10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(7):705-716. doi:10.7150/ijbs.18997 This issue Cite
Research Paper
Eaf1 and Eaf2 mediate zebrafish dorsal-ventral axis patterning via suppressing Wnt/β-Catenin activity
1. College of Fisheries, Key Laboratory of Freshwater Animal Breeding, Ministry of Agriculture, Huazhong Agricultural University, Wuhan, 430070, P. R. China.
2. Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, 430072, P. R. China.
Received 2016-12-31; Accepted 2018-3-17; Published 2018-5-12
Abstract
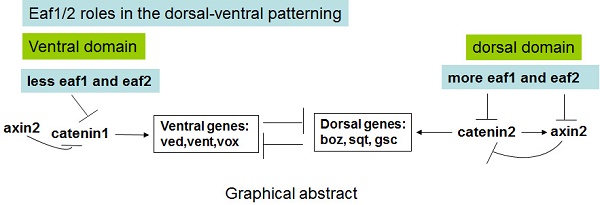
During early vertebrate embryogenesis, maternal Wnt/β-catenin signaling is thought to locally initiate expression of dorsal-specific genes. Here, eaf1 and eaf2 were identified as important maternal and zygotic modulators of Wnt signaling to initiate and specify ventral genes. Expression of ventral ved, vent, and vox was all obviously enhanced in either maternal or zygotic eaf1/2 morphants, and in both eaf1 heterozygous and homozygous mutants, but their expression was suppressed in embryos with over-expression of eaf1/2. Additionally, eaf1/2 were revealed to suppress ventral fates in embryos via Wnt/β-catenin1/Tcf signaling, complimentary to their roles in suppressing dorsal fates via Wnt/β-catenin2 signaling. Moreover, eaf1/2 were also revealed to obviously suppress the expression of axin2 induced by β-catenin2 rather than by β-catenin1, and the dorsal expression of axin2 in embryos was obviously suppressed by ectopic expression of eaf1/2. This study uncovers a novel dorsal-ventral patterning pathway, with eaf1 and eaf2 inhibiting ventral cells via suppressing Wnt/β-catenin1/Tcf signaling and inducing dorsal cells indirectly via suppressing β-catenin2-induced-axin2 on the dorsal side of embryos.
Keywords: eaf1, eaf2, β-catenin, axin2, dorsal-ventral patterning
Introduction
Vertebrate embryos form their axes by dorsal-ventral patterning and anterior-posterior patterning. Maternal factors are required to induce zygotic expression of ventral or dorsal genes to establish dorsal-ventral polarity during the first step of axis formation. In zebrafish, the dorsal-ventral specification starts at approximately 128-cell stage when maternal β-catenin accumulates in the nuclei of blastomeres in the dorsal domain (1), initiating the expression of dorsal organizer genes, such as sqt, boz, and gsc (2, 3). Lower expression of maternal β-catenin2 and reduced expression of dorsal genes are observed in zebrafish ich mutants (2, 3), where maternal β-catenin2 rather than maternal β-catenin1 has been found to be essential for the specification of dorsal fates in early stage embryos (4, 5). Reciprocal activities are needed to induce the expression of ventral genes. Maternal TGF-β/Rdr, Wnt/Ca2+ pathways and Pou2 complementarily specify ventral bmp2b and bmp4 expression (6-8). Moreover, maternal runx2 is the only maternal determinant identified to initiate the zygotic ventral factors of vent, vox, and ved (9). Despite observations of nuclear accumulation of β-catenin protein in both dorsal and ventral cells of axin-depleted embryos at early blastula stage (10) as well as ubiquitous expression of both β-catenin genes in early stage embryos (4), it still remains unknown whether maternal Wnt/β-catenin signaling functions in initiation and specification of ventral cells.
After the specification of dorsal-ventral polarity, zygotic Wnt/β-catenin signaling is activated and required to antagonize the dorsal organizer genes for proper dorsal-ventral axis formation. Zygotic Wnt/β-catenin signaling is activated by Wnt ligands after mid-blastula transition (MBT) and maintains high expression of vox, ved, and vent on the ventral side, thereby antagonizing expression of dorsal genes sqt, boz, and gsc (11). Consistently, zebrafish homozygous mutants for wnt8 show significant expansion of shield markers and almost absent expression of ventral-lateral mesoderm markers (12). Additionally, two dominant mutants of negative regulators of Wnt/β-catenin signaling, dntcf and tcfBD, can also induce ventral-lateral ectopic expression of dorsal-specific genes in the marginal region (13).
Axin2, a target of Wnt/β-catenin signaling, is part of a degradation complex that functions to phosphorylate β-catenin and negatively regulates the signaling pathway (14-16). In Xenopus, embryos exhibit severe ventralization with ectopic expression of axin but anterior/dorsalization after depleting maternal axin mRNA (10).
Eaf1 and Eaf2 are originally identified as partners of ELL (Eleven-nineteen lysine-rich leukemia), a fusion protein frequently associated with myeloid leukemia, and the two genes are found to suppress prostate cancer and leukemia (19-21). In our previous studies, during zebrafish embryogenesis, both eaf1 and eaf2 have been revealed to mediate effective convergence and extension movements (22), form a negative feedback loop with wnt4 (23), and function as novel factors antagonizing canonical Wnt/β-catenin signaling in forebrain induction (5) and as novel factors suppressing TGF-β signaling in germ layers specification (24). We have also found dorsal-ventral axis patterning defects in eaf morphants (5), but how maternal and zygote eaf1/2 act in patterning ventral cells is still unclear. In this study, maternal expression of eaf1/2 was revealed first. Then, eaf1 and eaf2 were revealed to suppress expression of ventral vent family genes by genetically modulating the Wnt/β-catenin1/Tcf pathway, complementing their roles of suppressing dorsal cell fates in the Wnt/β-catenin2 pathway (5). Eaf1 and eaf2 were also revealed to modulate the dorsal-ventral polarity of axin2 and promote the expression of dorsal genes indirectly by suppressing axin2 expression induced by β-catenin2 rather than by β-catenin1 in the dorsal region.
