10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(8):1051-1066. doi:10.7150/ijbs.19203 This issue Cite
Research Paper
Alkbh4 and Atrn Act Maternally to Regulate Zebrafish Epiboly
State Key Laboratory of Membrane biology, Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing 100084, China.
* These authors contributed equally to this work.
Received 2017-1-15; Accepted 2017-7-11; Published 2017-9-2
Abstract
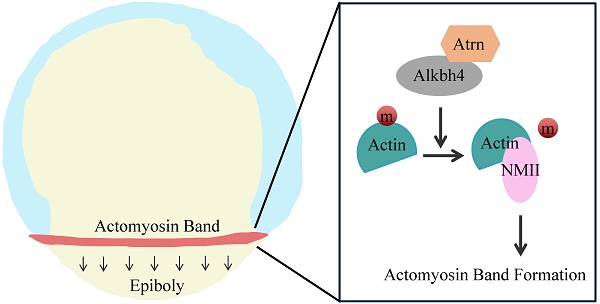
During embryonic gastrulation, coordinated cell movements occur to bring cells to their correct position. Among them, epiboly produces the first distinct morphological changes, which is essential for the early development of zebrafish. Despite its fundamental importance, little is known to understand the underlying molecular mechanisms. By generating maternal mutant lines with CRISPR/Cas9 technology and using morpholino knockdown strategy, we showed that maternal Alkbh4 depletion leads to severe epiboly defects in zebrafish. Immunofluorescence assays revealed that Alkbh4 promotes zebrafish embryonic epiboly through regulating actomyosin contractile ring formation, which is composed of Actin and non-muscular myosin II (NMII). To further investigate this process, yeast two hybridization assay was performed and Atrn was identified as a binding partner of Alkbh4. Combining with the functional results of Alkbh4, we found that maternal Atrn plays a similar role in zebrafish embryonic morphogenesis by regulating actomyosin formation. On the molecular level, our data revealed that Atrn prefers to interact with the active form of Alkbh4 and functions together with it to regulate the demethylation of Actin, the actomyosin formation, and subsequently the embryonic epiboly.
Keywords: alkbh4, atrn, epiboly, actomyosin, Actin, NMII
Introduction
In early vertebrate development, embryos undergo a series of evolutionarily conserved cell movements, including epiboly, emboly and convergent extension. Epiboly in zebrafish begins at the late blastula stage, and results in tissue spreading and thinning until it encloses the entire yolk cell at the end of gastrulation [1, 2]. At this time, a zebrafish embryo has three distinct cellular layers: the enveloping layer (EVL), deep cell layer (DEL), and yolk cell that consists of the yolk syncytial layer (YSL) and yolk cytoplasmic layer (YCL). The EVL is a single-cell-thick layer that covers the DEL and contacts the YSL at its vegetal end [3]. During the progression phase of epiboly, contractile actomyosin ring, composed of Actin and non-muscular myosin II (NMII), in the E-YSL is regarded as the major force driving epiboly, which is achieved by pulling on the EVL margin in the direction of the vegetal pole [2, 4-6]. However, the underlying molecular mechanism of actomyosin formation during epiboly remains to be elucidated.
Alkbh4 (alpha-ketoglutarate-dependent dioxygenase alkB homolog 4) belongs to the superfamily of Fe(II) and 2-oxoglutarate (2OG, alpha-ketoglutarate)-dependent dioxygenases, which can catalyse the demethylation of a variety of substrates [7]. Until now, nine mammalian AlkB homologs have been identified, ALKBH1-8 and FTO. ALKBH2 and ALKBH3 possess DNA and/or RNA repair activity [8-10]. ALKBH5 and FTO are two RNA demethylases that oxidatively remove the m6A (N6-methyl-adenosine) modification in mRNA [11, 12]. ALKBH8 exhibits tRNA methyltransferase activity [13-15]. ALKBH1 demethylates histone H2A K118 or K119 [16]. Recently, Li et al. reported that ALKBH4 functions to demethylate Actin at a mono methylated site (K84me1) and regulates actomyosin dynamics, and thereby the assembly of the contractile ring during mitosis [17]. Moreover, it is testified that Alkbh4 depletion in mice leads to spermatogenic defects and is lethal during early embryonic development [17, 18]. But it is unknown what role of Alkbh4 plays in zebrafish embryonic development.
Atrn (Attractin) is a member of CUB domain-containing protein family, which includes lots of cell adhesion and guidance proteins [19]. Previous reports show that there are two forms of ATRN proteins existing in both human and rat, a secreted form and a transmembrane form, as a result of alternative splicing. However, most of the species including zebrafish and mouse only express the transmembrane form of the ATRN protein [20, 21]. Secreted ATRN, an abundant serum glycoprotein, can function in monocyte/macrophage spreading and T cell clustering [19]. At the same time, secreted ATRN can promote glioma cell migration, implying ATRN is an important mediator of tumor invasiveness [22]. Transmembrane ATRN, a big single-pass transmembrane protein with about 1400 amino acids, plays important roles in monocyte function, pigmentation, neuronal degeneration and obesity [23-28]. As reported, transmembrane ATRN can act as a co-receptor for Agouti protein in MC1R and MC4R signaling pathway to regulate pigmentation and obesity [28, 29], as well as be involved in regulating the mitochondrial function [25]. However, nothing is known of Atrn functions during zebrafish embryonic development, especially in the epiboly process.
We show here for the first time that zebrafish maternal genes alkbh4 and atrn are both critical for the embryo epibolic morphogenesis. They are both required for the formation of actomyosin in the E-YSL, and that in either of them absence, the major force driving epiboly is perturbed. Finally, we found that Atrn interacts with Alkbh4 and promotes the binding affinity between Actin and NMII, subsequently regulates the formation of actomyosin band in the E-YSL during zebrafish epibolic movement.
Materials and Methods
Zebrafish strains
Tuebingen strains of zebrafish were ethical approved by the Animal Care and Use Committee of Tsinghua University. Zebrafish embryos were obtained from natural mating and were cultured in Holtfreter solution at 28.5°C. Developmental stages of embryos were determined according to Kimmel et al [30].
Constructs
alkbh4-5'UTR-GFP plasmid was generated by inserting a part of 5'UTR and coding sequence of alkbh4 into pEGFP-N3 (Clontech) with the forward primer 5'-GGAATTCAGGTCTTTGAGTCTCTGCTA ATGAGC-3' and the reverse primer 5'-CGGG ATCCACAACTTGGATTCATCATCAGAGC-3'. For constructing atrn-5'UTR-GFP plasmid, a DNA fragment including atrn-MO targeting sites was inserted into the pEGFP-N3 vector, the sequences were as follows: 5'-GCGGGCCCGGGATCCCT GTTTGTGCGCGAATGGCTGCGGAGGGATCCATCGCCACC-3'.
For genes' expression, alkbh4 and atrn coding sequences were amplified and ligated to pCS2-Flag and pCS2-HA vectors respectively. For pCS2-Flag-Alkbh4-Mut, H169A/D171A sites mutation primers were as follows: 5'-GAGCGGGGCTCTGCCATTGACCCCGCACTGGCTGACGCCTGGCTGTGGGGGGAGCGGC-3' (forward); 5'-GCCGCTCCCCCCACAGCCAGGCGTCAGCCAGTGCGGGGTCAATGGCAGAGCCCCGCTC-3' (reverse). For generating actin, actin-K84A and actin-K84R mRNA constructs, the following primers were used: 5'-GGAATTCCACCATGGATGAGGAAATCGCTGCCCTGGTC-3'(actin-F); 5'-CGCTCGAGCCGCCACCTCCGAAGCACTTCCTGTGAACGATGGATGGGC-3' (actin-R); 5'-GACCAACTGGGATGACATGGAGGCCATCTGGCATCACACCTTCTACAATGAGC-3' (actinK84A-midF); 5'-GCTCATTGTAGAAGGTGTGATGCCAGATGGCCTCCATGTCATCCCAGTTGGTC-3' (actinK84A-midR); 5'-GACCAACTGGGATGACATGGAGAGGATCTGGCATCACACCTTCTACAATGAGC-3' (actinK84R-midF); 5'-GCTCATTGTAGAAGGTGTGATGCCAGATCCTCTCCATGTCATCCCAGTTGGTC-3' (actinK84R-midR).
