10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(6):644-653. doi:10.7150/ijbs.25272 This issue Cite
Research Paper
MiR-152-5p as a microRNA passenger strand special functions in human gastric cancer cells
1. Department of Gastroenterology, the First Affiliated Hospital of Soochow University, Suzhou 215006, China.
2. GenoArray Biotech, Suzhou, China
3. Genex Health Co., Ltd, Beijing, China
Received 2018-1-31; Accepted 2018-3-8; Published 2018-4-30
Abstract
Gastric cancer (GC) is one of the most common malignancies with high mortality rate. MiR-152 may exert the function of tumor suppressor by regulating its target gene, including PIK3CA. Nevertheless, all of the described functions are referred explicitly to miR-152-3p, while miR-152-5p as a passenger strand is poorly realized and entirely ignored. We previously selected miR-152-5p as a candidate using cell migration inhibition screening for GC cells and predicted that miR-152-5p might also target PIK3CA. In this study, we found an abnormal proportion of miR-152-3p / miR-152-5p in GC (gastric cancer) tissues and cells and demonstrated that miR-152-5p had poorer stability in GC cells, revealing the possibility that miR-152-5p is abnormally "suppressed" in gastric cancer. We also investigated and confirmed the role of miR-152-5p in GC by a series of experiments, and found that miR-152-5p modulated cell viability, migration, invasion, and cell-cycle progression of human GC cells, and also inhibited tumor growth and metastasis in vivo partially by targeting PIK3CA. More interestingly, it was proved that miR-152-3p and miR-152-5p had synergistic effects on the inhibition of PIK3CA in GC cells. The results of this study suggest that miR-152-5p may act as a tumor suppressor in SGC-7901 gastric cancer cells via targeting PIK3CA. Further, the study provides a novel insight into the roles of miRNA* during carcinogenesis.
Keywords: Gastric cancer, miR-152-5p, miR-152*, PIK3CA
Introduction
In the development of gastric cancer, some specific genes often have abnormal expression[1]. MiRNA, as a post-transcriptional regulator, participates in cell proliferation, differentiation, and apoptosis, and may be involved in many aspects of carcinogenesis and development of gastric cancer. Many miRNAs could promote the development of gastric cancer by regulating oncogenes and tumor suppressor genes directly or indirectly. For example, the overexpression of miR-10a/b, miR-17, miR-18a/b, miR-19a, miR-21, miR-106a/b, miR-340, miR-421 or miR-658 is closely related to the down-regulation of tumor suppressor genes Rb and PTEN, which may promote the occurrence of gastric cancer[2-4]. Abnormal expression of miR-19a could promote the proliferation and tumorigenesis of gastric cancer by inhibiting MXD1[5]. Correspondingly, the let-7 family of miRNA plays a role in suppressing GC by regulating oncogenes RAS and HMGA2 [6]. MiR-9 inhibits the growth of GC cells by regulating the expression of NF-κB[7]. MiR-338-3p is designated as a tumor suppressor in gastric cancer, and oncogene SSX2IP is one of its target genes [8].
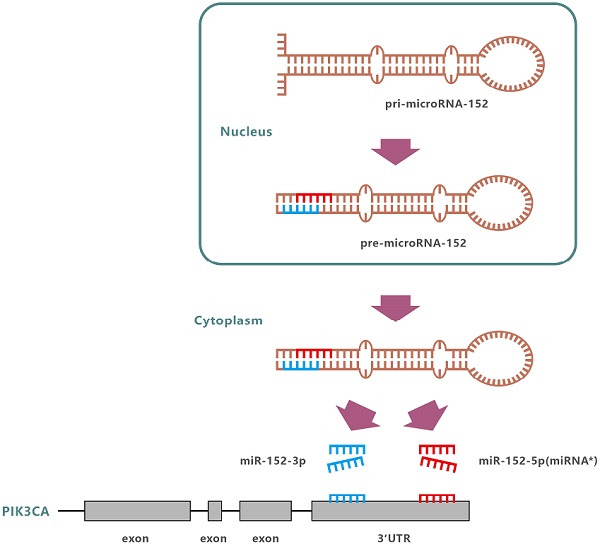
MiR-152 is abnormally expressed in a variety of diseases, including various cancers. Since miR-152 was first identified in mouse colon in 2002[9], more and more evidence suggests that miR-152 is a tumor suppressor, which are related to cell proliferation, migration, and invasion of human cancer[10-16]. In humans, the miR-152 gene is located at 17q21.32. Following transcription and undergoing nuclear cleavage, precursor miR-152 (pre-miR-152) is transported to the cytoplasm, and further cleaved by Dicer to form a miR-152 duplex. Finally, two different lengths and sequences of mature miRNAs are produced from opposite arms of the miR-152 duplex, named miR-152-3p and miR-152-5p (miR-152*). However, the detection frequency of miR-152-3p in each species was significantly higher than that of miR-152-5p. So far, the miR-152 reported involved in the regulation of development and progression of various cancers including gastric carcinoma, refers explicitly to miR-152-3p [17]. So, is miR-152-5p just garbage, only waiting to be degraded?