Materials and Methods
Fish stocks
Wild-type zebrafish (Danio rerio) (AB) maintenance, breeding, and staging were performed as described previously (5, 22).
Morpholino, CRISPR/Cas9, and mRNA synthesis
The translation-blocking morpholinos (ATG targeted), eaf1-MO1 and eaf2-MO1, and the splicing morpholinos, eaf1-MO3 and eaf2-MO3, have been described previously (5, 22). β-catenin1 and β-catenin2 antisense morpholinos have also been described previously (4, 5, 25). All morpholinos are purchased from Gene Tools, LLC (Philomath, Oregon, USA), and their sequences have been reported previously (5) and are shown in Supplemental Table 1. CRISPR/Cas9 genome editing system is reported as an effective tool for gene editing in organisms (26, 27). Cas9 system was used to constructed F0 fish for editing eaf1 or eaf2, and the sequences of gRNA for eaf1 or eaf2 were reported in our current published paper (24), but only eaf1 mutants were constructed successfully (24). Expression of ved, vent, or vox was tested in embryos from in-crossed eaf1 F1 heterozygous mutants, and the represented embryos with increased expression or normal expression were genotyped by primers in Table S2.
For mRNA preparation, capped mRNAs were synthesized using the AmpliCap SP6 High Yield Message Maker Kit (Epicentre Biotechnologies, Madison, WI, USA). The plasmids for zebrafish eaf1, eaf2, eaf1-N, eaf2-N, EnR-eaf1-full, EnR-eaf2-full, and dntcf mRNA were prepared as described previously (5, 22). Briefly, full-length zebrafish eaf1 and eaf2 were sub-cloned into PSC2-EnR to form fusion protein expression vectors EnR-eaf1-full and EnR-eaf2-full for mRNA synthesis, and exons1-3 of zebrafish eaf1 or eaf2 were sub-cloned into PSP64 vector for synthesizing mRNAs of eaf1-N or eaf2-N. The synthesized mRNAs and morpholinos were diluted into different concentrations and injected into one-cell stage embryos as described previously (5, 22).
Microscopy Observations
Zebrafish embryos at 30 hpf (hours post fertilization) were de-chorioned, and embryos at 5 dpf (days post fertilization) were side-viewed and photographed under light microscope (Leica M205FA, Germany). Morphological embryos from different groups were quantified using Adobe Photoshop CS5 software.
Whole-mount in situ hybridization
Probes for ved, vox, vent, axin2, wnt8a, bmp2, and fgf8 have been described previously (5, 22, 25). The procedure of whole mount in situ hybridization (WISH) was performed as described previously (5, 22, 25).
RNA extraction and semi-quantitative RT-PCR
Total RNA was isolated from 30 whole embryos at indicated developmental stages using TRIZOL reagent (Invitrogen). Oligo-dT-primed cDNA was synthesized using RevertAid First Strand cDNA Synthesis Kit (Fermentas, Burlington, Ontario). The primers used for amplifying zebrafish ved, vox, vent, bmp2b, wnt8a, and 18s (used as an internal control) were listed in Supplemental Table 2. Semi-quantitative RT-PCR (qRT-PCR) was performed as described previously (5, 24, 25). Each sample was run in triplicate and repeated at least three times. Differences were calculated by the △△Ct comparative quantization method using 18s as an internal control, and the data were analyzed with one-way analysis of variance (ANOVA) and post hoc Tukey's test (*** represents P < 0.001; ** represents P < 0.01; * represents P < 0.05) (24, 28, 29).
Plasmid construction
Plasmids of HA-zeaf1, HA-zeaf2, Myc-zeaf1, Myc-zeaf2, HA-zcatenin1 and HA-zcatenin2 have been described previously (5). Plasmids of Flag-zcatenin1, Flag-zcatenin2, Myc-zcatenin1, Myc-zcatenin2, Myc-ztcf, HA-ztcf were constructed using the primers listed in Supplemental Table 3. The axin2 promoter-driven luciferase reporter was a generous gift from Dr. Frank Costantini (Columbia University, New York, USA).
Luciferase reporter assay
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (HyClone). Luciferase reporter assays were performed as described previously (5, 25). The luciferase activities were determined at 24-30 hours post-transfection using the Dual-luciferase Reporter Assay System (Promega). The relative light units were measured using a luminometer (Sirius, Zylux Corporation, Oak Ridge, TN). Data were normalized by pTK-Renilla luciferase and were reported as the mean ± SEM of three independent experiments performed in triplicate. The statistical analysis (paired t-test) was performed using GraphPad Prism 5.
Immunoprecipitation assays
For Co-immunoprecipitation (Co-IP) assays, 293T cells were transfected with the indicated plasmids. After 28-36 hours of transfection, the cells were washed with ice-cold PBS (phosphate-buffered saline) buffer and then lysed with modified radio- immune- precipitation (RIPA) buffer (50 mM Tris, pH 7.5; 1% Triton X-100, 0.5% deoxycholate;10 mM EDTA;150 mM NaCl; 50 mM NaF; 1 mM PMSF; 20 μg/ml Aprotinin; 1 mM Na3VO4) with a protease inhibitor cocktail (Sigma). After incubation on ice for 1 hour, cell lysates were centrifuged for 15 min at 10000 g (4 °C), and supernatants were incubated with HA conjugated-agarose beads (SC-7392AC, Santa Cruz) for 6 h or over-night at 4°C. The immune-precipitates were washed 3 times with RIPA buffer and left with about 50 µL liquid. Immuno-precipitates (IP) and whole cell lysates (WCL) were boiled with SDS sample buffer, separated on SDS-PAGE and transferred to PVDF membrane (Millipore). For detecting the interaction abilities of Eaf1/Eaf2 with β-catenin1/2, cells were transfected with HA-catenin1, Myc-eaf1/2, and gradient Flag-β-catenin2 together, or HA-catenin2, Myc-eaf1/2, and gradient Flag-β-catenin1 together, and incubated with conditioned media from mouse L cells. For detecting the interaction abilities between Tcf and β-catenin1/2, cells were transfected with the indicated combination of plasmids and then incubated with conditional media. Western blot analysis was performed as described previously (5, 24) using the following antibodies: anti-Myc antibody (Santa Cruz) with a 1:2000 dilution, anti-HA antibody (Covance) with a 1:5000 dilution, anti-Flag antibody (Santa Cruz) with a 1:2000 dilution, anti-β-actin antibody (Abcam) with a 1:2000 dilution. The blots were photographed using a FujiFilm LAS4000 mini luminescent image analyzer.
