10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(7):824-835. doi:10.7150/ijbs.14408 This issue Cite
Research Paper
A Novel Malaria Pf/Pv Ab Rapid Diagnostic Test Using a Differential Diagnostic Marker Identified by Network Biology
1. Department of Bioinformatics, College of Natural Sciences, Chungbuk National University, Cheongju, Chungbuk, Republic of Korea
2. Department of Biochemistry, College of Natural Sciences, Chungbuk National University, Cheongju, Chungbuk, Republic of Korea
3. Laboratory Medicine, Chungbuk National University Hospital, Cheongju, Chungbuk, Republic of Korea
4. Hospital Kuala Lumpur, Jalan Pahang, Malaysia
5. GenBody Inc., Dankook Biotech Business IC, Cheonan, Chungnam, Republic of Korea
6. Department of Infection Biology, Wonkwang University School of Medicine, Iksan, Jeonbuk, Republic of Korea
* Sung Jin Cho and Jihoo Lee contributed equally to this work
Received 2015-11-13; Accepted 2016-5-6; Published 2016-5-25
Abstract
Rapid diagnostic tests (RDTs) can detect anti-malaria antibodies in human blood. As they can detect parasite infection at the low parasite density, they are useful in endemic areas where light infection and/or re-infection of parasites are common. Thus, malaria antibody tests can be used for screening bloods in blood banks to prevent transfusion-transmitted malaria (TTM), an emerging problem in malaria endemic areas. However, only a few malaria antibody tests are available in the microwell-based assay format and these are not suitable for field application. A novel malaria antibody (Ab)-based RDT using a differential diagnostic marker for falciparum and vivax malaria was developed as a suitable high-throughput assay that is sensitive and practical for blood screening. The marker, merozoite surface protein 1 (MSP1) was discovered by generation of a Plasmodium-specific network and the hierarchical organization of modularity in the network. Clinical evaluation revealed that the novel Malaria Pf/Pv Ab RDT shows improved sensitivity (98%) and specificity (99.7%) compared with the performance of a commercial kit, SD BioLine Malaria P.f/P.v (95.1% sensitivity and 99.1% specificity). The novel Malaria Pf/Pv Ab RDT has potential for use as a cost-effective blood-screening tool for malaria and in turn, reduces TTM risk in endemic areas.
Keywords: Malaria antibody test, MSP1, Transfusion-transmitted malaria, Blood screening
Introduction
Malaria is a mosquito-borne infectious disease that is caused by five species of human infective parasites belonging to the genus Plasmodium: P. falciparum, P. vivax, P. ovale, P. malariae and P. knowlesi. In total, 104 countries and territories have been considered endemic for malaria in 2013 [1]. WHO estimated 207 million cases of malaria with 627,000 deaths globally, especially in Africa among children under 5 years of age [1]. P. falciparum, the most deadly form, predominates in Africa whereas P. vivax shows a wider distribution. It grows in the vector mosquito, Anopheles, even at lower temperatures. In addition, it can continue its life cycle as a hypnozoite, the dormant liver-stage form, during winter when Anopheles is not present.
Rapid and accurate diagnosis is essential for efficient prevention and prompt treatment of malaria. If reliable diagnostic testing were thoroughly applied to every suspected case, the number of malaria cases requiring treatment can be determined and effective treatment can be administered while avoiding the unnecessary use of expensive anti-malarial drugs [1]. Rapid diagnostic tests (RDTs) offer accurate and timely diagnosis of malaria in endemic areas and are easily accessible, cost effective, and give timely results [2]. Currently, more than 200 malaria RDTs, especially antigen-based RDTs, are available on the market [3]. Malaria antigen-based RDTs detect specific antigens (proteins) produced by malaria parasites that are present in the blood of the infected individual. Here, the targets used for malaria antigen-based RDTs include P. falciparum histidine-rich protein 2 (PfHRP2) and two enzymes involved in Plasmodium glycolysis; plasmodial lactate dehydrogenase (pLDH) and aldolase [4]. PfHRP2- and pLDH-based antigen RDTs are most widely used and their limit of detections are approximately 50-100 parasites/μL [5, 6].
RDTs specific for detecting antibodies against Plasmodium-derived antigens also exist. Antibodies against parasite antigens appear within days to weeks after infection and persist for month or even years [7, 8]. Therefore, malaria antibody tests can detect parasite infection even at low parasitemia (< 50 parasites/μL). Malaria antibody tests, thereby, is useful in malaria endemic areas, in which light infection and/or re-infection of parasites are common. They are also helpful in testing patients who have recently been treated for malaria but their diagnosis is questioned as the microscopic examination of this case can result in a false-negative. Owing to these characteristics of malaria antibody tests, they are used for the screening of donated bloods in blood banks to prevent transfusion-transmitted malaria (TTM).
TTM is acquired through transfusion of whole blood and cellular blood components. It is responsible for most transfusion-transmitted diseases worldwide [9, 10]. Since the first case of TTM was described in 1911 [11], rare but significant number of TTM cases were reported worldwide, predominantly from non-endemic areas of malaria. The real incidence of TTM in malaria endemic areas is unknown as individuals residing in the areas often have asymptomatic parasitemia and it is difficult to ascertain whether TTM leads to clinical malaria [12]. However, individuals presenting asymptomatic malaria present a risk for TTM especially to vulnerable populations such as young children, pregnant women, and immunocompromised patients, as they are the predominant users of blood transfusions in endemic areas such as Africa [13]. To decrease the risks of TTM, WHO recommends pre-screening of all donated bloods for malaria with quality-assured tests [14]. Currently, a high throughput assay that is sensitive, practical, and cost-effective is not available. Microscopic examination of the blood film is considered the 'gold standard' for detecting parasites, especially in endemic areas, where serological tests are not helpful due to the semi-immune status of most residents [15, 16]. However, it requires extensive training for acquiring skill and is labor intensive and time consuming. In addition, microscopy may provide false-positives (artefacts) and false-negatives at low parasite density [17, 18]. The sensitivity varies by the skill of the microscopist, and the limit of detection is approximately 50 parasites/μL for an experienced microscopist [19].
Malaria antigen-based RDTs are not suitable to control TTM because of decreased sensitivity at low parasite density. This notion is supported by previous works reporting that two commercial malaria antigen-based RDTs using pLDH or circulating P. falciparum antigen were less sensitive and specific [10, 20]. Therefore, potential blood donors must be tested for antibodies against Plasmodium-derived antigens. However, only a few malaria antibody tests (Lab21 Healthcare Ltd., United Kingdom; Cellabs, Australia; Newmarket Laboratories Ltd., United Kingdom) are now commercially available in an ELISA (enzyme-linked immunosorbent assay) format rather than a RDT [21], and most of them are still under evaluation for use in blood screening, especially in non-endemic areas. This is because TTM has been neglected until recently because of the main interest in blood safety against HIV (human immunodeficiency virus) [10, 12]. To increase accessibility in field application, simple and cost-effective malaria antibody RDTs with high sensitivity and specificity are necessary. In this study, malaria antibody RDT has been developed using a differential diagnostic marker identified by data mining approaches.
