hypoganglionosis in mice. Gastroenterology. 2008;134:1104-1115
13. Zhu J, Garcia-Barcelo MM, Tam PKH. et al. HOXB5 cooperates with NKX2-1 in the transcription of human RET. PloS one. 2011;6:e20815
14. Arighi E, Borrello MG, Sariola H. RET tyrosine kinase signaling in development and cancer. Cytokine & growth factor rev. 2005;16:441-467
15. Wu Y, Moser M, Bautch VL. et al. HoxB5 is an upstream transcriptional switch for differentiation of the vascular endothelium from precursor cells. Mol Cell Biol. 2003;23:5680-5691
16. Winnik S, Klinkert M, Kurz H. et al. HoxB5 induces endothelial sprouting in vitro and modifies intussusceptive angiogenesis in vivo involving angiopoietin-2. Cardiovasc Res. 2009;83:558-565
17. Volpe MAV, Chung E, Ulm JP. et al. Aberrant cell adhesion molecule expression in human bronchopulmonary sequestration and congenital cystic adenomatoid malformation. Am J Physiology-Lung Cell Mol Physiol. 2009;297:L143-L152
18. Guo S, Colbert LS, Fuller M. et al. Vascular endothelial growth factor receptor-2 in breast cancer. Biochim Biophys Acta. 2010;1806:108-121
19. Imanishi Y, Hu B, Jarzynka MJ. et al. Angiopoietin-2 stimulates breast cancer metastasis through the α5β1 integrin-mediated pathway. Cancer Res. 2007;67:4254-4263
20. Zutter MM, Santoro SA, Staatz WD. et al. Re-expression of the alpha 2 beta 1 integrin abrogates the malignant phenotype of breast carcinoma cells. Proc Natl Acad Sci U S A. 1995;92:7411-7415
21. Kaplan-Meier Plotter. http://kmplot.com/analysis
22. Wu X, Chen H, Parker B. et al. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66:9527-9534
23. Pathiraja TN, Nayak SR, Xi Y. et al. Epigenetic reprogramming of HOXC10 in endocrine-resistant breast cancer. Sci Transl Med. 2014;6:229ra241
24. Bayraktar S, Glück S. Molecularly targeted therapies for metastatic triple-negative breast cancer. Breast Cancer Res Treat. 2013:1-15
25. Arpino G, Wiechmann L, Osborne CK. et al. Crosstalk between the estrogen receptor and the HER tyrosine kinase receptor family: molecular mechanism and clinical implications for endocrine therapy resistance. Endocr Rev. 2008;29:217-233
26. Morandi A, Plaza-Menacho I, Isacke CM. RET in breast cancer: functional and therapeutic implications. Trends Mol Med. 2011;17:149-157
27. di Pietro M, Lao-Sirieix P, Boyle S. et al. Evidence for a functional role of epigenetically regulated midcluster HOXB genes in the development of Barrett esophagus. Proc Natl Acad Sci U S A. 2012;109:9077-9082
Author contact
![]() Corresponding author: E-mail: mhkim1ac; Tel: +82-2-2228-1647, Fax: +82-2-365-0700
Corresponding author: E-mail: mhkim1ac; Tel: +82-2-2228-1647, Fax: +82-2-365-0700

 Global reach, higher impact
Global reach, higher impact