Whole-mount in situ hybridization (WISH)
Digoxigenin-UTP-labeled antisense RNA probes were synthesized in vitro using a linearized plasmid or PCR product as template. The templates for dlx3b and ntla were linearized as previously reported [31], while those for alkbh4, atrn, mpi and ranbp10 were amplified with the following primers:
alkbh4:
5'-CTCCAGAAGAATGATCTGATTC-3' (forward) and
5'-TAATACGACTCACTATAGGGCACTTTACATTTGTGCAATTGAAC-3' (reverse);
atrn:
5'-GTGGCATTGGAGACGGACGAGGAGC-3' (forward) and
5'-TAATACGACTCACTATAGGGCAGTTAGCGCACCAAACATGCACAC-3' (reverse);
mpi:
5'-TCTGTCCGGAGACTGTGTGGAGTGTATGGC-3' (forward) and
5'-TAATACGACTCACTATAGGGGAAGCGTCTCCTACAGTAAGCAGCACGCTC-3' (reverse);
ranbp10:
5'-CTCATTTGCACAGCACAGGCGCAGACAGTC-3' (forward) and
5'-TAATACGACTCACTATAGGGGCTTCTGGCAAGCGGCACACCAATTC-3' (reverse).
Whole-mount in situ hybridization was performed as previously described [31]. Embryos after whole-mount in situ hybridization were immersed in glycerol and photographed using the Ds-Ri1 CCD camera under a Nikon SMZ1500 stereoscope.
Semi-quantitative PCR
Zebrafish cDNA at indicated stages were prepared by reversely transcribed enzyme M-MLV reverse transcriptase (Promega). For alkbh4 PCR, the primer pairs were as follows: 5'-ATGATGCTTTTGTCGCTACTGTCGAC-3' and 5'-CCGCTCGAGTATAGAGGAGCACCCTGGAAGCTC-3'. For atrn PCR, the primer pairs were as follows: 5'-GCACACAGGTGGAGGTAGAAGAGTTC-3' and 5'-GATATCAGATCTCTCGAGGAATTCCTACTGCCAGACTGACTGTCC-3'. For β-actin PCR, the primer pairs were as follows: 5'-ATGGATGATGAAATTGCCGCAC-3' and 5'-ACCATCACCAGAGTCCATCACG-3'. Relative alkbh4 and atrn mRNA levels are measured by ratio of alkbh4 or atrn band intensity to that of β-actin band.
Morpholinos and microinjection
The morpholino oligonucleotides (MOs) were synthesized by Gene Tools, LLC, and the sequences were as follows: alkbh4-MO1, 5'-ACACCTCATTTAGATCATCAGCTCA-3'; alkbh4-MO2, 5'-AAGTAGGCAAAAAGCACCACAAGCA-3'; atrn-MO, 5'-TCCGCAGCCATTCGCGCACAAACAG-3'; std-MO, 5'-CCTCTTACCTCAGTTACAATTTATA-3'. MPPI-2 quantitative injection equipment (Applied Scientific Instrumentation Co.) was used for microinjection. About 1 nl morpholino solution was injected into the yolk of each embryo at 1-cell stage. The injection dose was the amount of a morpholino received by a single embryo.
Generation of mutant lines with CRISPR/Cas9
gRNAs for making alkbh4 and atrn mutant lines were designed to target the third exon of alkbh4 and second exon of atrn respectively. Their target sequences were as follows: 5'-CCATGTGGGAA GCTTCAGCGGCC-3' (alkbh4) and 5'-GGCTACCTCT CTGATGGACCAGG-3' (atrn). According to previous work, 100 pg gRNA with 300 pg Cas9 mRNA was co-injected into one-cell stage embryos. PCR primer pairs for identifying alkbh4 mutant embryos were 5'-CATGAGACATCTGTCTATCCAAAGGAC-3' and 5'-TAGAGGAGCACCCTGGAAGC-3'; while for atrn mutant embryos were 5'-CACTCTGTGCTTGTG GTTTC-3' and 5'-CGATACTGGTATTGGTGCAT CTC-3'. Then the PCR products were cut by T7 Endonuclease I firstly and then sequenced to identify their genotypes.
Whole-mount Immunofluorescence
Zebrafish embryos at indicated stages were fixed by 4% paraformaldehyde at 4°C and washed by PBST (0.1% Triton X-100) for 3 times. Then the embryos were incubated in PBST (0.5% Triton X-100) for 1 hr at RT and washed by PBST (0.1% Triton X-100) for 3 times, followed by blocking in block buffer (10% normal goat serum, 2% BSA, 1% DMSO, 0.1% Triton X-100 in PBS) for 2 hrs. Thereafter, phalloidin and/or primary antibodies in block buffer were added overnight at 4°C, and then secondary antibodies were used for another 5 hrs at RT. The following primary antibodies and dilutions were used: mouse anti-NMII with 1:100 dilution (ab55456, Abcam), mouse anti-HA with 1:100 dilution (sc-7392, Santa Cruz), rabbit anti-GFP with 1:100 dilution (ab290, Abcam). Embryos were imaged by Zeiss LSM 710 confocal microscope.
Yeast two hybridization assay
Zebrafish alkbh4 was cloned into pGBKT7 vector and used for bait. The cDNA library was derived from a mixture of zebrafish embryos at 1-cell, 40% epiboly, 5-somite and 24 hpf stages, and was described in our previous study [32, 33]. For a detailed description of the protocol, please refer to Matchmaker Two-Hybrid System 3 & Libraries User Manual from Clontech.
Immunoprecipitation and Western blot
Immunoprecipitation and Western blot experiments were performed as previously described [32, 33]. For Alkbh4 and Atrn interaction, Flag-alkbh4/alkbh4-mut and HA-atrn were transiently expressed in HEK293T cells. Cells were collected and lysed after 40 hrs transfection. For endogenous Actin and NMII interaction, wild-type zebrafish embryos were collected at 75% epiboly stage, while the MZalkbh4 and MZatrn mutant embryos were collected at similar morphological stages. Embryonic yolk was first removed by pipetting repeatedly with 200 μl tips, and then the embryonic cells were collected by centrifuge and lysed in TNE buffer. Mouse anti-Flag (1:1000; F1804, Sigma), mouse anti-NMII (1:100 for IP and 1:1000 for WB; ab55456, Abcam), goat anti-Actin (1:2000; sc-1615, Santa Cruz), mouse anti-α-Tubulin (1:100; T6199, Sigma) and rabbit anti-phospho-Histone H3 (Ser10) (1:100; #9701, CST) antibodies were used.
Measurements and Statistical analysis
The epiboly percentage, body length, actomyosin band width, cell length and cell width were all measured by Image J software. All the data were averaged from three times of independent experiments at least, and expressed as mean plus s.d. Student's T-tests (two-tailed, unequal variance) were used to determine p-values, and *, p<0.01 indicated the significance levels.
Rhodamine B isothiocyanate-Dextran endocytosis assay
The dechronized embryos at 50% and 75% epiboly stages were incubated in Rhodamine B isothiocyanate-Dextran solution (Sigma, R8881) at 10 mg/ml in holfreter water for 30 minutes [5]. After washing 2 times, embryos were imaged by Zeiss LSM 710 confocal microscope.
Results
The spatio-temporal expression of alkbh4 in zebrafish development
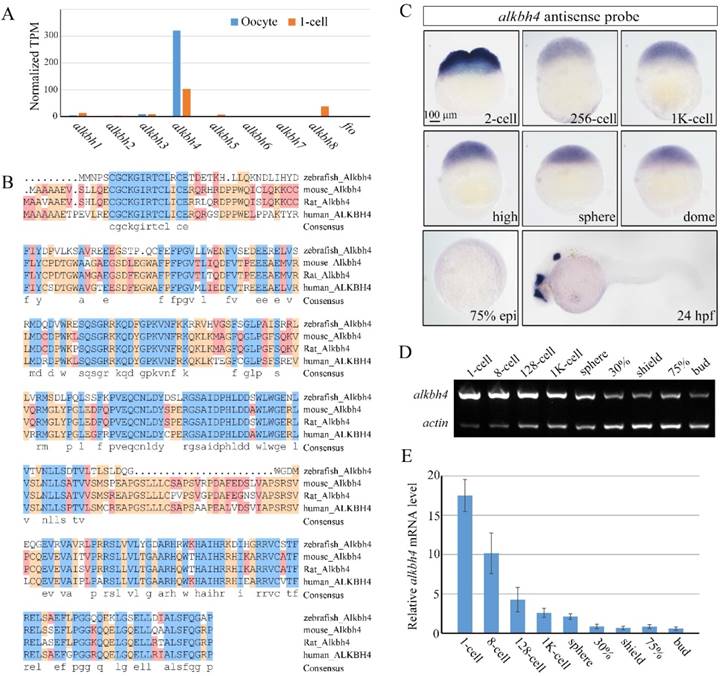
The earliest stages of embryonic development rely on maternal genes [34]. However, this maternal regulation has been little studied in vertebrates, owing to the difficulty in manipulating maternal gene function. With the development of CRISPR/Cas9-mediated genome engineering, it becomes convenient to generate mutant embryos for specific genes, and therefore identify their maternal effects. In our previous work, we carried out RNA-seq analysis to detect maternally enriched genes in zebrafish early development. It was found that alkbh4 mRNA level was extremely much higher than other alkbh family members, especially in the oocytes (Figure 1A). The zebrafish alkbh4 has an ORF that encodes a peptide of 268 amino acids and exhibits 48.3%, 49.2%, and 50.3% identities in amino acid sequence with mouse, rat and human ALKBH4 respectively (Figure 1B). Being interested in its functions in the embryonic early development, we first examined its expression pattern by whole-mount in situ hybridization with digoxin-labeled antisense probe. It was shown that alkbh4 mRNA is present in fertilized eggs, suggesting that it is maternally distributed. From the 2-cell stage to dome stage, alkbh4 mRNA can be continuously detected in all blastodermal cells (Figure 1C). Notably, alkbh4 maternal mRNA decreases gradually and becomes almost undetectable after dome stage with in situ hybridization. At 24 hrs postfertilization (hpf), its zygotic mRNA was determined to emerge in the forebrain, midbrain and otic vesicles (Figure 1C). Using β-actin as an internal control, semi-quantitative RT-PCR for comparison of alkbh4 mRNA expression level at different stages was carried out (Figure 1D). The statistical data demonstrated that expression of maternal alkbh4 mRNA decreased quickly during the very early stage of embryogenesis (Figure 1E), which raising the possibility that Alkbh4 might be an important maternal factor to regulate zebrafish early embryonic development.