Results
Eaf1 and Eaf2 are maternally expressed factors
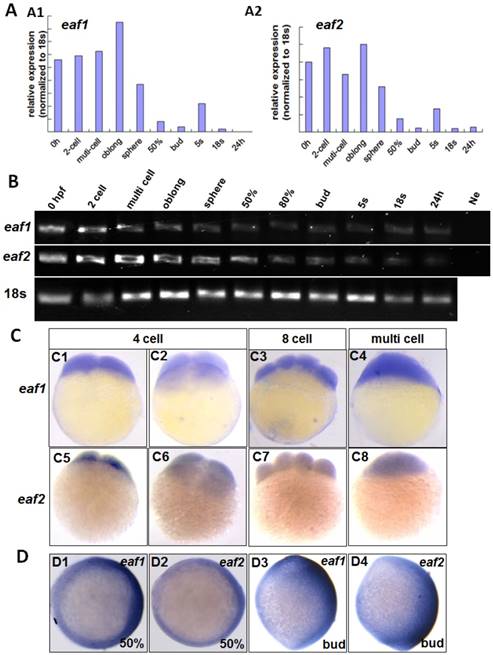
qRT-PCR was used to test eaf1 and eaf2 expression from 0 hpf to 24 hpf (hour post fertilization) in zebrafish embryos. Abundant maternal eaf1 and eaf2 (Figs.1A2 and B) transcripts were observed in embryos at 0 hpf, with their expression at high levels from 0 hpf to sphere stage, sharply reduced from 50% epiboly stage, and at relatively low levels at 24 hpf in embryos (Fig.1).
The distribution of eaf1 and eaf2 in embryos at cleavage stages was also detected by WISH. Maternal and ubiquitous distribution of eaf1 was observed in embryos at 4 cell, 8 cell, and multi-cell stages (Figs.1C1-C4), similar to its ubiquitous distribution in embryos at early blastula stage (5). Additionally, abundant eaf2 transcripts were also uniformly distributed in each cell of embryos at 4 cell to multi-cell stage (Figs.1C5-C8), but their slightly different spatial distribution was observed in the dorsal region of embryos at gastrula stage (Figs.1D1 and D2) and tail bud stage (Figs.1D3 and D4).
Eaf1 and Eaf2 suppress the initiation of early ventral zygotic genes
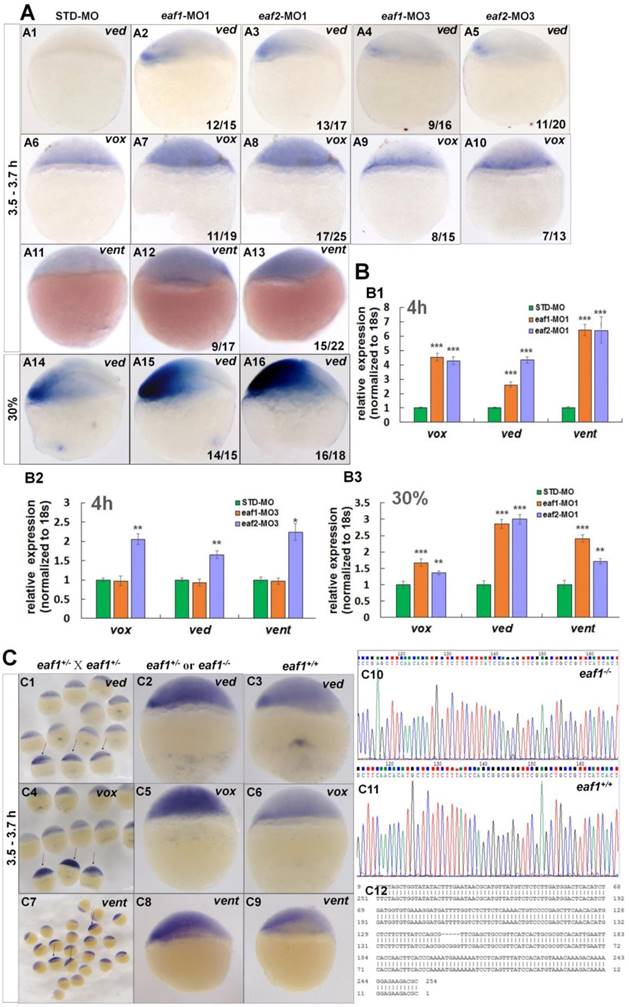
Expression of early zygotic ventral genes ved, vox, and vent was tested in eaf1 or eaf2 loss-of-function embryos first. Transcripts for ventral genes ved (Figs.2A1-A3, the number 12/15 in the right hand corner in A2 indicated that 12 embryos exhibited increased ved expression in a total of 15 tested eaf1-MO1 morphants, and similar indicating numbers were used in the following panels), vox (Figs.2A6-A8) and vent (Figs.2A11-A13) were enhanced in embryos injected with either zebrafish eaf1 (11/19 and 9/17 respectively) or eaf2 (17/25 and 15/22 respectively) ATG morpholinos (eaf1-MO1 and eaf2-MO1, respectively) at the oblong stage (3.5 hpf to 3.7 hpf). Additionally, increased expression of ved was also observed in eaf1/2 ATG morphants at 30% epiboly stage (Figs.2A14-A16) (14/15 and 16/18 respectively). To detect whether zygotic eaf1 or eaf2 patterned the dorsal-ventral cell fates, we targeted eaf1 or eaf2 with splice-blocking MO (eaf1-MO3 and eaf2-MO3, respectively) (5, 22, 24). Consistently, the splicing morpholinos induced slightly increased expression of ventral cell markers (Figs.2A4, A5, A9, and A10). Expression of ventral genes ved (Figs.2A4 and A5) and vox (Figs.2A9 and A10) was slightly up-regulated in embryos injected with eaf1-MO3 (9/16 and 8/15 respectively) or eaf2-MO3 (11/20 and 7/13) at oblong stage.