Since the currently used parasite antigens are still limited to blood screenings, a new diagnostic target was identified using data mining approaches. In particular, a protein-protein interaction network was utilized to understand protein functions and to identify hub proteins that have central roles in protein complexes in most biological processes [22, 23]. A dynamic interconnection among proteins is a fundamental component encoding complexity of intracellular events within a living cell. Thus, understanding and decomposing proteins based on modular organization of a protein network is a powerful method to identify drug targets and/or diagnostic markers [23-29]. Furthermore, as the Plasmodium protein network is significantly diverse from those of other eukaryotes [30, 31], dissecting a protein network can provide valuable diagnostic markers unique to the malaria parasites. Interestingly, Plasmodium MSP1 (merozoite surface protein 1), one of leading malaria vaccine candidates, was identified as a diagnostic marker [32, 33]. Here, the potential of P. falciparum and P. vivax MSP1 as diagnostic antigens was tested by developing and clinically evaluating PfMSP1- and PvMSP1-based antibody tests (both ELISA and RDT). The developed PfMSP1- and PvMSP1-based antibody tests were found to be highly sensitive and specific for differential diagnosis of falciparum and vivax malaria. The Malaria Pf/Pv antibody RDT developed in this study can be used to screen blood donors for malaria in blood transfusion services and blood banks.
Materials and Methods
Extraction and network establishment of Plasmodium falciparum proteins
Total genes of Plasmodium falciparum 3D7 were extracted from PlasmoDB (http://plasmodb.org/plasmo) [34] by using data files containing total gene information table such as gene or protein interaction, and gene ID (PlasmoDB-9.1_Pfalciparum3D7Gene.txt and PlasmoDB-9.1_Pfalciparum3D7_GeneAliases.txt). Protein list and interaction data were extracted from PlasmoDB-9.1_Pfalciparum3D7Gene.txt and the IDs (PlasmoDB type) were uniformly converted to ones used in PlasmoDB-9.1_Pfalciparum3D7_GeneAliases.txt to make them compatible with other data files. This process was performed by using a programming language, python (http://www.python.org).
Protein-protein interaction (PPI) network was established based on a total of 1,304 proteins and protein interaction information extracted from PlasmoDB-9.1_Pfalciparum3D7Gene.txt. Overlapping data were excluded and finally 2,745 PPI network data of 1,304 proteins were obtained. The PPI network was visualized to a graph with Cytoscape, an open source software platform (http://www.cytoscape.org). One protein is shown as a node and one interaction as a link in the graph.
Decomposition of PPI network to select core proteins
PPI network was simplified by using k-core algorithm. k-core decomposition is proceeded by sequentially removing all nodes with less links from the graph [35]. For example, in the first round, all nodes with one or less links were removed from the graph and in the second round, all nodes with two links or less were removed from the graph, and finally their links were deleted. Removal of a node is done reflexively. A program processed by programming language, perl was applied to k-core decomposition.
Screening of target proteins by functional analysis based on gene ontology
In total, 154 core proteins were selected through k-core decomposition. Target proteins were further screened based on their functions. Protein function was analyzed, sorted, and listed in order of importance (expressed by a degree in the final version of list) by using AmiGO (http://amigo.geneontology.org/cgi-bin/amigo/go.gi) [36]. Biological/pathogenic pathways and cell components were focused. In particular, proteins present in extracellular region, cell surface, and membrane of parasite were targeted, as the study aims to select a protein candidate for diagnosis.
Construction of recombinant PfMSP119 and PvMSP119
Plasmodium DNA was extracted from Pf- or Pv-infected patient plasma specimens that were obtained from General Hospital Kuala Lumpur, Malaysia using QIAamp Genomic DNA Kit (Qiagen, ICI Americas Inc., USA). Gene fragment of PfMSP119 was amplified by PCR with a forward primer (5'-CTG CTC GAG ATG AAC ATT TCA CAA CAC-3') and a reverse primer (5'-GGA TCC GTT AGA GGA ACT GCA GAA AAT-3'). The primers contain upstream XhoI and downstream BamHI restriction sites (underlined). Gene fragment of PvMSP119 was amplified by PCR using the following primers: PvMSP119ΔC - forward (5'-TCT GGA TCC CTG GAA AAA TTG ATG-3`) and reverse (5'-CTC GAG CCT ACA GAA AAC GCC CTC-3'), PvMSP119ΔN - forward (5`-GGA TCC GAA AAC GAG TCC AAG GAA -3`) and reverse (5`-CTC GAG AAG CTC CAT GCA CAG GAG-3`). The forward primers contain BamHI site and the reverse primers contained XhoI site (underlined). Primers were designed based on the sequences of the Uganda-Palo Alto strain for P. falciparum (GenBank Accession No. M37213) [37] and the Belem strain for P. vivax, respectively (GenBank Accession No. M60807) [38]. The amplified fragments were first inserted into pGEM-T Easy Vector (Promega, Madison, WI, USA) and then subcloned into the pET15b expression vector (PfMSP-119) and pET21a expression vector (PvMSP119ΔC and PvMSP119ΔN) (Novagen, Merck Millipore, Darmstadt, Germany). The resultant constructs were confirmed by sequencing.
Expression of recombinant PfMSP119 and PvMSP119
Laboratory scale: The recombinant construct was transformed subsequently into E. coli BL21(DE3). Transformed E. coli BL21(DE3) was cultured in 1 l LB media supplemented with 50 μg/ml ampicillin. When an OD600 reached 0.8~1.0 at 37°C, induction was performed as follows: PfMSP119; 0.2 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside) for 4 h at 30°C, PvMSP119; 0.5 mM IPTG for 16 h at 24°C. The bacterial cells were cold-centrifuged at 6,000~8,000 rpm for 15-30 min and the pellet was stored at -20°C before use.
Bulk scale: Transformed E. coli BL21(DE3) was cultured in a sterilized 500 l fermenter (FR-903, Fermentec, Cheongju, Korea) including 300 l LB media supplemented with 50 ml antifoam A and 50 μg/ml ampicillin. The fermenter was controlled at 100 rpm, 50 vvm airflow, 37°C, and pH 6.15. Induction was performed when OD600 reached 0.8~1.0. For PfMSP119, 0.2 mM IPTG was added and cultured at 30°C for 4 h with adjusted control by 110 rpm agitation, 55 vvm airflow, and pH 7.5. For PvMSP119, 0.5 mM IPTG was added and cultured at 24°C for 16 h without adjustment of cascade control. The bacterial cells were centrifuged by tubular type continuous separator (U-160H, Hanil Science Medical, Daejeon, Korea) at 8,000~15,000 rpm for 6 h and the pellet was stored at -20°C before use.
Purification of recombinant PfMSP119 and PvMSP119
Wet biomass (10 g) prepared from laboratory and bulk scale expression was resuspended in cell lysis buffer (20 mM Tris-Cl, 500 mM NaCl, 5 mM Imidazole): PfMSP119; 10 ml including 10% (v/v) Triton X-100, PvMSP119; 20 ml including 0.1 g lysozyme, 10 mM HEPES and 2.5% Triton X-100. Cells were lysed by sonication (Vibra-CellTM VCX130, Sonics & Materials Inc., USA) and cold-centrifuged at 13,000 rpm for 30 min. Impurities were filtered off by a 0.45-μm syringe filter. The filtrate was introduced to pre-equilibrated Ni-NTA column (Qiagen, ICI Americas Inc., USA). The bound proteins were washed (20 mM Tris-Cl, 500 mM NaCl, 30~60 mM imidazole) and eluted with buffer solution (20 mM Tris-Cl, 500 mM NaCl, 500 mM imidazole). The eluted protein was dialyzed using 100 mM carbonate (pH 9.0) at 4°C for 16 h by changing the buffer several times and concentrated with centrifugal filter device (Centricon, Merck Millipore, Darmstadt, Germany) at 3,000 rpm for 20 min. Proteins were quantified and used for the subsequent experiments.