Alkbh4 depletion affects epibolic morphogenesis
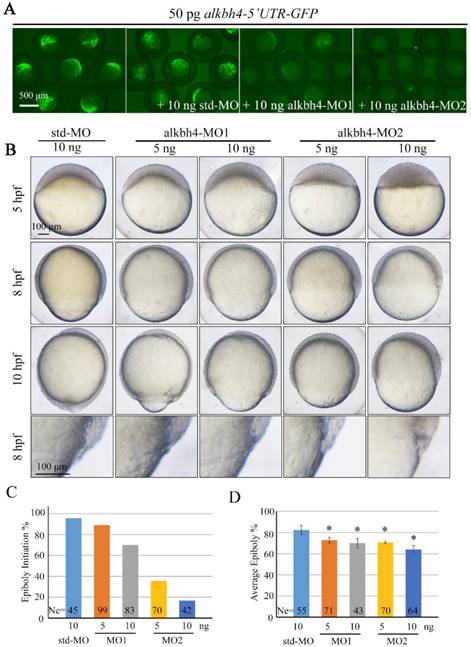
In order to investigate the functions of endogenous alkbh4 in zebrafish embryonic development, we synthesized two specific antisense morpholino oligonucleotides (MO), alkbh4-MO1 and alkbh4-MO2, which target different sequences in the 5' untranslated region (5' UTR) of alkbh4, to block its translation. Co-injection of 10 ng alkbh4-MO1 or alkbh4-MO2 with alkbh4-5'UTR-GFP reporter plasmid, which encodes a fusion Alkbh4-GFP protein, showed that both morpholinos could effectively block the translation of alkbh4 mRNA, and alkbh4-MO2 was more effective (Figure 2A). Compared to the embryos injected with standard MO (std-MO), those injected with alkbh4-MO1 or alkbh4-MO2 alone displayed slower epiboly in a dose-dependent manner (Figure 2B). At the same time, we noticed that the epiboly defects caused by injection of alkbh4-MO2 was much severe than that of alkbh4-MO1, which was consistent with their efficacy as shown in Figure 2A. Close examinations at higher magnifications also revealed that the cell movements in EVL, DEL and YSL were all slower in the morphant embryos (Figure 2B, last panel). When most of the std-MO injected embryos developed to 30% epiboly stage (5 hpf), embryos injected with 10 ng alkbh4-MO2 almost didn't initiate the epiboly process (Figure 2B and 2C). While at 80% epiboly stage (8 hpf), the embryos injected with 10 ng alkbh4-MO2 only reached about 60% epiboly on average (n=64) (Figure 2B and 2D). Therefore, it is possible that the observed epibolic defects might be due to the requirement of alkbh4 for cell motility during zebrafish embryonic development.
Spatiotemporal expression pattern of alkbh4 in zebrafish embryos. (A) mRNA levels of alkbh family members in zebrafish oocyte and 1-cell embryo from RNA-seq data. Normalized transcripts number per million of alkbh1-8 and fto transcripts are shown. (B) Protein sequences alignment of Alkbh4 among different species. (C) alkbh4 mRNA expression pattern in wild-type zebrafish embryos were detected by whole-mount in situ hybridization at indicated stages. (D, E) Semi-quantitative RT-PCR detection of zebrafish alkbh4 mRNA level at indicated stages. The relative expression ratio was calculated from the band intensity between alkbh4 and β-actin (internal control). Scale bars: 100 μm in (C).
To further validate and distinguish the functions of maternal mRNA and zygotic mRNA of alkbh4 in embryonic epiboly process respectively, we generated corresponding maternal and zygotic mutants using the CRISPR/Cas9-mediated approach [35]. By targeting the third exon of the alkbh4 gene, we obtained three kinds of deletion mutations, including alkbh4cd4 with -5/+1 bp alteration, alkbh4cd8 with 8 bp deletion, and alkbh4cd20 with 20 bp deletion, all of which led to a premature stop codon and therefore the lack of the catalytic domain of the protein (Figure 3A). An initial observation indicated that the zygotic alkbh4cd4/cd4, alkbh4cd8/cd8, and alkbh4cd20/cd20 homozygous mutants appeared normally without obvious morphological defects during embryogenesis (Figure 3B and data not shown). Then these Zalkbh4 mutant embryos were raised to adulthood, thus allowing generating maternal Malkbh4 mutant embryos by outcrossing female homozygous to the male wild-type fish, and then obtaining the maternal-zygotic MZalkbh4 mutants by intercrossing homozygous fish directly. Our results indicated that embryonic epiboly initiation and progression were significantly delayed in Malkbh4cd4/cd4 and MZalkbh4cd4/cd4 mutant embryos, although there was no obvious defect in Zalkbh4cd4/cd4 mutant (Figure 3B-3D). Because the three different lines of Malkbh4 and MZalkbh4 mutants have similar phenotype (data not shown), we only focused on MZalkbh4cd4/cd4 mutation in subsequent analyses. Consistently, the morphological defects of mutant embryos were quite similar to those of alkbh4 morphant embryos (Figure 2B-2D). To examine the exact marginal positions of EVL and deep cells in MZalkbh4 mutant embryos, we performed phalloidin and DAPI staining. Our analyses indicated that, compared with control embryos, movements of EVL and deep cells to the vegetal pole were largely postponed in the mutant embryos (Supplementary Figure S1A). To validate the lack of wild-type alkbh4 mRNA transcripts in the MZalkbh4cd4/cd4 mutant embryos, we performed whole in situ hybridization at 2-cell and 24 hpf stages, following with genotyping using the same in situ hybridized embryos. Our results showed that alkbh4 mRNA was almost disappeared in MZalkbh4 mutant embryos at both 2-cell and 24 hpf stages, meanwhile alkbh4 mutant mRNA was also undetectable probably due to the nonsense-mediated mRNA decay (Figure 3E). Taking into account that embryonic epiboly was only significantly delayed in maternal, but not zygotic alkbh4 depleted embryos, we claimed that the contribution of alkbh4 to gastrulation cell epibolic movements is a strictly maternal-effect.
Besides epiboly, we also paid attention to the convergent extension (CE) movement in both MZalkbh4 and Malkbh4 mutants. To investigate whether the CE movements are affected or not, we examined the expression of two marker genes, the midline marker ntla and the neural plate boundary marker dlx3b in mutant embryos [31]. Due to the existence of epiboly delay in mutant embryos, we focused on comparing the results at morphologically comparable stage, but not at the same time point. We found that at bud stage, alkbh4 depletion resulted in wider neural plate (marked by dlx3) and notochord (marked by ntla) in 93% (n=45) of MZalkbh4 and 91% (n=35) of Malkbh4 mutant embryos (Figure 3F). These observations further support the notion that alkbh4 participates in CE movements during zebrafish embryogenesis.