Expression of the ventral genes in eaf1/2 ATG or in eaf1/2 splice morphants was further detected by qRT-PCR. Transcripts of vox, ved, and vent were increased significantly in eaf1/2 ATG morphants (Fig.2B1), with 4.5-fold, 2.6-fold, and 6.4-fold up-regulation in eaf1 ATG morphants and 4.3-fold, 4.4-fold, and 6.4-fold up-regulation in eaf2 ATG morphants at 4 hpf (Fig.2B1). Their expression was found almost normal in embryos injected with eaf1 splice MOs (Fig.2B2), and was up-regulated by 2-fold, 1.7-fold, and 2.2-fold in embryos injected with eaf2 splicing MOs (Fig.2B2). Moreover, significantly increased expression of genes vox, ved, and vent was also observed in eaf1/2 ATG morphants at 30% epiboly stage (Fig.2B3).
Maternal expression of eaf1 and eaf2 was detected by qRT-PCR (A), semi quantitative PCR (B), and whole mount in situ hybridization (C). (A, B) Embryos from 0 hpf, 2-cell stage (2-cell), multi-cell stage, oblong stage, sphere stage, 50% epiboly stage (50%), 80% epiboly stage (80%), bud stage, 5 somite stage (5s), 18 somite stage (18s), and from 24 hours post fertilization (24 h) were used for qRT-PCR (A) and semi quantitative PCR (B). Ne: negative control. (C) Maternal and uniform distributions of eaf1 and eaf2 in embryos at 4 cell stage, 8-cell stage and multi-cell stage were revealed by WISH. (D) Spatial distribution of eaf1 or eaf2 in embryos at gastrula stage (D1, D2) and at bud stage (D3, D4), and more eaf1 or eaf2 was distributed in the dorsal region of embryos. C1, C3, C4, C5, C7, C8, D3, D4, lateral view, anterior to the up; C2, C6, D1, D2, animal view, dorsal to the right for D1 and D2.
Eaf1 mutants were constructed using CRISPR/gRNA technology, and the sequences of gRNAs for eaf1 were reported in our current study (24). Embryos from in-crossed eaf1 F1 heterozygous mutants (24) all exhibited wild type likeness phenotypes at both 30 hpf and 5pdf, however, they were eaf1 homozygous mutants, heterozygous, or wild type after genotyping (we have genotyped 10 embryos from in-crossed eaf1 F1 heterozygous mutants at 30 hpf and 5dpf respectively) (Fig.S1). Expression of ved, vox, and vent was tested in aforementioned embryos at both sphere stage (Fig.2C) and 30% epiboly stage (Fig.S2), and the embryos with increased expression of ved (Figs.2C1, C2, and Fig.S2), vox (Figs.2C4 and C5), or vent (Figs.2C7 and C8) were genotyped, and almost of them were homozygous mutants (5 bp deletion, indicated in Fig.2C12) and heterozygous (Figs.2C10 and C12) (we have genotyped nearly 50 embryos with increased ved expression, and the ratio of homozygous and heterozygous was nearly 1:2). However, embryos with normal expression of ved (Fig.2C3), vox (Fig.2C6), and vent (Fig.2C9) were all wild type (Fig.2C11). We also injected eaf1-MO1 to embryos from in-crossed eaf1 F1 heterozygous mutants, 52 embryos exhibited wild type similar phenotype in totally injected 100 in-crossed embryos. 15 injected larvae with wild type phenotype at 4 pdf were genotype, all are heterozygous or homozygous eaf1 mutants.
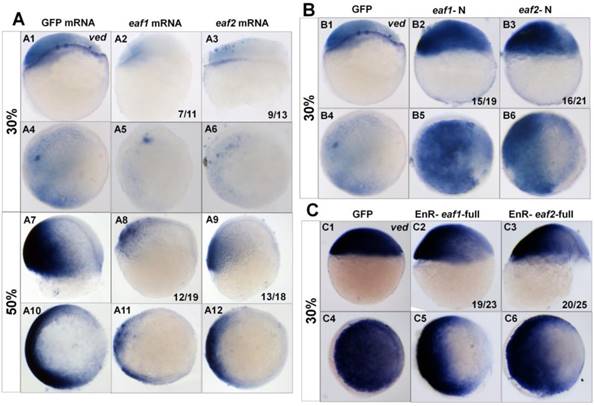
Additionally, we tested the transcripts of the ventral zygotic genes in embryos with ectopic expression of different forms of eaf mRNA using ved as an indicator. Transcripts of ved in embryos with ectopic expression of eaf1 or eaf2 were reduced at both 30% epiboly (7/11 and 9/13 respectively) (Figs.3A1-A6) and 50% epiboly stage (12/19 and 13/18 respectively) (Figs.3A7-A12). The expression of ved was further tested in embryos with ectopic expression of eaf-N mRNA, a dominant negative form of the full-length eaf1/2 (5, 24). In embryos with ectopic expression of eaf1-N or eaf2-N at blastula stage, ventral ved (Figs.3B1-B6) exhibited enhanced expression (15/19 and 16/21 respectively). When tested in embryos injected with EnR-eaf1/2-full, the ved expression was obviously down-regulated in embryos injected with EnR-eaf1-full (19/23) (Figs.3C2 and C5) or EnR-eaf2-full (20/25) (Figs.3C3 and C6).