In our previous study, we performed cell migration screening using the SAMcell (self-assembled cell microarray) method[18] in the highly metastatic SGC-7901 GC cell line and selected several candidate miRNAs which could inhibit cell migration[19], including miR-152-5p. Previously, Ge et al.[20]have reported that miR-152-3p could target PIK3CA in breast cancer as a tumor suppressor. Bioinformatics analysis revealed that PIK3CA might also be a potential target for miR-152-5p. However, whether miR-152-5p is also participating in the regulation of PIK3CA expression is unknown. In this study, we investigated and confirmed the role of miR-152-5p in GC by a series of experiments, and found that miR-152-5p significantly modulated cell viability, migration, invasion, angiogenesis, and cell-cycle progression of Human GC cells, and also inhibited tumor growth and metastasis in vivo by targeting PIK3CA. These results suggest that miR-152-5p, as a passenger strand of miR-152, might have potential functions beyond common sense.
Materials and Methods
Cell culture and transfection
The rat gastric epithelial cell line RGM-1 and human gastric epithelial cell line GES-1 were purchased from Procell Life Science & Technology Co,.Ltd. China. Human gastric cancer cell lines SGC-7901, MKN-45 and AGS were obtained from the Cell Resource Center, Peking Union Medical College. Cells were cultured in DMEM medium (Sigma-Aldrich, UK) with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C in 5 % CO2. Transfection experiments were performed using vectors, 20nM siRNA or 20nM miRNA using Lipofectamine 2000 (Invitrogen, USA) according to manufacturer's protocol when cells reached 70% confluence.
The miRNA mimic and siRNA were as follows:
MiR-152-3p mimic, 5′-UCAGUGCAUGACAGAACUUGG -3′ (sense); MiR-152-5p mimic, 5′-AGGUUCUGUGAUACACUCCGACU -3′ (sense). PIK3CAsiRNA, 5′-GGAUCUUCCACACAAUUAA-3′ (sense).
Clinical specimens
A total of 15 paired GC and adjacent non-tumor gastric tissues were collected at the First Affiliated Hospital of Soochow University (Suzhou, China). This study was approved by the First Affiliated Hospital of Soochow University. The written informed consent was obtained from all participants.
Quantitative real-time PCR
Total RNA was isolated from cells with TRIzol reagent (Invitrogen). Relative miRNA or mRNA levels were determined by RT-qPCR using 7500 Real-Time PCR System (Applied Biosystems, UK). U6 was used as the internal control for miRNA, and GAPDH was used as the endogenous control for mRNA.
Primers for RT-qPCR sequences are as follows: PIK3CA: 5′-GTCAATCGGTGACTGTGTGG-3′ (forward), 5′-AGGTTCTGTGATACACTCCGACT-3′ (reverse). GAPDH: 5′-ACAACTTTGGTATCGTGGAAGG-3′ (forward), 5′-GCCATCACGCCACAGTTTC-3′ (reverse). U6: 5′-CTCGCTTCGGCAGCACA-3′ (forward), 5′- GCGAGCACAGAATTAATACGAC-3′ (reverse). MiR-152-3p: 5′-GCAGTCAGTGCATGACAGA-3′ (forward), 5′-GTCCAGTTTTTTTTTTTTTTTCCAAG-3′ (reverse). MiR-152-5p: 5′-CAGAGGTTCTGTGATACACTC-3′ (forward), 5′-GGTCCAGTTTTTTTTTTTTTTTAGTC-3′ (reverse).
MicorRNA stability assay
Cells were seeded in 12-well plates and cultured at 37 ℃ under 5% CO2 for 24 h. Total RNA was isolated from cells treated with actinomycin D (10 mg/L) at 0, 4, 8, 12 and 24 hours respectively. Relative abundance of miRNAs was detected by RT-qPCR.
Western blot analysis
Total cell proteins were isolated with the ProteoJET™ Mammalian Cell Lysis Reagent (Thermo Scientific, USA). Proteins (50 μg) were separated using 10% SDS-PAGE and transferred to nitrocellulose membranes. The membranes were probed with the specific primary antibodies against PIK3CA (#4249, Cell Signaling, USA), or Tubulin (#5666, Cell Signaling, USA). Finally, protein bands were visualized using enhanced chemiluminescent substrate, according to the manufacturer's protocol (Thermo). The images were quantified by ImageJ software (http://imagej.nih.gov/ij/).
Plasmids
The human PIK3CA 3′ UTR (GenBank accession NM_006218.3) containing the predicted miR-152-5p binding site was PCR amplified using genomic DNA from SGC-7901 cells. Then, the PCR product was cloned into the pGL3 Vector (Promega, Madison, WI, USA) located between XbaI and EcoRI restriction enzyme sites downstream of the firefly luciferase reporter gene and named PIK3CA-wt-3′ UTR. The miR-152-5p target binding site was muted using MutanBEST mutation kit (Takara, Japan), and also cloned into the pGL3 Vector, named PIK3CA-mut-3′ UTR. Primers for cloning (bold italic for restriction sites) are as follows:
PIK3CA wt-3'UTR Forward: 5′-CTAGTCTAGACAGAGAACTGTGTTTTACCCG -3′. PIK3CA wt-3'UTR Reverse: 5′-CCGGAATTCCAGATCTTAGTCACCCATGTAG -3′.
PIK3CA mut-3'UTR Forward: 5′-TACATCCCCCCCTCATCCACACCAACCTCCACCATTAAAATGCACA -3′. PIK3CAMut-3'UTR Reverse: 5′-GTTGGTGTGGATGAGGGGGGGATGTAACTATCTGGACTGATAGATA -3′.
Luciferase assay
The firefly luciferase reporter plasmids containing wt or mutated 3'UTRs (100ng), the control renilla luciferase plasmid (pRL-CMV, 5ng) (Promega) and miRNA mimics were co-transfected into cells using Lipofectamine 2000, according to the manufacturer's protocol. After 48 h, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega).