Morpholino knockdown of alkbh4 leads to epiboly initiation and progression delays. (A) Effectiveness of alkbh4 morpholinos. 50 pg alkbh4-5'UTR-GFP plasmid DNA was injected alone or with 10 ng std-MO, alkbh4-MO1 or alkbh4-MO2 at 1-cell stage. The injected embryos were observed for GFP expression at late gastrulation stages. (B) Morpholino knockdown of alkbh4 leads to epiboly delay in a dose dependent manner. Embryos were observed and photographed at indicated stages. Magnified observations of the embryonic marginal cell layers are shown at the last panel. (C) Epiboly initiation in (B) was measured by the percentage of embryos with yolk cell doming at 5 hpf. (D) Epiboly progression in (B) was measured by the average epiboly percentage of the embryos at 8 hpf. Ne, the number of observed embryos. *, p<0.01. Scale bars: 500 μm in (A); 100 μm in (B).
Maternal alkbh4 mutation leads to epiboly delay. (A) CRISPR/Cas9-mediated deletion of alkbh4. Zebrafish alkbh4 contains 3 exons, and gRNA was designed to target the third exon. A specific genomic region was amplified from F2 individual embryo and sent for sequencing. The sequencing results of target sites are shown below, including the wild-type form and three kinds of alkbh4 mutant forms. (B) Maternal but not zygotic alkbh4 mutation results in epiboly initiation and progression delays. Wild-type, Zalkbh4, Malkbh4 and MZalkbh4 mutant embryos were collected after fertilization at the same time and then imaged at the indicated time points. (C) Epiboly initiation in (B) was measured by the percentage of embryos with yolk cell doming at 5 hpf. (D) Epiboly progression in (B) was measured by the average epiboly percentage of the embryos at 8 hpf. (E) In situ hybridization results of alkbh4 mRNA expression in wild-type and MZalkbh4 embryos at 2-cell and 25 hpf stages are shown. (F) CE movements are defective in MZalkbh4 and Malkbh4 mutant embryos. The mutant embryos were collected not only at the same time point but also at the morphologically comparable stages compared with wild-type embryos. dlx3b and ntla probes were used simultaneously for in situ hybridization. The ratios of embryos with representative pattern are indicated at the right corner of each picture. (G) The body length decreases in MZalkbh4 and Malkbh4 mutant larvae. Representative pictures of wild-type and mutant larvae at 5 dpf are shown. (H) The body lengths of wild-type and MZalkbh4 mutant larvae at 4 dpf, 5 dpf, 6 dpf and 7 dpf were measured by Image J software. The statistical results are shown here. Ne, the number of observed embryos. *, p<0.01. Scale bars: 100 μm in (B, E, F); 1 mm in (G).
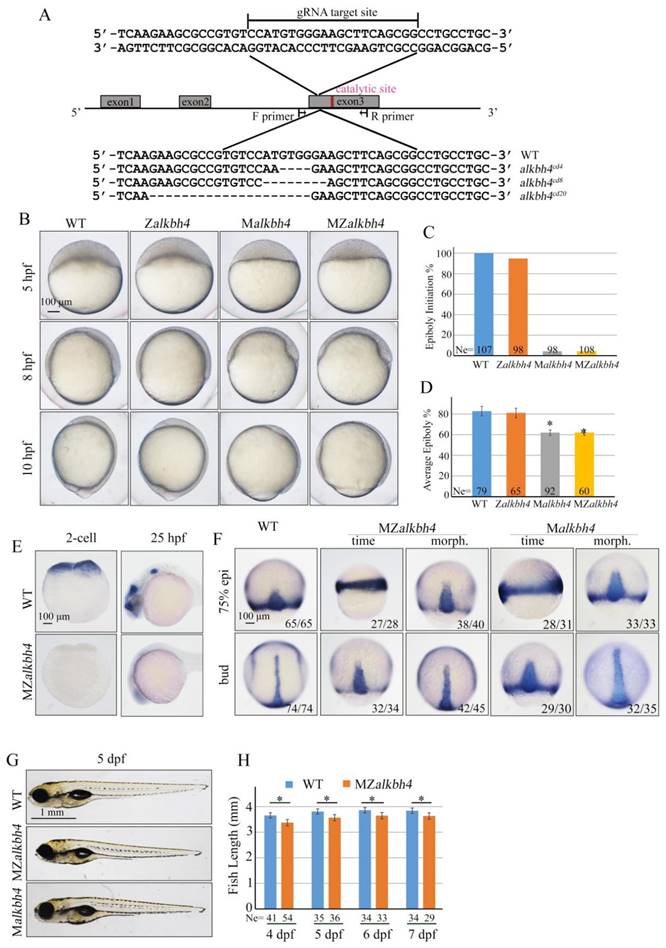
Taken together, these data suggest that maternally, but not zygotically expressed alkbh4 is critical for embryonic epiboly and CE movements during gastrulation. Additionally, we found that the average body lengths of MZalkbh4 and Malkbh4 mutants were reduced obviously compared with those of wild-type larvae in corresponding stages (Figure 3G, 3H, and data not shown), suggesting the maternal effect of alkbh4 can last to larval stages.
alkbh4 is required for Actin and NMII recruitment in the E-YSL and EVL cell-shape changes
Previous analyses indicated that during epiboly of the zebrafish embryo, the actomyosin ring, composed of Actin and NMII, within the yolk cytoplasm along the margin of the EVL is critical for the EVL marginal cells constriction, and subsequently the movement of the EVL over the yolk cell surface [6, 36]. In order to address whether the formation of actomyosin within the E-YSL is affected in MZalkbh4 mutant embryos or not, we first stained the embryos with fluorescent labeled phalloidin and observed by confocal fluorescent microscopy. Our data showed that at 75% epiboly, Actin became enriched at the EVL-YSL interface in wild-type embryos, and the average width of the band was 14.4 μm (n=32) approximately (Figure 4A and 4B). However, only a relatively thin band of Actin with an average width of 7.0 μm (n=20) was detected in MZalkbh4 mutant embryos at the same time point (Figure 4A and 4B). Even when the mutant embryos developed to the morphologically comparable stage, the staining of Actin was still distinctly thinner (6.2 μm, n=24) than the wild-type embryos (Figure 4A and 4B). Similarly, the expression of NMII was also examined by its specific antibody against NMII heavy chain. At 75% epiboly, the average width of NMII declined from 13.1 μm (n=20) in wild-type embryos to 6.1 μm (n=20) at the same time point and 5.3 μm (n=19) at the same morphologically comparable stage in MZalkbh4 mutant embryos respectively (Figure 4A and 4B). After confirming the similar results in Malkbh4 mutant embryos (Supplementary Figure S2A and S2B), we concluded that the formation of actomyosin ring was largely inhibited in maternal alkbh4 depleted embryos.
alkbh4 regulates actomyosin band formation in MZalkbh4 embryos. (A) Confocal images of phalloidin stained embryos at 50% epiboly, and phalloidin and anti-NMII double stained embryos at 75% epiboly. phalloidin and anti-NMII co-staining showing the defects of actomyosin band formation and marginal EVL cells morphology in MZalkbh4 embryos. The mutant embryos were collected not only at the same time point but also at the morphologically comparable stages compared with wild-type embryos. Embryos were laterally viewed with animal pole to the top and vegetal pole to the bottom. (B) Quantitative measurements of actomyosin band widths in (A) at 75% epiboly stage, which were represented by the widths of Actin and NMII staining in the E-YSL separately. (C) Quantitative measurements of the marginal EVL cell length/width ratios in (A) at both 50% and 75% epiboly. Ne, the number of observed embryos; Nc, the number of observed cells. *, p<0.01. (D) Epiboly defects caused by YSL-injection of alkbh4-MO2. At 4 hpf, 10 ng std-MO and alkbh4-MO2 were injected into the yolk cell of embryos and epiboly progression were observed at 50% and 80% epiboly stages. (E) Epiboly progression in (D) was measured by the average epiboly percentage of the embryos. Ne, the number of observed embryos. *, p<0.01. Scale bars: 50 μm in (A); 100 μm in (D).