Expressions of signaling ligands in embryos with gain-of-function or loss-of-function of Eaf
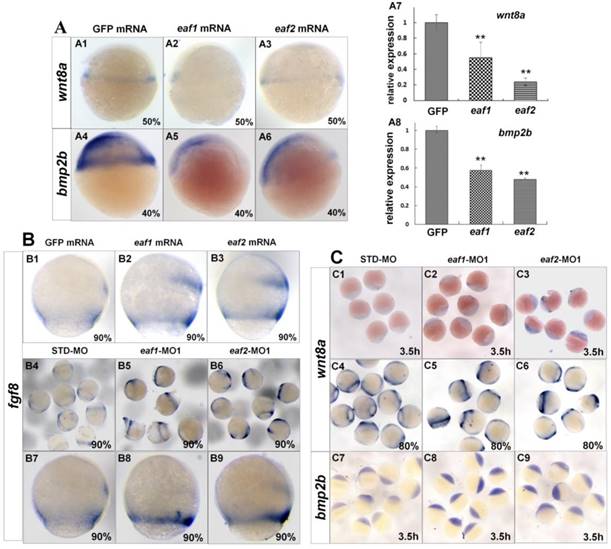
Several ligands and signaling pathways are reported to regulate dorsal-ventral patterning during early zebrafish development, such as Wnt signaling, Fgf signaling, and Bmp signaling (30, 31). Therefore, we further detected the expression of those ligands in eaf1/2 loss-of-function and eaf1/2 gain-of-function embryos. Wnt8a exhibited obviously reduced expression in embryos injected with eaf1 or eaf2 mRNA as detected by in situ and qRT-PCR (Figs.4A1-A3, A7). Similarly, bmp2b expression was also reduced obviously in embryos with ectopic expression of eaf1 or eaf2 (Figs.4A4-A6, A8). However, the expression of fgf8 was increased in embryos with ectopic expression of eaf1 or eaf2 (Figs.4B1-B3), and its increased expression was observed in eaf1 or eaf2 morphants (Figs.4B4-B9). Additionally, wnt8a exhibited increased expression in eaf1 and eaf2 morphants at both blastula and gastrula stages (Figs.4C1-C6), and bmp2b was also up-regulated in eaf1/2 morphants (Figs.4C7-C9), opposite to its expression in embryos with ectopic expression of eaf1 or eaf2.
Eaf1 and Eaf2 suppress β-Catenin2-induced axin2 expression
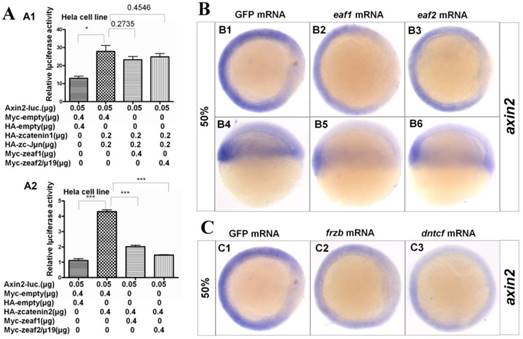
Axin2, a target and negative regulator of Wnt/β-catenin signaling (14-16), was also reported to pattern the axis during early development (10). Thus, whether eaf1 or eaf2 could modulate axin2 expression was further investigated in this study. Both β-catenin1 and β-catenin2 activated the axin2 promoter in Hela cells, but with higher inductive activity for β-catenin2 (Fig.5A2). Meanwhile, eaf1 and eaf2 only suppressed axin2 promoter activities induced by β-catenin2 (Fig.5A2) but not by β-catenin1 (Fig.5A1). Additionally, ectopic expression of eaf1 and eaf2 strongly abolished axin2 expression in the dorsal domain, but showed less suppressive effect on its expression in the ventral region of embryos (Figs.5B1-B6). Axin2 expression was also obviously abolished by Wnt antagonists frzb and dntcf in both dorsal and ventral domains (Figs.5C1-C3), and embryos injected with frzb or dntcf exhibited obvious developmental defects (Fig.S3). We also tested the binding abilities of zebrafish Eaf1/2 proteins to zebrafish β-Catenin1 and β-Catenin2 proteins. It was revealed that Catenin1 bound more Catenin2 but not Eaf with the increased transfected Catenin2 protein in the co-transfected system (Fig.S4). In addition, it was revealed that Tcf bound more β-Catenin1 rather than β-Catenin2 by Co-IP assays in this study (data not shown).
Eaf1 and Eaf2 suppress specification of ventral cells through maternal Wnt/β-catenin1/Tcf signaling
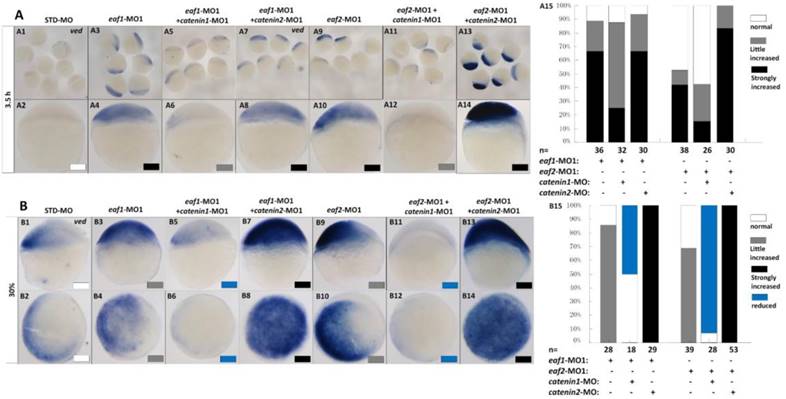
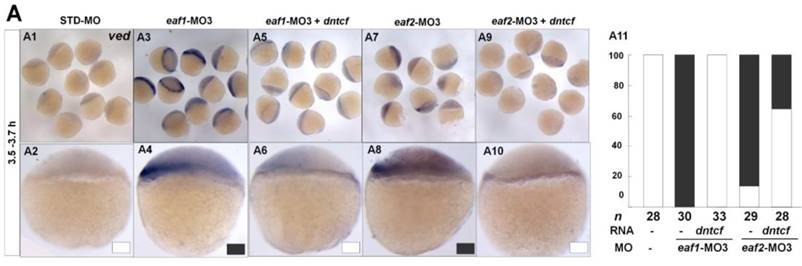
It has been reported that vent family genes are direct targets of Wnt8/β-catenin signaling and mediate the repression activities of wnt8 on the vertebrate organizer (11). Whether β-catenin-MO could recover enhanced expression of ventral genes to normal level in eaf1 or eaf2 blastula morphants was tested by combined injection of β-catenin1-MO or β-catenin2-MO into eaf1 or eaf2 morphants and using the ventral gene ved as an indicator. In the current study, it was β-catenin1-MO (Figs.6A5, A6, A11, A12 and A15) rather than β-catenin2-MO (Figs.6A7, A8, A13, A14 and A15) that could successfully rescue ved expression at oblong stage (3.5 hpf) in eaf1 or eaf2 morphants, suggesting that eaf1/2 might suppress ved expression via β-catenin1 rather than β-catenin2. At 30% epiboly stage, β-catenin1-MO nearly abolished the increased ved expression in embryos injected with eaf-ATG morpholinos (Figs.6B5, B6, B11, B12 and B15). While β-catenin2-MO enhanced ved expression in eaf morphants, the increased expression of ved was more apparent in embryos injected with eaf-MO and β-catenin2-MO together at 30% epiboly (Figs.6B7, B8, B13, B14 and B15). Additionally, dntcf could also recover the increased expression of ventral mesoderm genes to nearly normal level in embryos injected with eaf-splice-blocking morpholinos at sphere stage (Fig.7).