Cell proliferation assay
Cell Counting Kit-8 (CCK-8) assay: Cell viability was assessed indirectly by CCK-8 assay. SGC-7901 cells were seeded in a 96-well plate at a density of 3000 cells/well. After 24 h, cells were transfected with the indicated miRNAs or siRNAs. Forty-eight hours later, cells were incubated with 100 µL of fresh medium containing 10% CCK-8 reagent (DoJinDo Laboratories, Japan) for 1h at 37℃. Then, the absorbance values of the wells at 450 nm were detected using an automatic spectrometer (Multimode Reader, PerkinElmer, USA). This procedure was repeated at 1, 3, 4, 5, 6, and seven days after cell seeding. EdU (5-ethynyl-2'-deoxyuridine) Cell Proliferation Assay: The cells were seeded in 96-well plates at 5×103 cells/well. Forty-eight hours after transfection, EdU (50 μM; Thermo Fisher Scientific, USA) labeling media was added to the plates, and further incubated for two hours at 37℃ in 5% CO2. After being treated with 4% paraformaldehyde and 0.5% Triton X-100, the cells were stained with the anti-EdU working solution. DAPI stained the cell nuclei. Count the numbers both of EdU-positive and negative cell nuclei in five randomly areas and calculated the percentage of EdU-positive cells.
Transwell assay
For transwell migration assays, cells were plated into the upper chambers of transwell inserts (8μm pores; Costar, USA) in 100 μL FBS-free DMEM and the lower chamber was filled with DMEM (500μL) with 10% FBS. Non-migratory cells remaining on the upper surface of the transwell membranes were then removed with cotton swabs, and migratory cells on the lower surface were fixed with 4% formaldehyde. DAPI stained cell nuclei for quantification. In the invasion assays, cells were seeded into Matrigel-coated chambers and pre-incubated at 37℃ for 30 min. In each experiment, the number of cells in five random fields on the underside of the filter was calculated, and three independent filters were analyzed.
Flow-cytometric analyses
The transfected cells (at a density of 3×106) were collected at 48 h posttransfection and stained using Annexin V/PI Apoptosis Kit (Thermo). The stained cells were analyzed using a flow cytometer (FACScan; BD Biosciences, CA) and the relative ratios of apoptotic cells were compared with control. Cell cycle analysis was used PI staining and flow cytometry.The percentages of cells at different stages (G0/G1, S, and G2/M) were counted and compared. All samples were analyzed in triplicate.
In vitro angiogenesis assay
The original concentration of Matrigel (10 mg/ml) was placed in a 4℃ freezer overnight. The Matrigel was layered into 96-well plates (40 μl /well) and then allowed to cure in an incubator at 37℃ for 2 hours. Human umbilical vein endothelial cells (HUVEC) were serum starved for 24 hours, suspended in medium preconditioned with SGC-7901 cells, and added dropwise to the surface of the Matrigel (5×104 cells/well). After incubation for 18 hours at 37℃, angiogenic activity was detected and quantified by Image J Angiogenesis Analyzer.
Animal Model and In vivo Tumorigenesis Assay
All BALB/C (nu/nu) nude mice were purchased from the Animal Centre of Peking University. SGC-7901 cells were stably transfected with lentivirus-Flu-NC and lentivirus-Flu-miR-152-5p vectors (OriGene, USA). For tumor implantation, luciferase-labeled SGC-7901 cells (5×106) were suspended in PBS and performed intraperitoneal or tail vein injection. After four weeks, tumor growth and metastasis status were examined by the In-Vivo Imaging System (IVIS Spectrum CT, PerkinElmer).
Statistical analysis
The data were expressed as means ± standard deviations (SD). Data analysis was performed using SPSS Statistics 20.0 (IBM, SPSS, USA). When comparing two groups, the two-tailed Student's t-test was used to evaluate the significance. Groups with values of P< 0.05 were considered to be statistically significant.
Results
Special characteristics of miR-152-5p in gastric cancer tissues and cells
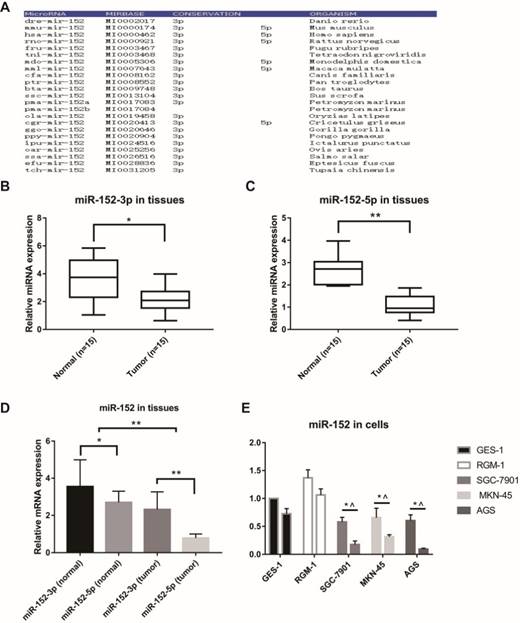
MicroRNAs are often highly conserved across multiple species, but there are exceptions. Firstly, we analyzed the conservatism of miR-152, and the results indicated that the miR-152-3p sequence was extremely conservative in many species. However, the conservatism of miR-152-5p was relatively poor (Figure 1A). Next, we examined the relative content of miR-152-3p and miR-152-5p in 15 paired GC and adjacent non-tumor tissues. The results showed that both miR-152-3p and 5p were down-regulated in gastric cancer (Figure 1B, C). However, it was noteworthy that we found that the proportion of miR-152-3p and 5p in GC tissues was significantly lower than that in normal tissues (Figure 1D). In other words, miR-152-5p appears to be deliberately reduced in tumors. Subsequently, we compared the expression rates of miR-152-3p and 5p in normal gastric epithelial cells (GES-1 and RGM-1) and gastric cancer cells (SGC-7901, MKN-45 and AGS), which results were similar to those in the tissues (Figure 1E).