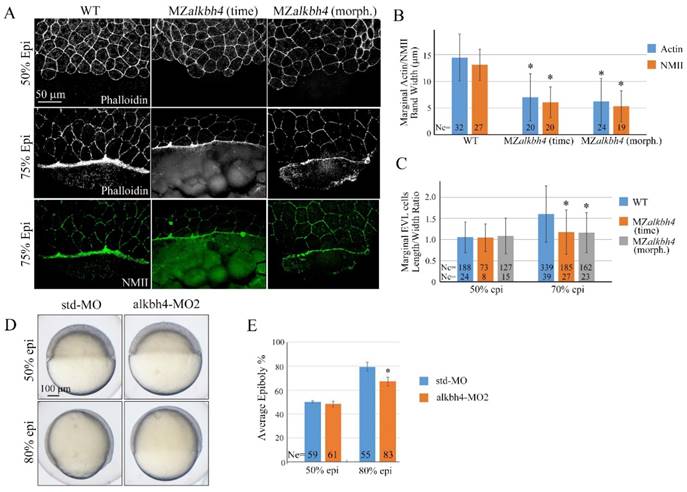
Given the evidence that the contractile actomyosin ring within the E-YSL drives the late phase of epiboly, possibly through coordinated EVL marginal cell shape changes [5, 6, 36], we analyzed MZalkbh4 and Malkbh4 mutant embryos at 50% and 75% epiboly stages by phalloidin staining. As shown in Figure 4A and Supplementary Figure S2A, at 50% epiboly stage, marginal cells of wild-type, MZalkbh4 and Malkbh4 mutant embryos were typically oval shaped. Thereafter, at 75% epiboly stage, the staining results revealed a delay of the elongation of the marginal EVL cells along the animal-vegetal axis in MZalkbh4 and Malkbh4 mutant embryos. To quantitatively compare the differences in EVL marginal cell constriction between wild-type and mutant embryos, the length to width ratios of individual border cells for about 10 cells per embryo were measured. Compared to the wild-type embryo, no significant difference of this ratio at 50% epiboly, but a remarkable declining at 75% epiboly was detected in both mutant embryos (Figure 4C and Supplementary Figure S2C), revealing a delay in the coordinated EVL marginal cell shape changes in the absence of maternal alkbh4. Taken together, it is indicated that alkbh4 is involved in the formation of actomyosin ring within the E-YSL and therefore regulates the epibolic morphogenesis in zebrafish.
To directly address the role of alkbh4 in the YSL, we injected its morpholino into this tissue at 4 hpf as described by Mathias Köppen et al [6]. Embryos injected with 10 ng alkbh4-MO2 displayed no obvious defect until 50% epiboly (Figure 4D). However, when the control group developed to 80% epiboly, the embryos injected with alkbh4-MO2 only reached about 67% epiboly on average (n=83), meanwhile showing a delay in epiboly of the DEL and the EVL cells (Figure 4D and 4E). Therefore, it is indicated that alkbh4 plays a direct role in the E-YSL to regulate cell mobility.
Alkbh4 associates and co-localizes with Atrn
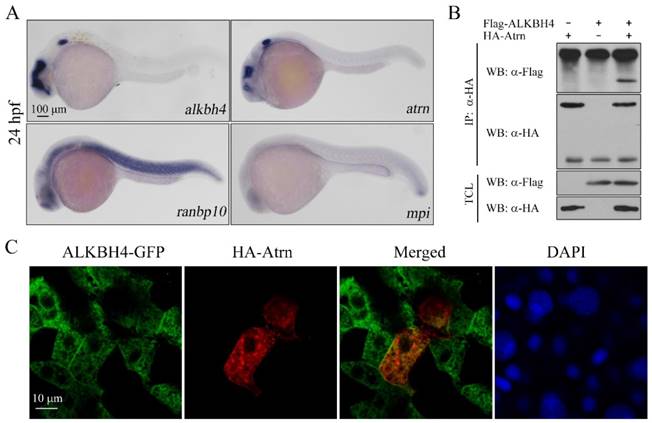
Protein-protein interactions play fundamental roles in many biological processes. In order to better understand how Alkbh4 activity is regulated in embryonic epiboly process, we devised a comprehensive yeast two-hybrid screen using Alkbh4 as the bait protein to look for Alkbh4-binding partners expressed in zebrafish embryos. By screening library, several proteins, such as Atrn, Ranbp10 and Mpi, were identified as the partners of Alkbh4 (data not shown). What draws more of our attention is that the spatio-temporal expression of atrn is quite similar to alkbh4, while the other two genes are not (Figure 5A). To confirm that there is a real physical association between Alkbh4 and Atrn, we carried out a coimmunoprecipitation (Co-IP) experiment. As shown in Figure 5B, immunoprecipitation of HA-tagged Atrn in HEK293T cells indeed retrieved Flag-tagged human AlLKBH4, implying an existing interaction between them. Additionally, to reveal the subcellular localization of Atrn and ALKBH4 proteins, we injected HA-tagged atrn plasmid DNA and GFP-tagged ALKBH4 mRNA into one cell at 4-cell stage of zebrafish wild-type embryos, and then detected their expression by immunofluorescence. We found that overexpressed HA-Atrn was well co-localized with ALKBH4-GFP in zebrafish embryonic cells, where they were distributed throughout the cells with prominent cytoplasmic and plasma membrane localization (Figure 5C), also supporting a possible interaction between them.
atrn plays a similar role to alkbh4 in epibolic morphogenesis
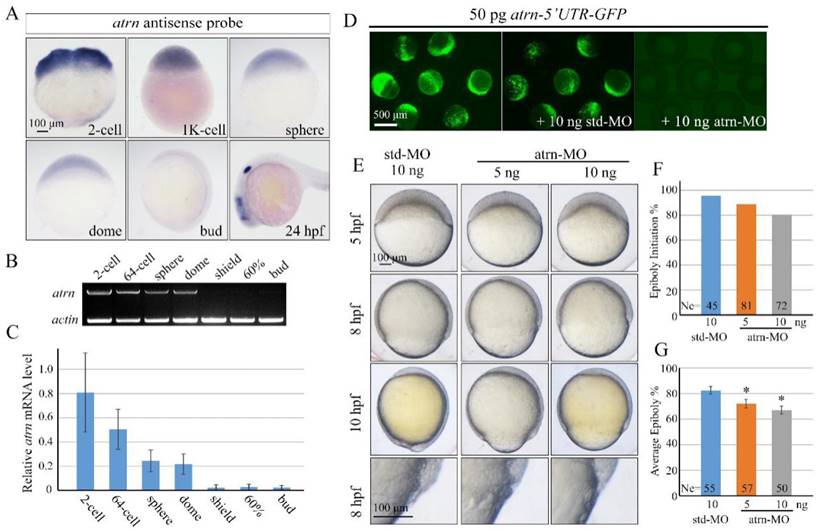
Different from human and rat, zebrafish atrn gene only encodes the transmembrane form of the protein with 1,345 amino acids, as in the case of mouse. Zebrafish Atrn protein has 68.04%, 67.55%, and 67.67% identities with human, rat and mouse transmembrane form of Atrn. We first examined the expression pattern of zebrafish atrn in detail and found that it was extremely similar to that of alkbh4. The results showed that atrn mRNA is maternally distributed and continuously exists in all blastodermal cells until dome stage (Figure 6A). Then, atrn maternal mRNA decreases and its zygotic mRNA appears in the forebrain, midbrain and otic vesicles at 24 hpf (Figure 6A). Semi-quantitative RT-PCR was also carried out to confirm the expression level of atrn during early developmental stages, showing that the expression of maternal atrn mRNA decreased obviously after dome stage (Figure 6B and 6C).
In order to explore functions of Atrn in embryonic development, we synthesized atrn-MO, which targeted the 5'UTR region of atrn mRNA, to block its translation. We first confirmed the efficacy of atrn-MO by its ability to block the expression of the corresponding 5'UTR-GFP reporter construct (Figure 6D). Compared to std-MO, atrn-MO injected embryos displayed slower epiboly in a dose dependent manner, which was quite similar to the defects of alkbh4 morphant or mutant embryos (Figure 6E, 2B and 3B). A large number of statistical results showed that when most of embryos injected with std-MO developed to 30% epiboly stage (5 hpf), about 20% (n=72) embryos injected with 10 ng atrn-MO did not initiate the epiboly process (Figure 6F). While at 80% epiboly stage (8 hpf), the embryos injected with 10 ng atrn-MO only reached about 60% epiboly on average (n=50) (Figure 6G). Thus, it is suggested that atrn might be involved in regulating the epiboly process of zebrafish embryos.