Eaf1 and eaf2 inhibited the expression of early ventral vent family genes at blastula stage. (A) Initial expression of ved (A1-A3), vox (A6-A8) and vent (A11-A13) was increased obviously in embryos injected with eaf-ATG-MO [eaf1-MO1 (8 ng/embryo) and eaf2-MO1 (8 ng/embryo)] or with eaf-splicing-MO (A4, A5, A9, A10) [eaf1-MO3 (8 ng/embryo) and eaf2-MO3 (8 ng/embryo)] at 3.5-3.7 h, and increased expression of ved was also observed in eaf1/2 ATG morphants at 30% epiboly stage (A14-A16). (B) The significantly increased expression of the ventral genes in ATG morphants at 4 h (B1) and at 30% epiboly stage (B3) was revealed by qRT-PCR, and their slightly increased expressionin eaf2 splice morphants but not in eaf1 splice morphants was revealed by qRT-PCR (B2). (C) Expression of ventral ved (C1-C3), vox (C4-C6), and vent (C7-C9) in embryos from in-crossed eaf1 F1 heterozygous mutants, and black arrows indicated the embryos with increased expression of ved (C1), vox (C4), and vent (C7). The genotypic results for embryos with increased expression (C2, C5, C8) or normal expression (C3, C6, C9) were indicted in C10, C11, and C12 respectively. eaf1-MO1: eaf1 ATG morpholino; eaf2-MO1: eaf2 ATG morpholino; eaf1-MO3: eaf1 splicing morpholino; eaf2-MO3: eaf2 splicing morpholino. A1-A16, C1-C9, lateral view, dorsal to the right.
Expression of ved in embryos with ectopic expressions of different forms of eaf1 or eaf2. (A1-A12) Expression of ved was obviously reduced in embryos with ectopic expression of eaf1 or eaf2 at 30% epiboly stage (A1-A6) and at 50% epiboly stage (A7-A12). (B1-B6) Ved expression was obviously increased in embryos injected with eaf1/2-N, dominant forms of eaf1/2. (C1-C6) Expression of ved was obviously reduced in embryos injected with EnR-eaf1/2-full. A1-A3, A7-A9, B1-B3, C1-C3, lateral view, dorsal to the right; A4-A6, A10-A12, B4-B6, C4-C6, animal view, dorsal to the right.
Expression of ligands wnt8a, bmp2b, and fgf8 in eaf1 or eaf2 gain-of-function or loss-of-function embryos. (A) wnt8a (A1-A3, A7) and bmp2b (A4-A6, A8) exhibited reduced expression in eaf1 or eaf2 gain-of-function embryos as detected by in situ and qRT-PCR. (B) Expression of fgf8 was increased in both eaf1/2 gain-of-function (B1-B3) and loss-of-function embryos (B4-B9). (C) Expression of wnt8a (C1-C6) and bmp2b (C7-C9) was increased in eaf1/2 gain-of-function embryos. A1-A6, B1-B9, C1-C9, lateral view.
Discussion
Eaf1 and eaf2 are novel maternal suppressors of ventral cell fates
The vent family genes pattern the dorsal-ventral axis during embryogenesis by acting as the ventralizing transcriptional repressors (11, 32, 33). Maternal runx2, the only identified maternal regulator, initiates expressions of zygotic vox, vent, and ved at the onset of zygotic transcription (9). In this study, we unexpectedly find that ventral marker genes ved, vent, and vox exhibit enhanced expression in both eaf morphants, eaf homozygous or heterozygous mutant embryos, and embryos injected with dominant-negative forms of zebrafish eaf, eaf1-N or eaf2-N. However, their expression is almost abolished in embryos injected with mRNA of eaf1, eaf2, and eaf fused with EnR. All these observations suggest that eaf1/2 act as repressors in specification of ventral cells, while exons1-3 of eaf1 and eaf2 act as dominant negative isoforms in dorsal-ventral patterning, similar to their roles in forebrain specification (5).
Eaf1 and eaf2 obviously suppressed axin2 expression. (A) Eaf1 and eaf2 suppressed axin2 promoter activity induced by β-catenin2 (A2) but not by β-catenin1 (A1). (B) Expression of axin2 was obviously abolished in the dorsal domain of eaf1/2 gain-of-function embryos. (C) Expression of axin2 was obviously abolished in both dorsal and ventral regions of embryos injected with Wnt antagonist frzb or dntcf. B1-B3, C1-C3, animal view, dorsal to the right; B4-B6, lateral view, dorsal to the right.
However, eaf1 mutants at 30 hpf and 5 dpf exhibited wild type similar phenotypes. The data here is similar as some reports that embryos morphants exhibit more severe defective phenotypes compared with their mutants (34), and the data here also suggests that the genetic compensation might occur in eaf1 mutants as previous study reported (35). Additionally, it is reported that TGF-beta factor Fascin1 mutants exhibit wild type similar phenotype at later developmental stage but reduced expression of mesoderm and endoderm marker genes at gastrula stage although its morphants exhibit defective phenotypes at all developmental stages (36). Thus, we speculate that genetic compensation might contribute to no obviously defective phenotypes observed in eaf1 homozygous mutants in this study. Certainly, it is possible that other unrevealed causes occurred in eaf1 mutants or in morphants, which might also contribute to the data that eaf1 mutants could not totally phenocopy the eaf1 morphants in this study. Moreover, lower percentage of defective phenotype occurred in group of embryos from in-crossed eaf1 F1 heterozygous injected with eaf1 morpholino than group of wild type embryos injected with eaf1 morpholino (21), and the genotypes of wild type similar embryos were all heterozygous or homozygous (data not shown). The data also suggest that genetic compensation might occur in eaf1 homozygous or heterozygous mutants.