Special characteristics of miR-152-5p in gastric cancer tissues and cells. A. MiR-152-3p and miR-152-5p show different conservative in the phylogeny. B, C. MiR-152-3p and miR-152-5p expression levels in GC tissues compared with non-cancerous tissues were analyzed using qRT-PCR. U6 served as an internal control. D. The bar graph showed the different ratio of miR-152-3p/5p between GC and adjacent non-tumor tissues. *P<0.05 and ** P<0.01. Data are representative of three independent experiments (means ±s.d.). E. qRT-PCR assays were performed to detect the expression ratios of miR-152-3p/5p in normal gastric epithelial cell lines (GES-1 and RGM-1) and various gastric cancer cell lines (SGC-7901, MKN-45 and AGS). * vs. GES-1 and ^ vs. RGM-1.
MiR-152-3p and miR-152-5p stabilities in gastric epithelial cells and GC cells.Total RNA was isolated from cells treated with actinomycin D (10 mg/L) at 0, 4, 8, 12 and 24 hours respectively. Relative abundance of miRNAs was detected by RT-qPCR. U6 served as an internal control. A, B, C and D. The line charts showed the stabilities of miR-152-3p and 5p in GES-1, SGC-7901, MKN-45 and AGS cells, respectively. The data represent the mean ±s.d. from three independent experiments.
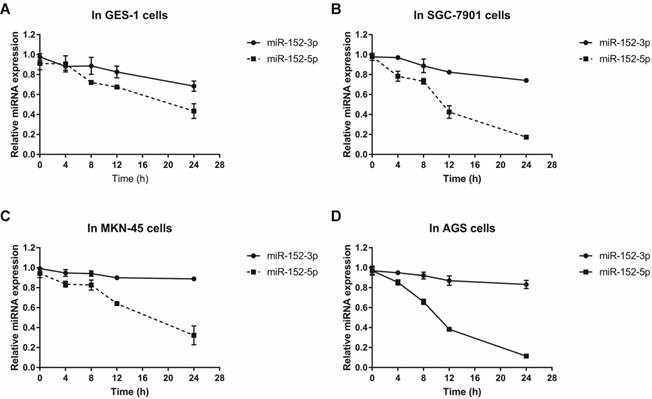
Why does this happen? Since both miR-152-3p and miR-152-5p are processed from pre-miR-152, this proportional difference is theoretically independent of miRNA transcriptional level. We suspect that it is more likely to be the difference in the stability of miRNA itself during or after processing. Using actinomycin D to inhibit cellular RNA transcription, we detected the stabilities of miR-152-3p and miR-152-5p in normal and tumor cells by qRT-PCR. The results showed that the stabilities of miR-152-3p in normal cells and tumor cells were not significantly different, but the stabilities of miR-152-5p in tumor cells were striking lower than in normal cells (Figure 2A-D). Taken together, these results indicated that although both miR-152-3p and 5p are derived from the same transcript, the processed miRNAs may face different fates in normal cells and tumor cells, resulting in the differences of miR-152-5p stabilities.
Mir-152-5p Specifically Targets PIK3CA in Human Gastric Cancer Cells
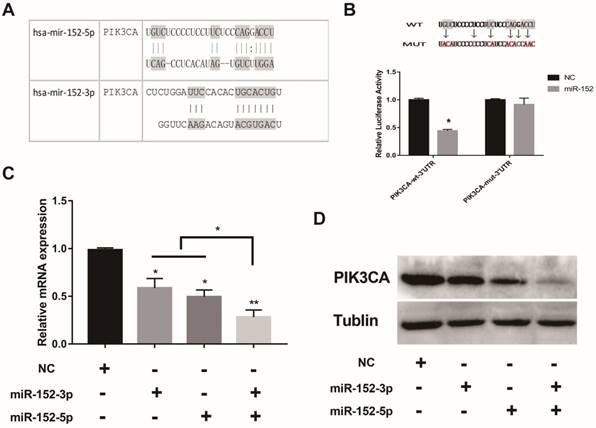
What is the function of a miRNA that is deliberately "suppressed" in gastric cancer cells? Interestingly, bioinformatics predictions showed that both miR-152-5p and 3p could potentially target PIK3CA (Figure 3A). The regulation of PIK3CA by miR-152-3p has been reported[20].To confirm the direct regulation of PIK3CA by miR-152-5p, luciferase reporter assays were performed by using a series of luciferase reporter vectors carrying the predicted miRNA-152-5p target sites or mutant (PIK3CA-mut-3′UTR) in SGC-7901 cells. As shown in Figure 3B, miR-152-5p markedly suppressed the luciferase activity of the reporter vectors with the predicted target sites but had little impact on the muted group. When miR-152-5p or 3p was overexpressed in SGC-7901 cells, both mRNA and protein levels of PIK3CA were decreased as expected (Figure 3C, D). It was important to note that when miR-152-3p and 5p were co-overexpressed, the inhibitory effect on PIK3CA was stronger than that of a single miRNA (Figure 3C, D). All the results implied that miR-152-5p could directly act on the predicted target sites in the PIK3CA 3′ UTR and may play a synergistic role with miRNA-152-3p.