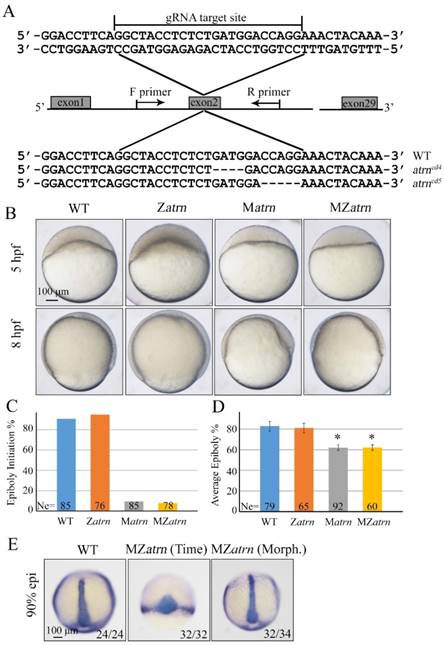
To substantiate the phenotype of atrn morphant is true, we generated its mutants using the CRISPR/Cas9 technology. By targeting second exon of atrn, two kinds of deletion mutants, atrncd4 and atrncd5 were obtained, both of which were lack of C-terminal part of the protein (Figure 7A). After confirming that both zygotic atrncd4/cd4 and atrncd5/cd5 homozygous mutants developed normally without obvious morphological defect during embryogenesis (Figure 7B and data not shown), these Zatrn mutants were raised to adulthood to generate Matrn and MZatrn mutant embryos. Similar to atrn morphants, embryonic epiboly initiation and progression were significantly delayed in Matrn and MZatrn mutants (Figure 7B-7D), which is also extremely similar to the defects of alkbh4 morphant and mutant embryos (Figure 3B-3D), suggesting that the maternally expressed atrn is important for embryonic epiboly process. Additionally, we examined the CE movements in MZatrn and found that although the ntla/dlx3b expression pattern of mutant embryos looked very different from that of wild-type embryos at the same time point after fertilization (9 hpf), but they finally became similar at the same morphological stage (90% epiboly) (Figure 7E). Therefore, it is suggested that atrn has little effect on the CE movements of gastrulation, which is different from alkbh4 (Figure 3F). Taken together, these data suggest that the maternal but not zygotical atrn is critical for embryonic epiboly during gastrulation.
atrn regulates actomyosin formation and EVL cell-shape changes
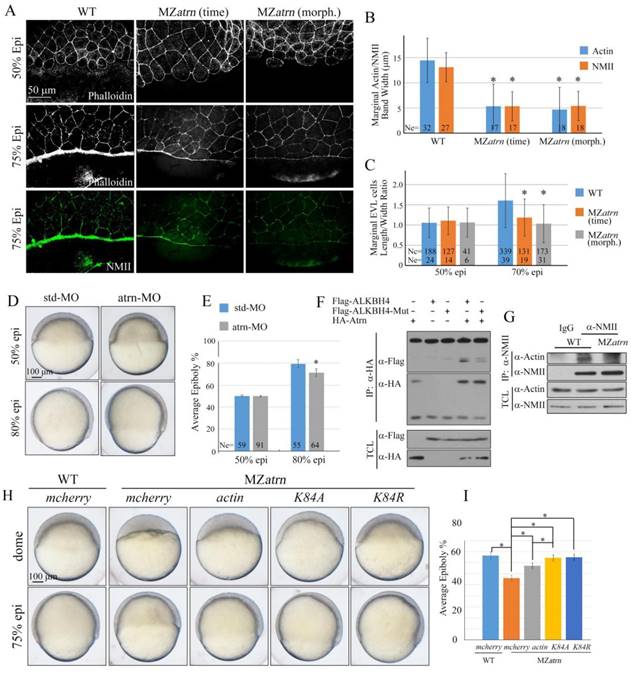
Because Alkbh4 was found to regulate actomyosin ring formation during embryonic epiboly, we wondered whether the Actin and NMII accumulation within the E-YSL is also affected in MZatrn mutant embryos or not. The phalloidin and NMII immunostaining results showed that compared to wild-type embryos at 75% epiboly, the accumulation of Actin and NMII within the E-YSL were inhibited obviously in MZatrn mutant embryos, not only at the same time point but also at the morphologically comparable stage (Figure 8A). The corresponding statistical data were obtained and shown in Figure 8B. Besides that, we also analyzed the changes in EVL cell shape in phalloidin-stained embryos, and found that the length to width average ratio of marginal cells in MZatrn mutant decreased significantly, which was consistent with the defects of actomyosin-based contractility in the mutant embryos (Figure 8A and 8C). Similarly, we performed morpholino YSL-injection assays to demonstrate that atrn acts directly on the mobility of EVL cells and DEL cells to affect zebrafish epibolic morphogenesis (Figure 8D and 8E).
Alkbh4 associates and co-localizes with Atrn. (A) Whole-mount in situ hybridization results of putative Alkbh4 binding partners, including atrn, ranbp10 and mpi. Only the expression pattern of atrn is similar to that of alkbh4 at 24 hpf. (B) Flag-ALKBH4 was co-expressed with HA-Atrn in HEK293T cells and their interaction was examined by immunoprecipitation. HA-Atrn was immunoprecipitated with anti-HA antibody and the precipitates were examined by western blotting for the presence of ALKBH4 with anti-Flag antibody. TCL, total cell lysate. (C) Co-localization of ALKBH4-GFP with HA-Atrn in zebrafish embryonic cells. HA-Atrn plasmid DNA and ALKBH4-GFP mRNA were injected into one cell at 4-cell stage of the embryos. Embryos were double stained with corresponding antibodies at 75% epiboly stage. Scale bars: 100 μm in (A); 10 μm in (C).
Alkbh4 and Atrn regulate the actomyosin formation by regulating the Actin demethylation
So far, we thought that alkbh4 and atrn might function together to regulate the formation of actomyosin, and subsequently the epiboly process in zebrafish early development. Given the evidences that Alkbh4 acts as an Actin demethylase to promote Actin-NMII interaction by erasing Actin K84 mono methylation [17], we constructed a catalytically inactive form of Alkbh4 (Alkbh4-Mut) with its catalytic domain mutated (H169A/D171A). We found that compared to the wild-type form of Alkbh4 (Alkbh4-WT), the catalytically inactive form of Alkbh4 (Alkbh4-Mut) showed a much lower affinity to interact with Atrn (Figure 8F). These data implied that Atrn might be involved in the catalytic function of Alkbh4. To validate our speculation and better understand the functional interplay between Alkbh4 and Atrn, we tested whether depletion of atrn would affect Actin-NMII interaction. Consistent with the fact that Alkbh4 regulates Actin-NMII interaction in mammalian cells, it was demonstrated in vivo that Actin-NMII interaction was also disrupted in MZatrn mutant embryos (Figure 8G). Taken together, we suspected that Atrn might play corporately with Alkbh4 to regulate the embryonic epiboly through promoting Actin-NMII interaction to form actomyosin ring in the E-YSL.
To further validate our speculation, we generated two different methylation-deficient actin mutants (K84A and K84R), and found that the morphological epiboly defects seen in MZatrn mutant embryos could be better rescued by either of these two mutant mRNAs than the wild-type mRNA (Figure 8H and 8I). Combined with our existing data that Atrn interacts with Alkbh4 and increases the binding affinity between Actin and NMII, we demonstrated that Atrn functions together with Alkbh4 to regulate the demethylation of Actin, and subsequently the actomyosin formation.
Morpholino knockdown of atrn leads to epiboly delay. (A) atrn mRNA expression pattern in wild-type embryos was detected by whole-mount in situ hybridization at indicated stages. (B, C) Semi-quantitative RT-PCR detection of zebrafish atrn mRNA level at indicated stages. The relative expression ratio was calculated from the band intensity between atrn and β-actin (internal control). (D) Effectiveness of atrn morpholino. 50 pg atrn-5'UTR-GFP plasmid DNA was injected alone or with 10 ng std-MO or atrn-MO at 1-cell stage. The injected embryos were observed for GFP expression at late gastrulation stages. (E) Morpholino knockdown of atrn results in epiboly delay in a dose dependent manner. Embryos were observed and imaged at indicated stages. Magnified observations of the embryonic marginal cell layers are shown at the last panel. (F, G) The quantitative data of epiboly initiation at 5 hpf and epiboly progression at 8 hpf in (E) are shown. *, p<0.01. Scale bars: 100 μm in (A, E); 500μm in (D).