Uniform distribution of eaf1 and eaf2 is observed in embryonic cells at early cleavage stages including 4 cell, 8 cell stage, etc., and their high expression is observed in embryos from 0 hpf to sphere stage (Fig.1). Additionally, expression of ventral vent family genes is significantly up-regulated in eaf1/2 ATG morphants, but slightly increased or normal in eaf1/2 splice morphants (Fig.2B), suggesting that eaf1 and eaf2 might function as maternal repressors of ventral ved, vent, and vox at the onset of zygotic transcription. Our data implies that besides runx2, eaf1 and eaf2 might be potential maternal determinants that regulate ventral cell fate specification. However, unlike maternal runx2 (9), eaf1 and eaf2 suppress, but do not induce the expression of ventral genes, and eaf1 and eaf2 also suppress the expression of dorsal genes at the onset of zygotic transcription (5). These differences suggest that eaf1 and eaf2 might act in a separate pathway to suppress the initiation of both ventral and dorsal genes.
Reduced expression of wnt8a and bmp2b is observed in embryos with ectopic expression of eaf1 and eaf2, and their increased expression is observed in eaf morphants in this study. Wnt8a is not only a ligand of Wnt signaling but also acts as a ventral-lateral marker during zebrafish embryogenesis to indicate the ventral mesoderm pattern (37). BMP family genes have also been reported as important for ventral genes (38, 39). Our data implicates that eaf1 and eaf2 might be general repressors for ventral cells in dorsal-ventral axis patterning. Fgf8 signaling is revealed to act in inducing ventral and posterior cells in zebrafish (31). In this study, the expression of fgf8 is found increased in both eaf1/2 morphants and embryos with ectopic expression of eaf1/2, suggesting that fgf8 might act parallel to eaf1/2 in regulating specification of ventral cells and is required for neutralizing the dorsal-ventral defects in both eaf1/2 morphants and embryos with ectopic expression of eaf1/2.
Eaf1 and eaf2 suppress the initiation of vent family genes via Wnt/β-catenin1/Tcf signaling
In this study, expanded expression of ved, vox, and vent is revealed in eaf morphants at blastula stage (Fig.2), and reduced expression of ved is observed in embryos with ectopic expression of eaf1 and eaf2 at blastula stage (Fig.3). Moreover, β-catenin1 knockdown or ectopic expression of dntcf is revealed to restore ved expression in eaf morphants at blastula stage in this study (Figs.6,7), suggesting that eaf1 and eaf2 modulate ventral ved expression through antagonizing Wnt/β-catenin1/Tcf signaling. Previous studies have shown that maternal XTcf3 functions as a repressor to prevent both dorsal and ventral cells from expressing dorsal genes (40, 41), and the vent family genes harbor β-catenin/Tcf binding sites in their promoters (42). Our data here suggest that tcf and β-catenin1 might suppress dorsal fates by inducing expressions of ventral vent genes. In the eaf morphants with combined injections of dntcf or β-catenin1-MO, almost no expression of ved was observed in several embryos (Figs.6,7), implying that tcf and β-catenin1 might act downstream of eaf1 and eaf2 and are essential for the induction of ventral vent family genes. Although we have tested the binding abilities of Eaf1/2 with Catenin1/2 by competitive Co-IP assays in this study, but it is revealed Catenin1 could bind more Catenin2 but not Eaf with the increased Catenin2 protein in the co-transfected system. However, a reasonable answer should be given for the Co-IP data in the further days.
Despite dorsal enrichment of endogenous β-catenin in embryos at blastula stage for the initiation of organizer genes, there is a certain amount of β-catenin present in the ventral region (43). We speculate that the ventrally active β-catenin protein might be β-catenin1, and its activity is suppressed by eaf1 and eaf2. When eaf1 or eaf2 is removed by morpholino-mediated-knockdown, transcriptional activity of β-catenin1 is sufficient to induce expression of the ventral genes, leading to the observation of enhanced expression of ved, vox, and vent in the eaf1 or eaf2 morphants at the blastula stage (Figs.2,6,7 and S2). Additionally, we observe the reduced transcripts of ventral gene ved in embryos with ectopic expression of eaf1 or eaf2 at the blastula stage. These observations in this study are consistent with previous studies in that Wnt/β-catenin target genes are abolished in blastula stage embryos with ectopic expression of Wnt inhibitors (3, 13).
This study reveals that Wnt/β-catenin1/Tcf signaling might initiate early zygotic expression of vent genes at the blastula stage. This finding compliments the role of Wnt/β-catenin2 signaling in the initiation of early zygotic expressions of dorsal genes (4, 5), and the induction activity of this signaling pathway in dorsal-ventral patterning is inhibited by eaf1 and eaf2 (Figs.6,7) (5). However, more studies need to be done to verify the finding in the present study, such as detecting active catenin proteins using catenin splicing morpholinos and detecting the distribution of nuclear catenin proteins in cells of eaf1/2 morphants at the initiation of dorsal-ventral patterning using specific antibodies for β-Catenin1 or for β-Catenin2.
This model could also be used to explain why ved expression is more obviously enhanced when β-catenin2-MO is co-injected into eaf1 or eaf2 morphants. β-Catenin2 is essential for the expression of dorsal genes (4, 5), and co-injection of β-catenin2-MO into eaf1 or eaf2 morphants results in significantly decreased expression of dorsal genes (5). Thus, the reduction of dorsal antagonistic activities might lead to highly increased expression of ventral ved in eaf morphants co-injected with β-catenin2-MO (Figs. 6,7).