MiR-152-5p Inhibits Cell Proliferation and Promotes Apoptosis of SGC-7901 Cells by Downregulating PIK3CA
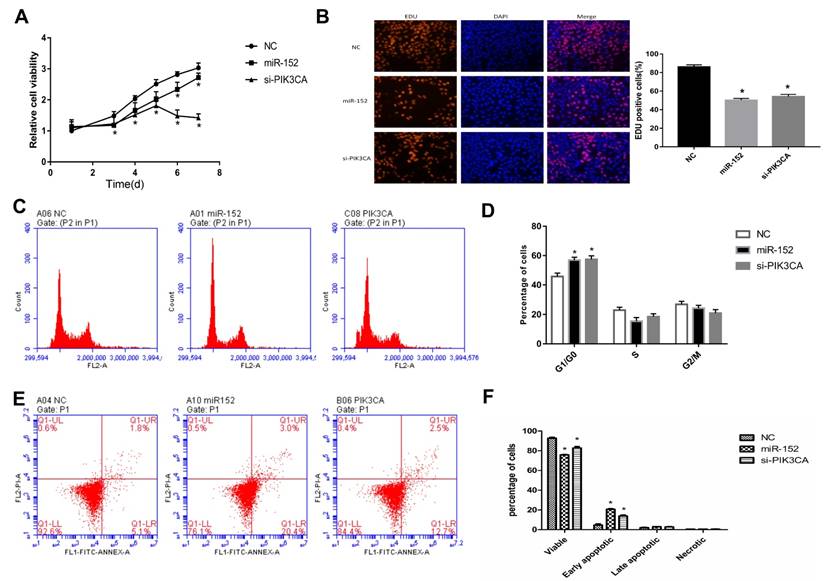
To explore the function of miR-152-5p, cell proliferation assays were performed in gastric cancer cell line SGC‑7901. SGC-7901 cells were transfected with miR-152-5p mimic, PIK3CAsiRNA (as a positive control) or NC mimic (scrambled control). The effects of miRNA on SGC-7901 cell viability were determined over a period of 7 days posttransfection by CCK-8assays (Figure 4A). Transfection with the miR-152-5p mimic resulted in a clear reduction in cell viability of SGC-7901 cells compared with the NC group. Similarly, these findings were confirmed by microscopic analysis of the EdU (red) / DAPI (blue) immunostaining assay (Figure 4B). To further explore whether miR-152-5p influences the proliferation of SGC-7901 cells through cell cycle regulation, cell cycle analysis was performed by PI staining and flow cytometry. The results showed that, compared with the control group, SGC-7901 cells overexpressed with miR-152-5p had obvious cell cycle arrest in G1/G0 phase (Figure 4C, D). Also, we performed flow cytometry analysis to determine whether apoptosis affected cell proliferation. The results indicated that knockdown of PIK3CA by miR-152-5p significantly induced SGC-7901 cell apoptosis (Figure 4E, F). In general, these data indicated that miR-152-5p might play an anti-cancer effect by suppressing the proliferation and cell cycle of SGC-7901 cells.
MiR-152-5p Inhibits Migration, Invasion, EMT and Angiogenesis Abilities of SGC-7901 Cells
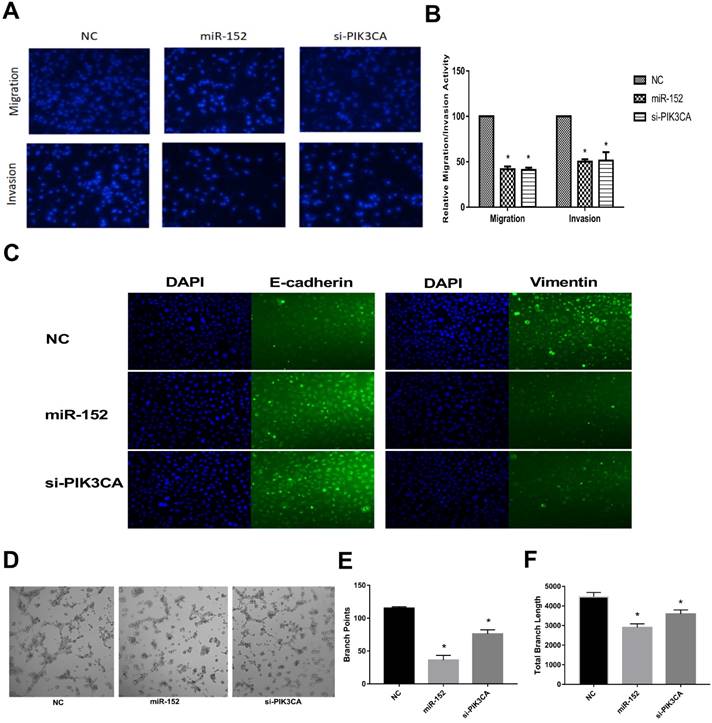
Next, we continued to explore the function of miR-152-5p in SGC-7901 cells. It was shown that the migration and invasive abilities of SGC-7901 cells with a high expression of miR-152-5p were lower than those of NC by transwell migration and invasion assay (Figure 5A, B). Given that EMT is an important early step in cancer metastasis, we investigated whether miR-152-5p could mediate this process. As shown by immunofluorescent staining in Figure 5C, overexpression of miR-152-5p resulted in up-regulation of E-cadherin expression and reduction of Vimentin expression. Further, the tube formation of endothelial cells was tested by treatment with the medium preconditioned by transfection of SGC-7901 cells with miR-152-5p. MiR-152-5p overxpression presented impaired tube formation, as defined by fewer branching points and less total branch length (Figure 5D-F). These data suggested that miR-152-5p could inhibit the malignant behavior of gastric cancer cells.