Clinical specimens
Plasmodium-positive and -negative blood samples were used for the in-house test for measuring the antigenicity of purified recombinant proteins: 10 Pv-positive samples from Korean Center for Disease Control, 6 Pf-positive samples and 20 Plasmodium-negative samples from Dankook University Hospital in Korea. For the independent evaluation of a novel RDT developed in this study, 102 Pf or Pv positive samples were provided by the Kuala Lumpur General Hospital, Malaysia and 340 Plasmodium negative samples were obtained from ChoongAng Hospital and Hankook Hospital in Korea. Malaria positivity of all samples was checked using microscopic analysis and PCR [39]. Malaria negativity was confirmed by ELISA using Pan Malaria Antibody CELISA Kit (Cellabs Pty Ltd., Australia). Ethical approvals were obtained from the Institutional Review Board of the Dankook University Hospital, ChoongAng Hospital and Hankook Hospital in Korea, and the Kuala Lumpur General Hospital in Malaysia with written informed consent of all subjects.
Antigenicity test of recombinant PfMSP119 and PvMSP119 by ELISA
Purified recombinant Plasmodium MSP119 proteins were coated on the microplate wells (50 ng/well). Pf- or Pv-infected patient blood samples were added after diluting with 1× PBS (1:10 v/v) and incubated for 30 min at 37°C. After washing with 1× PBS containing 0.05% Tween-20, goat anti-human IgG-HRP (horseradish peroxidase) conjugates (Caltag Lab., CA, USA) were added and incubated for 30 min at 37°C. TMB (3,3',5,5'-Tetramethylbenzidine) solution was added and incubated under the same condition to colorize the reaction. After final washing, the reaction was terminated by adding 0.5 N H2SO4 solution. The absorbance was measured by a microplate reader (Microplate spectrophotometer, Bio-Rad Laboratories Inc., CA, USA) at 450 nm.
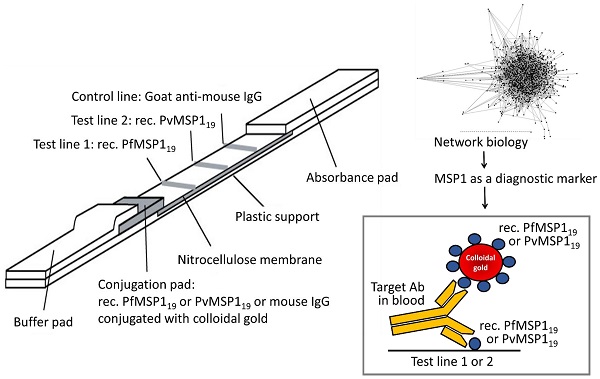
Preparation and evaluation of RDT strips
The strip was prepared as previously described [40, 41]. 1.0 mg/ml of the purified recombinant PfMSP119 and PvMSP119ΔC proteins, and 1.2 mg/ml of goat anti-mouse IgG were dispensed and immobilized to the test line and the control line onto a nitrocellulose membrane, respectively. The membrane was then dried for 1 day at room temperature. The recombinant PfMSP119 and PvMSP119ΔN proteins, and mouse IgG were conjugated with gold colloid as described [42] and then dried on a glass fiber to use as the conjugator pad. The cellulose paper was treated with 0.1 M Tris-Cl (pH 8.0) containing 0.5% PVA (polyvinyl alcohol) and 0.5% Tween-20, and used as the sample pad. The absorbance pad was prepared with cellulose paper without any treatment. The test strip was assembled by placing all pads partially overlapped to enable the migration of the sample solution along the strip.
Ten microliters (10 μl) of blood was dropped into the sample well of the test device and then, 100 μl of assay solution was added (0.1% casein, 0.05% Tween 20, 0.05% sodium azide, 50 mM Borax, pH 9.5). The test results were interpreted in 15 min. Control line should appear in all valid tests. If a red color band appears at the test line, it means that the specimen contains Plasmodium MSP119 specific antibodies.
Results
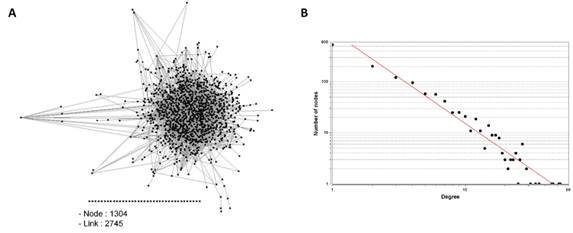
Scale-free network of P. falciparum protein-protein interaction (PPI)
PPI network was established based on the information extracted from PlasmoDB. The visualized network graph displaying PPI showed 1,304 nodes and 2,745 links (Fig. 1A). It had abundant nodes with few links (nodes having low degree), and few hubs (the highest-degree nodes) with a large number of links, suggesting that the P. falciparum PPI network is a scale-free network, the degree distribution of which follows a power law, as described previously [30, 43] (Fig. 1B).
Extraction of key proteins from PPI network
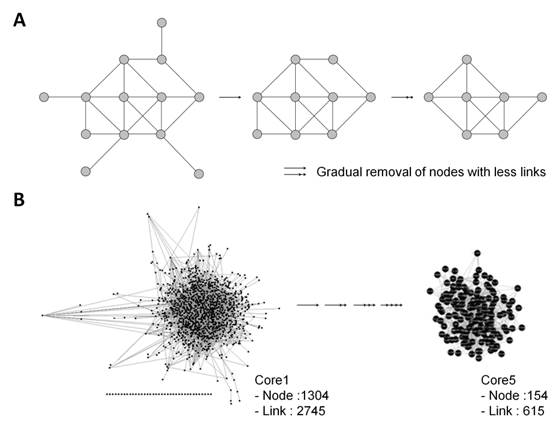
k-core algorithm was applied to simplify the PPI network. It is a very simple algorithm, originating from statistical mechanics for investigating graph properties, which removes all nodes with less links from the network graph, thus, simplifying the network (Fig. 2A). Based on the degree of nodes, removal of nodes and links was gradually carried out up to building Core5 from an original network (Core1) (Fig. 2B). Finally, Core5 had 154 nodes and 615 links. These 154 core proteins were further analyzed based on gene ontology to select target proteins that would be used as diagnostic markers (Fig. 3). Based on the cell component, 61 proteins were unknown and the others were mainly located in the cytoplasm, cell membrane and nucleus of parasite (Fig. 3A). Based on the function, the most proteins (90) were unknown (Fig. 3B). The rest were involved in biological/pathogenic pathways such as transcription, cell invasion including host cell adhesion, locomotion and transport, and cell communication (Fig. 3B). Three functional groups were screened to identify a target protein as they directly interact with external routes and thus, relate to the induction of immune response (specifically antibody production) in the host: membrane, extracellular region, and cell surface of parasite. Proteins in each group were listed in the order of degree, and as a result, merozoite surface protein 1 (MSP1) (PFI1475w) exhibited the highest degree in membrane group (Table 1 and Table S1). The degree of MSP1 was 24, which was the highest among other proteins in all functional groups. This suggests that compared to other protein, MSP1 has the largest number of links, acting as a key protein in the P. falciparum PPI network. Thus, MSP1, a membranous antigenic protein involved in pathogenesis, could be a good diagnostic marker.