MZalkbh4;MZatrn double mutants exhibit a similar phenotype as either of the single mutants
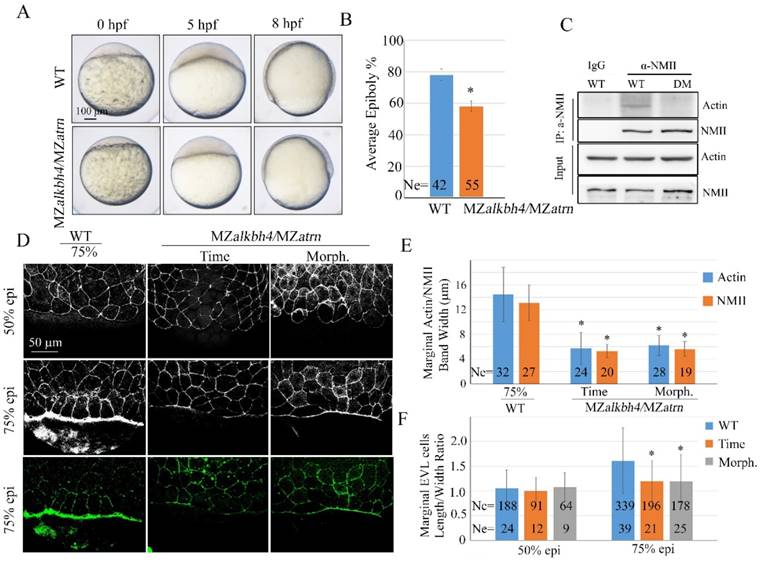
To investigate the genetic relationship between alkbh4 and atrn, we examined maternal-zygotic homozygous embryos for both mutations. Firstly, we got zygotic alkbh4-/-;atrn-/- homozygotes by intercrossing alkbh4+/-;atrn+/- heterozygous. With no obvious morphological defects, zygotic alkbh4-/-;atrn-/- homozygous embryos developed normally to adults, thus allowing generating maternal-zygotic MZalkbh4;MZatrn by intercrossing homozygous fish directly. As shown in Figure 9A and 9B, MZalkbh4;MZatrn double mutants exhibited a similar epibolic delay as either of the single mutants. Co-IP assay was performed to test whether knockdown of alkbh4 and atrn at the same time would affect the Actin-NMII interaction. As our suspected, it was revealed in vivo that the Actin-NMII interaction was also disrupted in double knockdown embryos (Figure 9C). Additionally, we stained the embryos with fluorescent labeled phalloidin and anti-NMII antibody, and found that at 75% epiboly, only a relatively thin band of Actin or NMII was detected in the double mutants at the same time point. Even when they developed to the morphologically comparable stage, the staining of Actin and NMII was still distinctly thinner than the wild-type (Figure 9D and 9E). On the other hand, we also demonstrated a delay of the elongation of the marginal EVL cells in the double mutants (Figure 9F). Thus, it is well demonstrated that the formation of actomyosin band was obviously inhibited in alkbh4 and atrn double mutants.
Maternal atrn mutation leads to epiboly delay. (A) CRISPR/Cas9-mediated deletion of atrn. Zebrafish atrn contains 29 exons, and gRNA was designed to target the second exon. Sequencing results of the target sites from several individual F2 embryos are shown below, including the wild-type form and two atrn mutant forms. (B) Maternal but not zygotic atrn mutation results in epiboly initiation and progression delays. Wild-type, Zatrn, Matrn and MZatrn mutant embryos were collected after fertilization at the same time and then imaged at indicated time points. (C, D) The quantitative data of epiboly initiation at 5 hpf and epiboly progression at 8 hpf in (B) are shown. (E) CE movements appear essentially normal in MZatrn mutant embryos. The mutant embryos were collected at both same time and morphologically comparable stages with wild-type embryos. dlx3b and ntla probes were used simultaneously for in situ hybridization. The ratios of embryos with representative pattern are indicated at the right corner of each picture. *, p<0.01. Scale bars: 100 μm in (B, E).
Atrn regulates actomyosin formation and Actin-NMII interaction in zebrafish embryos. (A) Confocal images of phalloidin stained embryos at 50% epiboly, and phalloidin and anti-NMII double stained embryos at 75% epiboly. The mutant embryos were collected at both the same time point and comparable morphological stages compared with wild-type embryos. Embryos were laterally viewed with animal pole to the top and vegetal pole to the bottom. (B) Quantitative measurements of actomyosin band widths in (A) at 75% epiboly stage, which are represented by the widths of Actin and NMII staining in the E-YSL separately. (C) Quantitative measurements of the marginal EVL cell length/width ratios in (A) at both 50% and 75% epiboly stages. Ne, the number of observed embryos; Nc, the number of observed cells. (D) Epiboly defects caused by YSL-injection of atrn-MO. 10 ng std-MO and atrn-MO were used. (E) Epiboly progression in (D) was measured by the average epiboly percentage of the embryos. (F) Atrn associated with less catalytically inactive form of ALKBH4 (ALKBH4-Mut), as revealed by co-IP in HEK293T cells. (G) Co-IP assay shows that endogenous Actin-NMII interaction is decreased in MZatrn mutant embryos. Embryos at 75% epiboly were harvested for lysis and Co-IP. (H) Rescue effects of wild-type and two methylation-deficient actin mRNAs in MZatrn embryos. 100 pg mcherry and different forms of actin mRNA were used for injection. Embryos injected with 100 pg mcherry mRNA were used as negative controls. Embryos were observed at dome and 75% epiboly stages, and shown as lateral views. (I) Statistical results of (H). *, p<0.01. Scale bars: 50 μm in (A); 100 μm in (D, H).
Discussion
Previous reports indicated that ALKBH4 is essential for the development of spermatocytes by using inducible Alkbh4 knockout mice, but there is no chance to study the functions of Alkbh4 in mouse embryonic development due to the lethality of Alkbh4 homozygous during early preimplantation stage [18]. Here, using zebrafish as animal model, we find that alkbh4 homozygous mutant embryos can develop to adult normally, which is quite different from the situation in mouse. It is perhaps due to the redundancy of other Alkbh family members or unknown compensatory effects, and thus allowing us to investigate the roles of Alkbh4 in early development of zebrafish embryos. In this study, we report that maternal mutation of alkbh4 results in embryonic epiboly delay, aberrant cell shape and impaired actomyosin band formation at the marginal region. Taking into account the facts that Alkbh4 can demethylate Actin and regulate Actin-NMII interaction, we speculate that Alkbh4 functions in zebrafish embryonic cell movements by regulating actomyosin band formation in the E-YSL.
Epiboly and actomyosin formation are defective in MZalkbh4;MZatrn double mutant embryos. (A) Epiboly initiation and progression delay in MZalkbh4;MZatrn double mutants. (B) The quantitative data of epiboly progression at 8 hpf are shown. (C) Co-IP assay shows that endogenous Actin-NMII interaction is decreased in alkbh4 and atrn double morphant embryos. Embryos at 75% epiboly stage were harvested for lysis. (D) Confocal images of phalloidin stained embryos at 50% epiboly, and phalloidin and anti-NMII double stained embryos at 75% epiboly. (E) Quantitative measurements of actomyosin band widths in (D) at 75% epiboly stage. (F) Quantitative measurements of the marginal EVL cell length/width ratios in (D) at both 50% and 75% epiboly stages. Ne, the number of observed embryos; Nc, the number of observed cells. *, p<0.01. Scale bars: 100 μm in (A); 50 μm in (D).
It has been proposed that endocytosis in the E-YSL and longitudinally organized microtubules in the YCL are also regarded as key regulating factors during epiboly progression [5, 37]. Endocytosis in E-YSL is critical for providing a pulling force as the membrane is internalized, and yolk cell microtubules contribute to the vegetal movement of the E-YSN. Here, we found that compared to wild-type embryos, MZalkbh4 and MZatrn mutants internalized Rhodamine-dextran dye in the E-YSL and assembled microtubules in the YCL normally, indicating that they are not the causes of the epiboly defect in our case (Supplementary Figure S3 and S4). Thus, it is believed that Alkbh4 and Atrn regulate embryonic epiboly by promoting Actin-NMII band formation in the E-YSL specifically. Additionally, we also excluded the possibility that the epibolic defects in MZalkbh4 and MZatrn mutants were ascribed to changes in amount of embryonic cells (Supplementary Figure S1) or cell proliferation (Supplementary Figure S5). However, it is noticed that Actin filaments distribution in cortical yolk cell were severely disrupted in MZalkbh4 mutants, but not in MZatrn mutants as shown in Supplementary Figure S1. As the previous study indicated that the formation of large patches appeared to be devoid of Actin filaments is likely due to the displacement of the cortical cytoplasm by the yolk mass in embryos [36], we speculate that alkbh4 but not atrn might be functional in keeping the integrity of yolk cortical cytoplasmic layer. It will be of great interest to figure out the detailed mechanisms in the future.
Furthermore, we identified Atrn as an Alkbh4 interacting protein, and found that its maternal depletion also causes similar defects in epibolic morphogenesis, as well as the actomyosin band formation during zebrafish early development. Although the molecular mechanisms of Atrn regulating actomyosin formation are not clear until now, our data suggested that Atrn might work together with Alkbh4 to participate in the same process. Based on the fact that Atrn shows a much lower affinity to interact with catalytically inactive form of Alkbh4, and methylation-deficient actin mRNA could rescue the epiboly defects of MZatrn mutant embryos, we speculate that Atrn might be involved in regulating the demethylation activity of Alkbh4. For the underlying mechanisms, there are several other possibilities. Because Atrn protein contains 7 Kelch repeats, which have been implicated in binding to Actin and subsequently regulating the organization of the cytoskeleton, it also can act as a scaffold protein to mediate the interaction between Alkbh4 and Actin, and then promote the actomyosin formation. Besides, we also can't exclude the possibility that Atrn can function in a parallel way to that of Alkbh4 behaves.