MiR-152-5p Inhibits Tumor Growth and Metastasis in vivo
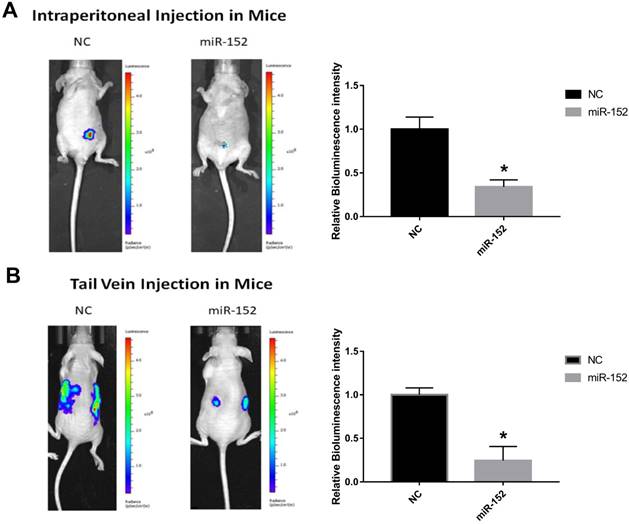
To confirm the effect of miR-152-5p function on tumorigenesis in vivo, xenograft mouse models were established. SGC-7901 cells stably transfected with lentivirus-Flu-miR-152-5p or a control vector were subcutaneously injected into male nude mice. Peritoneal implantation metastasis was evaluated after stably transfected cells were injected into the abdominal cavity of nude mice. Hematogenous metastasis was also observed after SGC-7901 cells were transplanted into nude mouse through the tail vein. The results showed that the bioluminescence intensity of miR-152-5p group was significantly lower than that of NC group (Figure 6A, B). That is, miR-152-5p inhibited tumor growth and metastasis capabilities.
MiR-152-5p specifically targets PIK3CA in SGC-7901cells. A. Schematic representation of miR-152-3p and miR-152-5p targets in the 3′ UTR of human PIK3CA; B. The artificial mutant 3′UTR without the miR-152-5p binding sites (top). Dual luciferase reporter assays were performed to test the interaction of miR-138 with wild-type predicted PIK3CA 3′UTR targeting sequences (PIK3CA-wt-3′ UTR) and mutated targeting sequences (PIK3CA-mut -3′ UTR) (bottom). C. QRT-PCR assays were performed to examine the effects of miR-152-5p and/or miR-152-3p mimics transfection on endogenous PIK3CA gene transcription. GAPDH served as an internal reference. *P<0.05, **P<0.01. Data are representative of three independent experiments (means ±s.d.). D. The effects of miR-152-5p and/or miR-152-3p overexpression on endogenous PIK3CA protein levels were examined by Western Blot analyses. Tubulin served as an internal control.
MiR-152-5p inhibits SGC-7901 cell proliferation, cell cycle and promotes apoptosis in-vitro. A. CCK-8 assays were performed 1, 3, 4, 5, 6, and 7days after transfection to determine the cell viability of the transfected SGC-7901cells. B. EdU (red)/DAPI (blue) assays were conducted 48h after transfection to determine the viability of the transfected SGC-7901 cells. C, D. Forty-eight hours after transfection, the cell cycle stages of SGC-7901cells were analyzed by flow cytometry. The bar chart represents the percentage of cells in the G1, S, or G2/M phases, as indicated. E, F. Related apoptotic cells were measured by flow cytometry 48h after transfection. The data represent the mean ±s.d. from three independent experiments. *, P< 0.05.
Discussion
Gastric cancer (GC) is a kind of complex disease related to multiple factors, including environmental exposure and genetic factors. Familial aggregation and different responses to the same environmental exposure suggested that individual diversity played a significant role in the risk of cancer. Searching for potential biomarkers for gastric cancer may provide essential clues to early diagnosis and personalized treatment. PIK3CA is an oncogene, located at 3q26.3. Under normal circumstances, PIK3CA regulates somatic cell proliferation, differentiation, survival and has other physiological functions. However, the mutation of PIK3CA will lead to PI3K-Akt pathway over-activation, increasing aberrant cell invasion and metastasis [21]. It is widely considered that PIK3CA mutations play an important role in the development of various solid tumors such as colon cancer, brain cancer, breast cancer and lung cancer [22-25]. Therefore, PIK3CA has potential clinical value as a new molecular target for tumor therapy. Similarly, the mutation, frequency, and hotspot pattern of PIK3CA gene in gastric cancer tissues are also specific topics. However, so far, there is no significant correlation between the mutation of PIK3CA gene and the clinicopathological features of gastric cancer[24, 26, 27]. Gene amplification is another primary mechanism of oncogene activation. In most cases, the increased copy number is closely related to the transcription level and the corresponding protein expression level of the oncogene. Related studies have shown that PIK3CA highly expressed highly expressed at mRNA and protein levels in a variety of GC cells and GC tissues [28, 29], which are related to tumor invasion ability. We also confirmed similar results from 15 paired GC and adjacent tissues (Supplementary Fig. 1). However, it remains unclear whether PIK3CA gene amplification occuring in GC is more important than mutation.