Diagnostic marker candidates extracted from PPI network
| Membrane | Extracellular region | Cell surface | |||
|---|---|---|---|---|---|
| Gene name | Degree* | Gene name | Degree | Gene name | Degree |
| PFI1475w (MSP1) | 24 | PF14_0678 | 16 | MAL13P1.269 | 6 |
| PFD0885c | 14 | PFB0340c | 12 | PF10_0155 | 5 |
| PFA0430c | 12 | PFA0110w | 10 | PF10_0348 | 5 |
| MAL7P1.7 | 9 | PF14_0344 | 8 | ||
| PF10_0242 | 9 | PFE0070w | 6 | ||
| MAL13P1.206 | 9 | PF14_0442 | 5 | ||
| PF14_0201 | 9 | ||||
| PF13_0197 | 9 | ||||
| PFA0410w | 8 | ||||
| PF07_0128 | 7 | ||||
| PF13_0198 | 7 | ||||
| PFB0300c | 6 | ||||
| PFA0125c | 6 | ||||
| PF10_0143 | 6 | ||||
| PF11_0127 | 6 | ||||
| PFC1045c | 5 | ||||
| MAL13P1.60 | 5 | ||||
| PF11_0341 | 5 | ||||
| MAL13P1.193 | 5 | ||||
| PF10_0348 | 5 | ||||
| PF08_0127 | 5 | ||||
| MAL8P1.95 | 5 | ||||
| PF14_0731 | 5 | ||||
| PFD0545w | 5 | ||||
| PF14_0315 | 5 | ||||
| PF10_0146 | 5 | ||||
| PF11_0165 | 5 | ||||
| PFL1395c | 5 | ||||
| PF10_0155 | 5 | ||||
*The larger degree means that the node (protein) has a large number of links in the PPI network.
Protein-protein interaction (PPI) network of Plasmodium falciparum. (A) Each node (shown by Black square) represents each protein and links represent the interconnection among nodes. (B) Distribution of nodes based on their degrees in the PPI network. R = 0.987, R2 = 0.938
(A) Schematic drawing of decomposing PPI network by k-core algorithm. All nodes with less links are gradually removed from the network graph. (B) Simplified P. falciparum PPI network by k-core algorithm.
Gene ontology of 154 core proteins extracted from Core5 PPI network was analyzed based on (A) cellular location and (B) function. Membrane and cell surface in (A) indicate those of parasite. The total number indicated in pie graphs (A and B) exceeds 154 as many proteins have multiple cellular locations and functions.

Production of antigenic Plasmodium MSP1 protein
P. falciparum MSP119 protein (PfMSP119) containing the 19-kDa C-terminal region was generated as the region including the EGF (epidermal growth factor) domain, which is known to retain high antigenicity. The most C-terminal region, GPI (glycosyl-phosphatidylinositol) anchor, was not included in the recombinant protein to increase antigenicity. The 19-kDa C-terminal region of P. vivax MSP1 protein was selected for the same reason. This region was further dissected and generated into two recombinant proteins: one missing 11 N-terminal amino acids (PvMSP119ΔN) and the other missing 22 C-terminal amino acids (PvMSP119ΔC).
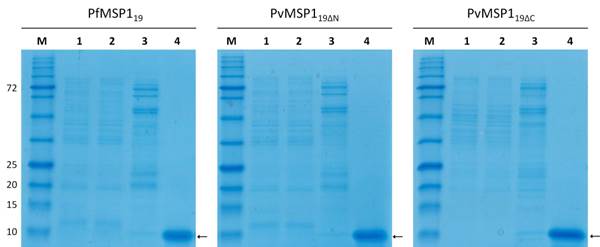
The recombinant PfMSP119 and PvMSP119 proteins were expressed and purified at laboratory scale and in bulk scale. Both purification processes successfully delivered approximately 10-kDa Plasmodium MSP1 recombinant proteins (Fig. 4). Laboratory scale produced 1.5 mg of PfMSP119 protein from 1 g of bacterial wet mass (3.75 mg/l) and bulk scale produced 4 mg of protein from 1 g of bacterial wet mass (20 mg/l) after final purification (Table 2). Laboratory scale production of recombinant PvMSP119ΔC and PvMSP119ΔN resulted in 0.37 mg and 0.36 mg of protein from 1 g of bacterial wet mass, respectively (0.63 mg/l and 0.72 mg/l). Whereas bulk scale production resulted in 0.4 mg and 0.36 mg of each protein from 1 g of bacterial wet mass, respectively (1.83 mg/l and 1.25 mg/l) (Table 2).
Protein yield from laboratory (Lab.) and bulk scale expression
| PfMSP119 | PvMSP119ΔC | PvMSP119ΔN | ||||||
|---|---|---|---|---|---|---|---|---|
| Lab. | Bulk | Lab. | Bulk | Lab. | Bulk | |||
| Culture size (L) | 4 | 300 | 4 | 300 | 4 | 300 | ||
| Cell pellet (g) | 10 | 1,500 | 6.8 | 1,372 | 8.01 | 1,043 | ||
| Purified protein per gram (mg) | 1.5 | 4.0 | 0.37 | 0.4 | 0.36 | 0.36 | ||
| Calculated yield (mg/L) | 3.75 | 20 | 0.63 | 1.83 | 0.72 | 1.25 | ||
Clinical evaluation of antigenic recombinant PfMSP119 and PvMSP119 proteins
To test the antigenicity of recombinant proteins, a direct ELISA was performed using clinical blood specimens collected from KCDC and Dankook University Hospital in Korea. Purified recombinant proteins were coated on the well as antigens, and Pf- or Pv-infected blood samples were applied to test whether antibodies present in patients' bloods can recognize these antigens. Plasmodium-negative blood samples were used as controls. A direct ELISA using purified recombinant PfMSP119 perfectly discriminated Pf-positive blood samples from Pv-positive and Plasmodium negative blood samples, thus achieving 100% sensitivity and specificity (Fig. 5A). Purified recombinant PvMSP119 was also tested against Pv-positive blood samples. Antibodies in Pv-positive patient samples specifically recognized both PvMSP119ΔC and PvMSP119ΔN proteins (100% sensitivity and specificity). Similar responses were observed for both PvMSP119ΔC and PvMSP119ΔN protein-based ELISAs (identical clinical samples were used in the same order for both ELISAs), and the absorbance was slightly higher in the case of recombinant PvMSP119ΔC protein (Fig. 5B and 5C). Results showed that purified recombinant PfMSP119 and PvMSP119 proteins were successfully and effectively recognized by antibodies in patient samples, suggesting the potential of these markers for use in a differential diagnosis of falciparum and vivax malaria.
Bulk scale expression and purification of PfMSP119 and PvMSP119 recombinant proteins. M, protein size marker; Lane 1, total cell lysate; Lane 2, unbound fraction (flow-through); Lane 3, eluted fraction with washing buffer containing 40 mM imidazole; Lane 4, eluted fraction with elution buffer containing 500 mM imidazole. Arrows indicate purified recombinant proteins.