As we known, Atrn is most famous as an accessory receptor for Agouti protein in MC1R and MC4R signaling pathway to regulate pigmentation and obesity [38]. In this study, we demonstrate that Atrn plays an important role in modulating actomyosin formation, and subsequently promoting the cell migration in zebrafish embryos for the first time. Actually, MC1R was also reported to play functional role in controlling both human and mouse melanoma cells migration via enhancement of syndecan-2 expression [39]. Whether there is a potential cross talk between Atrn-MC1R and Alkbh4-Atrn signaling need to be investigated further.
In conclusion, results from our studies indicate that Alkbh4 can interact with Atrn, and both genes maternally regulate zebrafish embryonic epiboly via affecting marginal actomyosin band formation. Our studies identified Atrn as a new regulator of actomyosin formation and reflect the importance of maternal genes in early embryonic development.
Supplementary Material
Supplementary figures.
Acknowledgements
We thank the members of the Meng laboratory for experimental assistance and help discussion. We also thank the Image Core Facility of Protein Technology Center in Tsinghua University. This work was financially supported by grants from the National Natural Science Foundation of China (#31522035, #31371460 and #31590832).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Warga RM, Kimmel CB. Cell movements during epiboly and gastrulation in zebrafish. Development. 1990;108:569-80
2. Bruce AE. Zebrafish epiboly: Spreading thin over the yolk. Developmental dynamics: an official publication of the American Association of Anatomists. 2016;245:244-58
3. Zalik SE, Lewandowski E, Kam Z, Geiger B. Cell adhesion and the actin cytoskeleton of the enveloping layer in the zebrafish embryo during epiboly. Biochemistry and cell biology = Biochimie et biologie cellulaire. 1999;77:527-42
4. Behrndt M, Salbreux G, Campinho P. et al. Forces driving epithelial spreading in zebrafish gastrulation. Science. 2012;338:257-60
5. Cheng JC, Miller AL, Webb SE. Organization and function of microfilaments during late epiboly in zebrafish embryos. Developmental dynamics: an official publication of the American Association of Anatomists. 2004;231:313-23
6. Koppen M, Fernandez BG, Carvalho L. et al. Coordinated cell-shape changes control epithelial movement in zebrafish and Drosophila. Development. 2006;133:2671-81
7. Ougland R, Rognes T, Klungland A, Larsen E. Non-homologous functions of the AlkB homologs. Journal of molecular cell biology. 2015;7:494-504
8. Duncan T, Trewick SC, Koivisto P. et al. Reversal of DNA alkylation damage by two human dioxygenases. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16660-5
9. Aas PA, Otterlei M, Falnes PO. et al. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature. 2003;421:859-63
10. Ringvoll J, Nordstrand LM, Vagbo CB. et al. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. The EMBO journal. 2006;25:2189-98
11. Zheng G, Dahl JA, Niu Y. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular cell. 2013;49:18-29
12. Jia G, Fu Y, Zhao X. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature chemical biology. 2011;7:885-7
13. Fu D, Brophy JA, Chan CT. et al. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Molecular and cellular biology. 2010;30:2449-59
14. Songe-Moller L, van den Born E, Leihne V. et al. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Molecular and cellular biology. 2010;30:1814-27
15. Zdzalik D, Vagbo CB, Kirpekar F. et al. Protozoan ALKBH8 oxygenases display both DNA repair and tRNA modification activities. PloS one. 2014;9:e98729
16. Ougland R, Lando D, Jonson I. et al. ALKBH1 is a histone H2A dioxygenase involved in neural differentiation. Stem cells. 2012;30:2672-82
17. Li MM, Nilsen A, Shi Y. et al. ALKBH4-dependent demethylation of actin regulates actomyosin dynamics. Nature communications. 2013;4:1832
18. Nilsen A, Fusser M, Greggains G. et al. ALKBH4 depletion in mice leads to spermatogenic defects. PloS one. 2014;9:e105113
19. Duke-Cohan JS, Gu J, McLaughlin DF. et al. Attractin (DPPT-L), a member of the CUB family of cell adhesion and guidance proteins, is secreted by activated human T lymphocytes and modulates immune cell interactions. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11336-41
20. Tang W, Gunn TM, McLaughlin DF. et al. Secreted and membrane attractin result from alternative splicing of the human ATRN gene. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6025-30
21. Kuramoto T, Nomoto T, Fujiwara A. et al. Insertional mutation of the Attractin gene in the black tremor hamster. Mammalian genome: official journal of the International Mammalian Genome Society. 2002;13:36-40
22. Khwaja FW, Duke-Cohan JS, Brat DJ, Van Meir EG. Attractin is elevated in the cerebrospinal fluid of patients with malignant astrocytoma and mediates glioma cell migration. Clinical cancer research: an official journal of the American Association for Cancer Research. 2006;12:6331-6
23. Gunn TM, Miller KA, He L. et al. The mouse mahogany locus encodes a transmembrane form of human attractin. Nature. 1999;398:152-6
24. Hida T, Wakamatsu K, Sviderskaya EV. et al. Agouti protein, mahogunin, and attractin in pheomelanogenesis and melanoblast-like alteration of melanocytes: a cAMP-independent pathway. Pigment cell & melanoma research. 2009;22:623-34
25. Paz J, Yao H, Lim HS. et al. The neuroprotective role of attractin in neurodegeneration. Neurobiology of aging. 2007;28:1446-56
26. Tang W, Duke-Cohan JS. Human secreted attractin disrupts neurite formation in differentiating cortical neural cells in vitro. Journal of neuropathology and experimental neurology. 2002;61:767-77
27. Wrenger S, Faust J, Friedrich D. et al. Attractin, a dipeptidyl peptidase IV/CD26-like enzyme, is expressed on human peripheral blood monocytes and potentially influences monocyte function. Journal of leukocyte biology. 2006;80:621-9
28. He L, Gunn TM, Bouley DM. et al. A biochemical function for attractin in agouti-induced pigmentation and obesity. Nature genetics. 2001;27:40-7
29. Overton JD, Leibel RL. Mahoganoid and mahogany mutations rectify the obesity of the yellow mouse by effects on endosomal traffic of MC4R protein. The Journal of biological chemistry. 2011;286:18914-29
30. Kimmel CB, Ballard WW, Kimmel SR. et al. Stages of embryonic development of the zebrafish. Developmental dynamics: an official publication of the American Association of Anatomists. 1995;203:253-310
31. Huang H, Lu FI, Jia S. et al. Amotl2 is essential for cell movements in zebrafish embryo and regulates c-Src translocation. Development. 2007;134:979-88
32. Liu X, Xiong C, Jia S. et al. Araf kinase antagonizes Nodal-Smad2 activity in mesendoderm development by directly phosphorylating the Smad2 linker region. Nature communications. 2013;4:1728
33. Zhang Y, Li X, Qi J. et al. Rock2 controls TGFbeta signaling and inhibits mesoderm induction in zebrafish embryos. Journal of cell science. 2009;122:2197-207
34. Abrams EW, Mullins MC. Early zebrafish development: it's in the maternal genes. Current opinion in genetics & development. 2009;19:396-403
35. Hwang WY, Fu Y, Reyon D. et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature biotechnology. 2013;31:227-9
36. Lachnit M, Kur E, Driever W. Alterations of the cytoskeleton in all three embryonic lineages contribute to the epiboly defect of Pou5f1/Oct4 deficient MZspg zebrafish embryos. Developmental biology. 2008;315:1-17
37. Solnica-Krezel L, Driever W. Microtubule arrays of the zebrafish yolk cell: organization and function during epiboly. Development. 1994;120:2443-55
38. Barsh GS, He L, Gunn TM. Genetic and biochemical studies of the Agouti-attractin system. Journal of receptor and signal transduction research. 2002;22:63-77
39. Chung H, Lee JH, Jeong D. et al. Melanocortin 1 receptor regulates melanoma cell migration by controlling syndecan-2 expression. The Journal of biological chemistry. 2012;287:19326-35
Author contact
![]() Corresponding author: Shunji Jia, E-mail: jiasjtsinghua.edu.cn
Corresponding author: Shunji Jia, E-mail: jiasjtsinghua.edu.cn

 Global reach, higher impact
Global reach, higher impact