MiR-152-5p inhibits migration, invasion, EMT and angiogenesis abilities of SGC-7901 cells in vitro. A. Migration and invasion assay of SGC-7901 cells following transfection with miR-152-5p mimic or PIK3CA siRNA using the transwell apparatus. Microscopic analysis of migrated or invaded cells stained with DAPI. B. Relative cell migration or invasion was determined by the number of the DAPI-stained cells. Cell migration or invasion is expressed as a percentage of that observed in control. C. Overexpression of miR-152-5p in SGC-7901 cells repressed epithelial-mesenchymal transition (EMT). E-cadherin and vimentin were used as EMT markers. DAPI was used in nucleus staining. D. Representation of tube formation assay shows the angiogenic capability of SGC-7901 cells transfected with NC, miR-152-5p or PIK3CA siRNA. E, F. Graphical representation shows that miR-152-5p impaired tube formation as a result of fewer branch points and total branch length as quantified by the Image J Angiogenesis Analyzer. Data are the means ±s.d. of three independent experiments. *P<0.05 versus control.
MiR-152 has been thought to be a tumor suppressor in a variety of solid tumors since it was identified. It has confirmed that miR-152 is abnormally downregulated in gastric cancer[30, 31], which may be closely related to the development of GC. However, all of the mentioned functions were specific to miR-152-3p, which were seemed unrelated to miR-152-5p. MiR-152-5p, like other miRNAs*, was ignored as a passenger strand. It is generally believed that the passenger strand of the miRNA is degraded in the process of miRNA biosynthesis, and only the guide strand of the miRNA duplex is chosen to become the mature functional miRNA. However, more and more evidence suggests that passenger strand miRNAs could also act on target mRNAs and have biological functions in cancer and other diseases[32-35]. For example, miR-17-3p, as a passenger strand microRNA, is involved in the induction of tumorigenesis and invasion by targeting TIMP3 and PTEN[32, 33]. Bang et al. found that cardiac fibroblasts secrete many passenger strand microRNAs through exosomes and regulate the relevant signaling pathways of cardiomyocytes in a paracrine manner[34].
MiR-152-5p inhibits the tumorigenesis of SGC-7901cells in vivo. A. Images showed primary tumor growth upon intraperitoneal implantation of 5×106 luciferase-labeled SGC-7901 cells infected with indicated vectors. Mice were imaged four weeks after tumor cell injection to observe the local tumor growth and metastasis. B. Images showed tumor metastasis upon tail vein implantation of 5×106 luciferase-labeled SGC-7901 cells infected with indicated vectors. Mice were imaged four weeks after tumor cell injection.
In this study, we provided experimental evidence that by directly targeting oncogene PIK3CA, miR-152-5p as a microRNA passenger strand inhibited cell proliferation, migration, invasion, and induced cell apoptosis in vitro and suppressed tumorigenesis and metastasis of SGC-7901 cells in vivo. Although the significance and function of PIK3CA coding region mutations in gastric cancer are still controversial, there are few reports of high-frequency mutation in PIK3CA 3′UTR, especially the target sites of miR-152-5p, suggesting that miR-152- 5p could be a therapeutic target for GC.
The expression of miR-152 is frequently downregulated in different solid tumors. Notably, the miR-152 gene is located within intron 1 of the COPZ2 gene, and several CpG islands are typically identified around its promoter region. As a result, the downregulation of miR-152 may be attributed to hypermethylation of its promoter in tumor cells [14, 36]. The results of this study also showed that the expression level of miR-152-5p in GC tissues and cells was much lower than that of miR-152-3p. Further, we found an abnormality in the proportion of miR-152-3p/5p in gastric cancer tissues and cells (Figure 1D, E). Preliminary experiments also showed that miR-152-5p was significantly less stable in gastric cancer cells than normal gastric mucosa cells (Figure 2 A-D), which may be the reason for the abnormal ratio of miR-152-3p/5p. More interestingly, in normal breast cells (MCF-10A) vs. breast cancer cells (MCF-7 and MDA-MB-231), ovarian epithelial cells (IOSE80) vs. ovarian cancer cells (SKOV3), and normal liver cells (L02) vs. hepatocellular carcinoma cells (HepG2 and Hep3B), we did not find the abnormality of miR-152-3p/5p (Supplementary Fig. 2). These results suggest the specificity of the role of miR-152-5p as well as its limitations. We are wondering whether there is a unique processing mechanism for miR-152 in GC, which will be an attractive research.
Although numerous genes have been predicted to be targets of miR-152, only PIK3CA is the common target of miR-152-3p and miR-152-5p. We also found that miR-152-3p and miR-152-5p had synergistic effects on the inhibition of PIK3CA in GC cells. Therefore, it will be attractive to analyze the mutual antagonism or synergistic effect between miRNAs and miRNAs*, which will be a great benefit to understanding the diversity of miRNA regulatory networks. We listed some miRNA-3ps and miRNA-5ps with common targets from bioinformatics analysis in Supplementary Table 1, which indicated that miRNAs and miRNAs* have common targets is not an accident. However, the mutual antagonism or synergistic effect between miRNAs and miRNAs* needs more experimental evidence.