Clinical evaluation of PfMSP119 and PvMSP119 antigens by ELISA. PfMSP119 (A), PvMSP119ΔC (B) and PvMSP119ΔN (C) antigens were coated on the well and reacted with Pf-positive (Pf Ab(+)), Pv-positive (Pv Ab(+)), and Plasmodium-negative clinical samples. The identical samples were used for PvMSP119∆C (B) and PvMSP119∆N (C) in the same order.
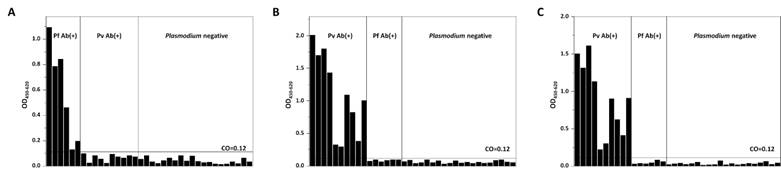
Clinical evaluation of PfMSP119- and PvMSP119-based rapid diagnostic tests (RDTs). (A) PfMSP119-based Ab RDT (B) Comparison of diagnostic performance between PvMSP119-based Ab RDT (Novel) and SD BioLine Malaria P.f/P.v test kit (Commercial). Absorbance in ELISA: high (nos. 1 and 3), medium (nos. 7 and 10) and low (nos. 5 and 6).

Construction and evaluation of a novel Malaria Ab RDT
Clinical evaluation was performed in the format of a RDT. Malaria Ab RDT strips were constructed using purified PfMSP119 or PvMSP119 proteins. Clinical test delivered PvMSP119ΔN protein as a conjugator and PvMSP119ΔC protein as a capturer in the strip (data not shown). Each PfMSP119 or PvMSP119ΔN protein was conjugated with colloidal gold to prepare a conjugator pad. The conjugator pad includes mouse-IgG conjugated with colloidal gold to present a valid test by observing the appearance of color band at the control line. Test line contains PfMSP119 or PvMSP119ΔC proteins. Identical clinical samples used in a direct ELISA were applied to strips and results were interpreted within 15 min. PfMSP119-based Ab RDT successfully detected Pf-positive blood samples but not Pv-positive or Plasmodium-negative samples, thus providing 100% sensitivity and specificity (Fig. 6A). The color intensity was measured to judge the positivity of two Pf samples (nos. 5 and 6) as each of them showed only faint band at the test line (Fig. 6A), making hard interpretation of test results. Analysis showed that those two Pf samples (nos. 5 and 6) are apparently positive (Fig. S1). The color intensity of Pf-positive samples at the test line was correlated with the absorbance observed in ELISA (Fig. S1). However, as the parasite density of samples is not available and a colorimetric assay is limited in its ability to accurately represent the quantity of antigens, this is not conclusive. PvMSP119-based Ab RDT also discriminated only Pv-positive blood samples, showing 100% sensitivity and specificity. In addition, the diagnostic performance of PvMSP119-based Ab RDT was compared to that of a commercial kit (SD BIOLINE Malaria P.f/P.v from Alere Inc). Comparison was conducted using three groups of Pv-positive samples; two showing high (nos. 1 and 3), two showing medium (nos. 7 and 10) and two showing low (nos. 5 and 6) absorbance, as determined in ELISA. Similar to the diagnostic results of Pf-positive samples, both RDTs exhibited plausible correlation between the color intensity at the test lines and the absorbance observed in ELISA. Results showed that the diagnostic performance was better or comparable in PvMSP119-based Ab RDT; the appearance of apparent lines at the control/test lines in all samples and low background signal (e.g. normal sample) including no appearance of a non-specific band (e.g. sample no. 5) were noted (Fig. 6B). This demonstrated the high sensitivity and specificity of the novel RDT developed in this study.
Malaria Pf/Pv Ab RDT strips were then constructed to contain both PfMSP119 and PvMSP119 proteins to differentially diagnose falciparum and vivax malaria in a single rapid test. This novel Malaria Pf/Pv Ab RDT was objectively evaluated in an independent facility (out-of-house evaluation) by using malaria-positive blood samples collected from Malaysia. The sensitivity and specificity of a novel Ab RDT were compared to those of SD BIOLINE Malaria P.f/P.v Ab rapid test. The results showed that the novel Malaria Pf/Pv Ab RDT developed in this study is superior to the commercial kit. The sensitivity and specificity of a novel Malaria Pf/Pv Ab RDT were 98% and 99.7%, respectively (Table 3). Both RDTs resulted in two false-negatives (sample nos. 66 and 92 in Table 4) and one false-positive (very weak Pv-positive signal by both) against identical specimens (sample no. 13; data not shown). The commercial kit displayed three more false-negatives (sample nos. 6, 21 and 44 in Table 4) and two more false-positives (weak Pf-positive signals) (sample nos. 26 and 40; data not shown), thus providing 95.1% sensitivity and 99.1% specificity (Table 3). Among the 97 samples positively diagnosed by both kits, only seven samples (sample nos. 1, 10, 23, 36, 49, 62 and 75) were differently diagnosed; the novel kit displayed both Pf- and Pv-positive signals while the commercial kit showed only Pf-positive signals (Table 4). The overall correlation ratio of both kits was 90.2% (Table S2). Interestingly, the novel kit was able to detect Pv-positive signals from 3 false-negatives diagnosed by the commercial kit (Table 4 and S2). These results suggest that the novel kit is efficient in detecting falciparum malaria but is better at detecting vivax malaria.
Comparative evaluation of a novel Malaria Pf/Pv Ab rapid test and SD BioLine Malaria P.f/P.v
| A novel Malaria Pf/Pv | SD BioLine Malaria P.f/P.v | ||||||
|---|---|---|---|---|---|---|---|
| + | - | + | - | ||||
| Malaria* | + (n=102) | 100 | 2 | 97 | 5 | ||
| - (n=340) | 1 | 339 | 3 | 337 | |||
| Sensitivity (%) | 98.0 (100/102) | 95.1 (97/102) | |||||
| Specificity (%) | 99.7 (339/340) | 99.1 (337/340) | |||||
*Malaria positivity was checked using microscopic analysis and malaria negativity was confirmed by ELISA (see Materials and Methods).
Test results of malaria positive samples using a novel Malaria Pf/Pv and SD BioLine Malaria P.f/P.v
| Sample no. | A novel Malaria Pf/Pv | SD BioLine Malaria P.f/P.v | |||||
|---|---|---|---|---|---|---|---|
| Pf | Pv | Negative | Pf | Pv | Negative | ||
| 1* | + | + | - | + | - | - | |
| 2 | + | + | - | + | + | - | |
| 3 | + | - | - | + | - | - | |
| 4 | - | + | - | - | + | - | |
| 5 | - | + | - | - | + | - | |
| 6*** | - | + | - | - | - | + | |
| 7 | - | + | - | - | + | - | |
| 8 | - | + | - | - | + | - | |
| 9 | + | - | - | + | - | - | |
| 10* | + | + | - | + | - | - | |
| 11 | + | + | - | + | + | - | |
| 12 | - | + | - | - | + | - | |
| 13 | - | + | - | - | + | - | |
| 14 | + | + | - | + | + | - | |
| 15 | + | - | - | + | - | - | |
| 16 | - | + | - | - | + | - | |
| 17 | - | + | - | - | + | - | |
| 18 | - | + | - | - | + | - | |
| 19 | - | + | - | - | + | - | |
| 20 | - | + | - | - | + | - | |
| 21*** | - | + | - | - | - | + | |
| 22 | + | - | - | + | - | - | |
| 23* | + | + | - | + | - | - | |
| 24 | + | + | - | + | + | - | |
| 25 | - | + | - | - | + | - | |
| 26 | - | + | - | - | + | - | |
| 27 | + | + | - | + | + | - | |
| 28 | + | - | - | + | - | - | |
| 29 | + | - | - | + | - | - | |
| 30 | - | + | - | - | + | - | |
| 31 | - | + | - | - | + | - | |
| 32 | - | + | - | - | + | - | |
| 33 | - | + | - | - | + | - | |
| 34 | - | + | - | - | + | - | |
| 35 | - | + | - | - | + | - | |
| 36* | + | + | - | + | - | - | |
| 37 | + | + | - | + | + | - | |
| 38 | + | + | - | + | + | - | |
| 39 | - | + | - | - | + | - | |
| 40 | + | - | - | + | - | - | |
| 41 | - | + | - | - | + | - | |
| 42 | - | + | - | - | + | - | |
| 43 | - | + | - | - | + | - | |
| 44*** | - | + | - | - | - | + | |
| 45 | + | + | - | + | + | - | |
| 46 | + | + | - | + | + | - | |
| 47 | + | - | - | + | - | - | |
| 48 | - | + | - | - | + | - | |
| 49* | + | + | - | + | - | - | |
| 50 | - | + | - | - | + | - | |
| 51 | - | + | - | - | + | - | |
| 52 | - | + | - | - | + | - | |
| 53 | - | + | - | - | + | - | |
| 54 | - | + | - | - | + | - | |
| 55 | - | + | - | - | + | - | |
| 56 | + | + | - | + | + | - | |
| 57 | + | + | - | + | + | - | |
| 58 | + | - | - | + | - | - | |
| 59 | + | - | - | + | - | - | |
| 60 | - | + | - | - | + | - | |
| 61 | - | + | - | - | + | - | |
| 62* | + | + | - | + | - | - | |
| 63 | + | + | - | + | + | - | |
| 64 | - | + | - | - | + | - | |
| 65 | - | + | - | - | + | - | |
| 66** | - | - | + | - | - | + | |
| 67 | + | - | - | + | - | - | |
| 68 | + | - | - | + | - | - | |
| 69 | + | + | - | + | + | - | |
| 70 | + | + | - | + | + | - | |
| 71 | + | - | - | + | - | - | |
| 72 | - | + | - | - | + | - | |
| 73 | - | + | - | - | + | - | |
| 74 | - | + | - | - | + | - | |
| 75* | + | + | - | + | - | - | |
| 76 | - | + | - | - | + | - | |
| 77 | - | + | - | - | + | - | |
| 78 | - | + | - | - | + | - | |
| 79 | - | + | - | - | + | - | |
| 80 | + | + | - | + | + | - | |
| 81 | + | - | - | + | - | - | |
| 82 | + | - | - | + | - | - | |
| 83 | + | - | - | + | - | - | |
| 84 | - | + | - | - | + | - | |
| 85 | + | + | - | + | + | - | |
| 86 | - | + | - | - | + | - | |
| 87 | - | + | - | - | + | - | |
| 88 | + | + | - | + | + | - | |
| 89 | + | + | - | + | + | - | |
| 90 | - | + | - | - | + | - | |
| 91 | - | + | - | - | + | - | |
| 92** | - | - | + | - | - | + | |
| 93 | + | + | - | + | + | - | |
| 94 | + | - | - | + | - | - | |
| 95 | - | + | - | - | + | - | |
| 96 | - | + | - | - | + | - | |
| 97 | - | + | - | - | + | - | |
| 98 | - | + | - | - | + | - | |
| 99 | - | + | - | - | + | - | |
| 100 | + | - | - | + | - | - | |
| 101 | + | + | - | + | + | - | |
| 102 | + | + | - | + | + | - | |
+ and - indicate positive and negative result, respectively. *Samples with Pf-positive for both kits but difference in the Pv diagnosis between two kits (Pv-positive only in a novel kit); **Samples with false-negative for both kits; ***Samples with Pv-positive in a novel kit but false-negative in a commercial kit.
Discussion
Here, we identified Plasmodium MSP1 as a useful marker to differentially diagnose falciparum and vivax malaria. Plasmodium MSP1 is a leading candidate for the malaria vaccine, especially a blood-stage vaccine, as it is abundantly expressed in the intraerythrocytic cycle (ring, trophozoite, and schizont stages) [44]. MSP1 is proteolytically cleaved into a 42-kDa C-terminal polypeptide at the time of invasion into red blood cells. It is further processed to 33-kDa and 19-kDa fragments. The 19-kDa C-terminal fragment remains on the surface of the merozoite and is highly immunogenic [21, 45-49]. Thus, a 19-kDa C-terminal fragment of MSP1 (MSP119) has been utilized to develop vaccines. Due to its ability to elicit antibody response in humans, MSP119 is used in commercial ELISAs as an antigen for predetermining antibody titers in vaccine recipients prior to vaccination. Hence, it is not surprising that Plasmodium MSP1 was chosen as a diagnostic marker. However, the novelty of this study is to elucidate Plasmodium MSP1 as a marker.
Target and biomarker discovery in disease treatment and diagnosis is conducted with two approaches. Systemic approach, a classical way, where the information derived from intensive in vitro and in vivo studies of diseases is used to select targets and markers. However, the post-genomic era opened a new strategy to uncover useful targets and markers as large-scale sequencings have not only provided complete genome sequence information but also allowed the development of organism-specific cellular networks based on the complex interconnectivity of thousands of proteins at the system level [22, 23, 50]. This network approach can facilitate the elucidation of novel targets and/or markers of diseases. In this study, with the generation of the Plasmodium-specific network and the hierarchical organization of modularity in the network, MSP1 was confirmatively identified as an important marker.