In conclusion, the results of this study suggested that miR-152-5p might act as a tumor suppressor in gastric cancer cells via targeting PIK3CA. Further, the study provides a novel insight into the roles of miRNA* during carcinogenesis.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 31270919). We also thank Suzhou Genepharma Biotech for the miRNA mimics and assistance with siRNA design and synthesis.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Stock M, Otto F. Gene deregulation in gastric cancer. Gene. 2005;360:1-19
2. Guo J, Miao Y, Xiao B. et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652-7
3. Strand MS, Lockhart AC, Fields RC. Genetics of Gastric Cancer. Surg Clin North Am. 2017;97:345-70
4. Yepes S, López R, Andrade RE. et al. Co-expressed miRNAs in gastric adenocarcinoma. Genomics. 2016;108:93-101
5. Wu Q, Yang Z, An Y. et al. MiR-19a/b modulate the metastasis of gastric cancer cells by targeting the tumour suppressor MXD1. Cell Death Dis. 2014;5:e1144
6. Ohshima K, Inoue K, Fujiwara A. et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247
7. Wan HY, Guo LM, Liu T. et al. Regulation of the transcription factor NF-kappaB1 by microRNA-9 in human gastric adenocarcinoma. Mol Cancer. 2010;9:16
8. Li P, Chen X, Su L. et al. Epigenetic silencing of miR-338-3p contributes to tumorigenicity in gastric cancer by targeting SSX2IP. PLoS One. 2013;8:e66782
9. Lagos-Quintana M, Rauhut R, Yalcin A. et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735-9
10. Zhou X, Zhao F, Wang ZN. et al. Altered expression of miR-152 and miR-148a in ovarian cancer is related to cell proliferation. Oncol Rep. 2012;27:447-54
11. Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881-90
12. Azizi M, Teimoori-Toolabi L, Arzanani MK. et al. MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol Ther. 2014;15:419-27
13. Kindrat I, Tryndyak V, de Conti A. et al. MicroRNA-152-mediated dysregulation of hepatic transferrin receptor 1 in liver carcinogenesis. Oncotarget. 2016;7:1276-87
14. Tsuruta T, Kozaki K, Uesugi A. et al. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 2011;71:6450-62
15. Ma J, Yao Y, Wang P. et al. MiR-152 functions as a tumor suppressor in glioblastoma stem cells by targeting Krüppel-like factor 4. Cancer Lett. 2014;355:85-95
16. Xiang Y, Ma N, Wang D. et al. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2014;33:378-86
17. Liu X, Li J, Qin F. et al. miR-152 as a tumor suppressor microRNA: Target recognition and regulation in cancer. Oncol Lett. 2016;11:3911-6
18. Zhang H, Hao Y, Yang J. et al. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun. 2011;2:554
19. You W, Xu L, Zhang X. et al. High-Throughput Screening Identifies miR-451 as a Pleiotropic Modulator That Suppresses Gastric Cancer Metastasis. SLAS Technol. 2017;22:136-43
20. Ge S, Wang D, Kong Q. et al. Function of miR-152 as a Tumor Suppressor in Human Breast Cancer by Targeting PIK3CA. Oncol Res. 2017;25:1363-71
21. Samuels Y, Diaz LA, Schmidt-Kittler O. et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561-73
22. Desmedt C, Zoppoli G, Gundem G. et al. Genomic Characterization of Primary Invasive Lobular Breast Cancer. J Clin Oncol. 2016;34:1872-81
23. Janiszewska M, Liu L, Almendro V. et al. In situ single-cell analysis identifies heterogeneity for PIK3CA mutation and HER2 amplification in HER2-positive breast cancer. Nat Genet. 2015;47:1212-9
24. Fang WL, Huang KH, Lan YT. et al. Mutations in PI3K/AKT pathway genes and amplifications of PIK3CA are associated with patterns of recurrence in gastric cancers. Oncotarget. 2016;7:6201-20
25. Kris MG, Johnson BE, Berry LD. et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998-2006
26. Ito C, Nishizuka SS, Ishida K. et al. Analysis of PIK3CA mutations and PI3K pathway proteins in advanced gastric cancer. J Surg Res. 2017;212:195-204
27. Harada K, Baba Y, Shigaki H. et al. Prognostic and clinical impact of PIK3CA mutation in gastric cancer: pyrosequencing technology and literature review. BMC Cancer. 2016;16:400
28. Liu JF, Zhou XK, Chen JH. et al. Up-regulation of PIK3CA promotes metastasis in gastric carcinoma. World J Gastroenterol. 2010;16:4986-91
29. Shi J, Yao D, Liu W. et al. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer. 2012;12:50
30. Song YX, Yue ZY, Wang ZN. et al. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Cancer. 2011;10:1
31. Chen Y, Song Y, Wang Z. et al. Altered expression of MiR-148a and MiR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg. 2010;14:1170-9
32. Yang X, Du WW, Li H. et al. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013;41:9688-704
33. Shan SW, Fang L, Shatseva T. et al. Mature miR-17-5p and passenger miR-17-3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J Cell Sci. 2013;126:1517-30
34. Bang C, Batkai S, Dangwal S. et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124:2136-46
35. Mah SM, Buske C, Humphries RK. et al. miRNA*: a passenger stranded in RNA-induced silencing complex. Crit Rev Eukaryot Gene Expr. 2010;20:141-8
36. Chen Y, Song YX, Wang ZN. The microRNA-148/152 family: multi-faceted players. Mol Cancer. 2013;12:43
Author contact
![]() Corresponding author: Department of Gastroenterology, the First Affiliated Hospital of Soochow University, Suzhou 215006, China. Email: sdfyyliruicom
Corresponding author: Department of Gastroenterology, the First Affiliated Hospital of Soochow University, Suzhou 215006, China. Email: sdfyyliruicom

 Global reach, higher impact
Global reach, higher impact