The potential of Plasmodium MSP1 for use in a diagnostic assay was validated by preparing recombinant proteins. The 19-kDa C-terminal fragments of P. falciparum and P. vivax MSP1 were found to be antigenic as described previously. In particular, two recombinant proteins of PvMSP119 (PvMSP119ΔN and PvMSP119ΔC) were prepared to generate highly antigenic proteins. This was based on the prediction of antigenicity. When antigenicity of PvMSP119 was predicted using various methods, and different results were delivered. Thus, two different recombinant proteins were expressed and evaluated. ELISA results showed that two recombinant proteins of PvMSP119 (PvMSP119ΔN and PvMSP119ΔC) induced similar antibody responses. Interestingly, the combination of two recombinant proteins in a strip delivered the better performance in diagnosis than the use of a single recombinant protein. Thus, two recombinant proteins of PvMSP119 (PvMSP119ΔN and PvMSP119ΔC) were used to construct a strip. Recombinant proteins were expressed in bulk level to test whether industrial-level protein production was feasible. In addition, purification process was conducted with no addition of denaturants to preserve the antigenicity of proteins and thereby build reliable malaria antibody tests. Clinical evaluation was performed by using Plasmodium-negative and -positive blood samples. Evaluation results suggest that purified PfMSP119 and PvMSP119 recombinant proteins are highly sensitive and specific for the differential diagnosis of falciparum and vivax malaria. PfMSP119- and PvMSP119-based antibody assays presented 100% sensitivity and specificity in both in-house ELISA and RDT evaluation. A novel Malaria Pf/Pv Ab RDT was subsequently constructed and comparatively evaluated with a commercial kit with a large number of clinical specimens. Results showed that the novel kit developed in this study is more sensitive (98%) and specific (99.7%) than is SD BioLine Malaria P.f/P.v from Alere Inc. (95.1% sensitivity and 99.1% specificity), which uses the same target as that used in the novel kit. The diagnostic performance of the novel Malaria Pf/Pv Ab RDT is comparable or better than that of others presented in previous reports [21, 48, 49, 51, 52], although most previous studies tested the reactivity of PfMSP119 or PvMSP119 recombinant proteins with ELISA and western blot analysis rather than RDT. The sensitivities ranged from 90~100% for vivax malaria [21, 49, 51] and 50~90.6% for falciparum malaria [21, 51]. The specificity was 50~98.3% for vivax malaria [21, 49, 51] and 50% for falciparum malaria [21]. One report described the development and evaluation of PvMSP1-based antibody RDT [52]. The sensitivity and specificity were 96.6% and 91.4%, respectively in the study. Considering that most attempts resulted in relatively high cross-reactivity of Plasmodium MSP1-based immunoassays among Plasmodium species, this study provides a valuable antibody test kit for differential diagnosis of falciparum and vivax malaria. However, further evaluation is needed using clinical samples positive with the other Plasmodium species such as P. malariae and P. ovale.
The novel Malaria Pf/Pv Ab RDT developed in this study provides potential for use as a blood-screening tool in blood transfusion services and blood banks for the control of TTM. The previous studies using Plasmodium MSP1 for blood screening substantiate this potential. A commercial malarial antibody EIA (enzyme immunoassay) utilizing four recombinant Plasmodium MSP1 antigens (1 PfMSP1, 2 PfMSP2 and 1 PvMSP1) has been applied to screen blood donors in UK [53] and Australia [20]. This 8-well strip microplate-type sandwich assay demonstrated 82.6% and 98.1% sensitivity for acute infection of P. falciparum in independent tests performed in the UK and Australia, respectively. It demonstrated 84.6% and 100% sensitivity for acute infection of P. vivax in each test. Considering that the novel kit exhibited the improved sensitivity in the detection of both P. falciparum and P. vivax compared to that achieved with the above-mentioned commercial EIA, and improved sensitivity in the detection of P. vivax compared to that achieved with the commercial RDT (SD BioLine Malaria P.f/P.v from Alere Inc.), it is significantly encouraging to use the novel Malaria Pf/Pv Ab RDT in blood screening. However, in-depth evaluation is required using a significant number of donated bloods in related facilities. The novel Malaria Pf/Pv Ab RDT may offer the most cost-effective strategy to screen blood donors for malaria and thereby lessen the risk of TTM in endemic areas. In addition, with this reliable and efficient screening test for malaria, unnecessary rejection of blood donations might be minimized.
Supplementary Material
Supplementary tables and figures.
Abbreviations
RDT: rapid diagnostic test; TTM: transfusion-transmitted malaria; Ab: antibody; MSP1: merozoite surface protein 1; PfHRP2: Plasmodium falciparum histidine-rich protein 2; pLDH: plasmodial lactate dehyhrogenase; ELISA: enzyme-linked immunosorbent assay; HIV: human immunodeficiency virus; PPI: protein-protein interaction; EGF: epidermal growth factor; GPI: glycosyl-phosphatidylinositol; IPTG: Isopropyl β-D-1-thiogalactopyranoside; HRP: horseradish peroxidase; TMB: 3,3',5,5'-Tetramethylbenzidine; PVA: polyvinyl alcohol; EIA: enzyme immunoassay.
Acknowledgements
This work was supported by Wonkwang University in 2015.
Competing Interests
The authors have declared that no competing interest exists.
References
1. WHO. World malaria report: 2013. 2013.
2. WHO. A rapid dipstick antigen capture assay for the diagnosis of falciparum malaria. WHO Informal Consultation on Recent Advances in Diagnostic Techniques and Vaccines for Malaria. Bull World Health Organ. 1996;74:47-54
3. WHO. Information note on recommended selection criteria for procurement of malaria rapid diagnostic tests. 2014.
4. Mouatcho JC, Goldring JP. Malaria rapid diagnostic tests: challenges and prospects. J Med Microbiol. 2013;62:1491-505
5. Proux S, Hkirijareon L, Ngamngonkiri C, McConnell S, Nosten F. Paracheck-Pf: a new, inexpensive and reliable rapid test for P. falciparum malaria. Trop Med Int Health. 2001;6:99-101
6. Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15:66-78
7. Drakeley CJ, Corran PH, Coleman PG, Tongren JE, McDonald SL, Carneiro I. et al. Estimating medium- and long-term trends in malaria transmission by using serological markers of malaria exposure. Proc Natl Acad Sci U S A. 2005;102:5108-13
8. Vinetz JM, Li J, McCutchan TF, Kaslow DC. Plasmodium malariae infection in an asymptomatic 74-year-old Greek woman with splenomegaly. N Engl J Med. 1998;338:367-71
9. Owusu-Ofori AK, Bates I. Impact of inconsistent policies for transfusion-transmitted malaria on clinical practice in Ghana. PLoS One. 2012;7:e34201
10. Seed CR, Kee G, Wong T, Law M, Ismay S. Assessing the safety and efficacy of a test-based, targeted donor screening strategy to minimize transfusion transmitted malaria. Vox Sang. 2010;98:e182-92
11. Woolsey G. Transfusion for permicious anaemia: two cases. Annals of Surgery. 1911;53:132-5
12. Nansseu JR, Noubiap JJ, Ndoula ST, Zeh AF, Monamele CG. What is the best strategy for the prevention of transfusion-transmitted malaria in sub-Saharan African countries where malaria is endemic? Malar J. 2013;12:465
13. Owusu-Ofori AK, Betson M, Parry CM, Stothard JR, Bates I. Transfusion-transmitted malaria in Ghana. Clin Infect Dis. 2013;56:1735-41
14. Okwa OO. The status of malaria among pregnant women: a study in Lagos, Nigeria. Afr J Reprod Health. 2003;7:77-83
15. Hassig A, Ambrois-Thomas P, Bruce-Chwatt LJ. Which are the appropriate modifications of existing regulations designed to prevent the transmission of malaria by blood transfusion, in view of the increasing frequency of travel to endemic areas? Vox Sang. 1987;52:138-48
16. Owusu-Ofori AK, Parry C, Bates I. Transfusion-transmitted malaria in countries where malaria is endemic: a review of the literature from sub-Saharan Africa. Clin Infect Dis. 2010;51:1192-8
17. Kilian AH, Metzger WG, Mutschelknauss EJ, Kabagambe G, Langi P, Korte R. et al. Reliability of malaria microscopy in epidemiological studies: results of quality control. Trop Med Int Health. 2000;5:3-8
18. Makler MT, Palmer CJ, Ager AL. A review of practical techniques for the diagnosis of malaria. Ann Trop Med Parasitol. 1998;92:419-33
19. Milne LM, Kyi MS, Chiodini PL, Warhurst DC. Accuracy of routine laboratory diagnosis of malaria in the United Kingdom. J Clin Pathol. 1994;47:740-2
20. Seed CR, Cheng A, Davis TM, Bolton WV, Keller AJ, Kitchen A. et al. The efficacy of a malarial antibody enzyme immunoassay for establishing the reinstatement status of blood donors potentially exposed to malaria. Vox Sang. 2005;88:98-106
21. Muerhoff AS, Birkenmeyer LG, Coffey R, Dille BJ, Barnwell JW, Collins WE. et al. Detection of Plasmodium falciparum, P. vivax, P. ovale, and P. malariae merozoite surface protein 1-p19 antibodies in human malaria patients and experimentally infected nonhuman primates. Clin Vaccine Immunol. 2010;17:1631-8
22. Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651-4
23. Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297:1551-5
24. Kitano H. Systems biology: a brief overview. Science. 2002;295:1662-4
25. Wolf YI, Karev G, Koonin EV. Scale-free networks in biology: new insights into the fundamentals of evolution? Bioessays. 2002;24:105-9
26. Lauffenburger DA. Cell signaling pathways as control modules: complexity for simplicity? Proc Natl Acad Sci U S A. 2000;97:5031-3
27. Rao CV, Arkin AP. Control motifs for intracellular regulatory networks. Annu Rev Biomed Eng. 2001;3:391-419
28. Holter NS, Maritan A, Cieplak M, Fedoroff NV, Banavar JR. Dynamic modeling of gene expression data. Proc Natl Acad Sci U S A. 2001;98:1693-8
29. Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64-8
30. LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR. et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103-7
31. Suthram S, Sittler T, Ideker T. The Plasmodium protein network diverges from those of other eukaryotes. Nature. 2005;438:108-12
32. Ellis RD, Martin LB, Shaffer D, Long CA, Miura K, Fay MP. et al. Phase 1 trial of the Plasmodium falciparum blood stage vaccine MSP1(42)-C1/Alhydrogel with and without CPG 7909 in malaria naive adults. PLoS One. 2010;5:e8787
33. Sheehy SH, Duncan CJ, Elias SC, Collins KA, Ewer KJ, Spencer AJ. et al. Phase Ia clinical evaluation of the Plasmodium falciparum blood-stage antigen MSP1 in ChAd63 and MVA vaccine vectors. Mol Ther. 2011;19:2269-76
34. Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B. et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539-43
35. Alvarez-Hamelin I, Dall'Asta L, Barrat A, Vespignani A. k-core decomposition: a tool for the visualization of large scale networks. arXiv (arXiv: cs/0504107). 2005
36. Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288-9
37. Chang SP, Kramer KJ, Yamaga KM, Kato A, Case SE, Siddiqui WA. Plasmodium falciparum: gene structure and hydropathy profile of the major merozoite surface antigen (gp195) of the Uganda-Palo Alto isolate. Exp Parasitol. 1988;67:1-11
38. del Portillo HA, Longacre S, Khouri E, David PH. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci U S A. 1991;88:4030-4
39. Brown AE, Kain KC, Pipithkul J, Webster HK. Demonstration by the polymerase chain reaction of mixed Plasmodium falciparum and P. vivax infections undetected by conventional microscopy. Trans R Soc Trop Med Hyg. 1992;86:609-12
40. Chong CK, Jeong W, Kim HY, An DJ, Jeoung HY, Ryu JE. et al. Development and clinical evaluation of a rapid serodiagnostic test for toxoplasmosis of cats using recombinant SAG1 antigen. Korean J Parasitol. 2011;49:207-12
41. Kim WS, Lee GC, Yoo JH, Kim HY, Yun YP, Chong CK. Development and diagnostic application/evaluation of pandemic (H1N1) 2009 influenza virus-specific monoclonal antibodies. Microbiol Immunol. 2012;56:372-7
42. Lee J, Kim YE, Kim HY, Sinniah M, Chong CK, Song HO. Enhanced performance of an innovative dengue IgG/IgM rapid diagnostic test using an anti-dengue EDI monoclonal antibody and dengue virus antigen. Sci Rep. 2015;5:18077
43. Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509-12
44. Good MF. Towards a blood-stage vaccine for malaria: are we following all the leads? Nat Rev Immunol. 2001;1:117-25
45. Soares IS, Levitus G, Souza JM, Del Portillo HA, Rodrigues MM. Acquired immune responses to the N- and C-terminal regions of Plasmodium vivax merozoite surface protein 1 in individuals exposed to malaria. Infect Immun. 1997;65:1606-14
46. Soares IS, da Cunha MG, Silva MN, Souza JM, Del Portillo HA, Rodrigues MM. Longevity of naturally acquired antibody responses to the N- and C-terminal regions of Plasmodium vivax merozoite surface protein 1. Am J Trop Med Hyg. 1999;60:357-63
47. Soares IS, Oliveira SG, Souza JM, Rodrigues MM. Antibody response to the N and C-terminal regions of the Plasmodium vivax Merozoite Surface Protein 1 in individuals living in an area of exclusive transmission of P. vivax malaria in the north of Brazil. Acta Trop. 1999;72:13-24
48. Cunha MG RM, Soares IS. Comparison of the immunogenic properties of recombinant proteins representing the Plasmodium vivax vaccine candidate MSP1(19) expressed in distinct bacterial vectors. Vaccine. 2001;20:385-96
49. Rodrigues MH, Cunha MG, Machado RL, Ferreira OC Jr, Rodrigues MM, Soares IS. Serological detection of Plasmodium vivax malaria using recombinant proteins corresponding to the 19-kDa C-terminal region of the merozoite surface protein-1. Malar J. 2003;2:39
50. Yang Y, Adelstein SJ, Kassis AI. Target discovery from data mining approaches. Drug Discov Today. 2009;14:147-54
51. Kim YM, Hwang HA, Yun WS, Kim SI, Lee KW, Park SK. et al. Efficacy of the merozoite surface protein 1 of Plasmodium vivax as an antigen for ELISA to diagnose malaria. Yonsei Med J. 2004;45:129-34
52. Park SK, Lee KW, Hong SH, Kim DS, Lee JH, Jeon BH. et al. Development and evaluation of an immunochromatographic kit for the detection of antibody to Plasmodium vivax infection in South Korea. Yonsei Med J. 2003;44:747-50
53. Kitchen AD, Lowe PH, Lalloo K, Chiodini PL. Evaluation of a malarial antibody assay for use in the screening of blood and tissue products for clinical use. Vox Sang. 2004;87:150-5
Author contact
![]() Corresponding authors: Hak-Yong Kim: Tel +82-43-261-2307, E-mail: hykimac.kr. Chom-Kyu Chong: Tel +82-41-523-8990, Fax +82-41-523-8991, E-mail: ckchongco.kr. Hyun-Ok Song: Tel +82-63-850-6972, E-mail: hosongac.kr.
Corresponding authors: Hak-Yong Kim: Tel +82-43-261-2307, E-mail: hykimac.kr. Chom-Kyu Chong: Tel +82-41-523-8990, Fax +82-41-523-8991, E-mail: ckchongco.kr. Hyun-Ok Song: Tel +82-63-850-6972, E-mail: hosongac.kr.

 Global reach, higher impact
Global reach, higher impact