Eaf1 and eaf2 suppress β-catenin2-induced axin2 expression
In eaf morphants, β-catenin1-MO efficiently rescues expression of ventral Wnt/β-catenin target genes (Figs.6,7), whereas β-catenin2-MO rescues expression of dorsal Wnt/β-catenin target genes (5). Our data suggest that β-catenin1 and β-catenin2 might differentially regulate Wnt/β-catenin target genes, depending on the spatial distributions of those targets. Promoter assays data indicates that β-catenin2 activates the axin2 promoter more effectively than β-catenin1 and that eaf1 and eaf2 can selectively suppress axin2 expression induced by β-catenin2 rather than by β-catenin1 (Fig.5). Additionally, more transcripts of eaf1 and eaf2 are observed on the dorsal side of wild-type embryos (Fig.1D), with axin2 expression almost abolished in the dorsal domain of the embryos with ectopic expression of eaf1 or eaf2 (Fig.5) and the dorsal organizer genes significantly expanded as detected by in situ and RT-PCR (5). All of our observations suggest that eaf1 and eaf2 might enhance the expressions of dorsal organizer genes indirectly by suppressing β-catenin2-induced axin2 expression in the dorsal domain.
A percentage of embryos have been reported to display ventralization defects, but most of them exhibit anterior/dorsal defects after ectopic expression of negative regulators of Wnt signaling, such as maternal Wnt negative regulators (25, 44-46). However, in our previous studies, almost all embryos injected with eaf1 or eaf2 mRNA exhibit anterior/ dorsal defects (5), which we speculate might be attributed to the distribution of more eaf1 and eaf2 in the dorsal region of embryos, possibly leading to their inhibition on β-catenin2 induced axin2 expression. Slight changes in axin2 expression can result in increased nuclear localization of β-catenin (10), thus, a slight spatial distribution and more dorsal distribution of eaf1 and eaf2 in the dorsal region of embryos also contributes to the almost all anterior/dorsal defects in embryos with ectopic expression of eaf1 or eaf2 in the present study.
Eaf1 or eaf2 regulated expression of ventral ved gene via Wnt/β-catenin1 signaling. (A, B) β-catenin1-MO (8 ng/embryo) rather than β-catenin2-MO (8 ng/embryo) could recover enhanced ved expression in eaf1 or eaf2 morphants to normal at both oblong stage (A) and 30% epiboly stage (B). Embryos were injected with STD-MO (A1, A2, B1, B2), eaf1-MO1 (A3, A4, B3, B4) or eaf2-MO1 (A9, A10, B9, B10). Embryos were injected with either a combination of eaf1-MO1 and β-catenin1-MO (A5, A6, B5, B6), eaf1-MO1 and β-catenin2-MO (A7, A8, B7, B8), eaf2-MO1 and β-catenin1-MO (A11, A12, B11, B12), or eaf2-MO1 and β-catenin2-MO (A13, A14, B13, B14). (A15, B15) The percentage of embryos exhibiting different expression level of ved was scored at the sphere stage (A15) and at 30% epiboly stage (B15). White box, normal; gray box, slightly increased; black box, strongly increased; blue box, reduced. All of the injections, including MO alone or eaf-MO combined with catenin-MO, were performed using the same batch of embryos produced by a select number of zebrafish to eliminate error caused by embryo variation. A1-A14, B1, B3, B5, B7, B9, B11, B13, lateral view, dorsal to the right; B2, B4, B6, B8, B10, B12, B14, animal view, dorsal to the right.
dntcf could effectively rescue enhanced ved expression to normal level in eaf1 or eaf2 morphants. White box, normal; black box, increased. Embryos were injected with STD-MO (A1, A2), eaf1-MO3 (A3, A4) or eaf2-MO3 (A7, A8). Embryos were injected with eaf1-MO3 and dntcf mRNA (10 pg) (A5, A6) or with eaf2-MO3 and dntcf mRNA (10 pg) (A9, A10). All of the injections, including MO alone or MO combined with mRNA, were performed using the same batch of embryos produced by a select number of zebrafish to eliminate error caused by embryo variation. A1-A10, lateral view, dorsal to the right.
Our study provided a novel dorsal-ventral patterning pathway: eaf1 and eaf2 suppress both maternal Wnt/β-catenin1 signaling and Wnt/β-catenin2 signaling and mediate the zebrafish dorsal-ventral axis patterning by inhibiting the initiation and specification of both ventral and dorsal genes via differential regulation of distinct targets of β-catenin1 and β-catenin2. However, it still remains to be elucidated why β-catenin1 and β-catenin2 differentially regulate their targets depending on the dorsal or ventral distribution of those genes due to their similar and ubiquitous distribution in early stage embryos (4). A possible explanation for their differential functions is that the two catenin proteins bind to different Tcf/Lef partners. It has been reported that different Tcf/Lef genes in Xenopus mediate specific responses to Wnt/β-catenin signaling during mesoderm development (40). In this study, we observed that Tcf3 could bind more β-Catenin1 rather than β-Catenin2 in the competitive Co-IP assays. The data is consistent with the observations that catenin1-MO and dntcf could rescue the increased expression of vent family genes in eaf morphants.
In summary, we demonstrate the maternal expressions of eaf1 and eaf2 as well as their differential and spatial distribution during zebrafish embryogenesis. We also uncovere the novel roles of the two factors in suppressing expressions of ventral genes via Wnt/β-catenin1/Tcf signaling. In the future, more research efforts should be focused on the roles of eaf1/2 in cancer and during embryogenesis as well as their molecular regulation network in cells.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We are grateful to Dr. Frank Costantini for his generous gift of reagents. J.X. L. is financially supported by an innovation project of the Chinese Academy of Science (KSCX2-EW-Q-12), Fundamental Research Funds for the Central Universities (Program No.2662015PY002), and Agricultural University Scientific & Technological Self-Innovation Foundation (42000481-7-2014RC007).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Weaver C, Kimelman D. Move it or lose it: axis specification in Xenopus. Development. 2004;131:3491-3499
2. Kelly C, Chin AJ, Leatherman JL, Kozlowski DJ, Weinberg ES. Maternally controlled (beta)-catenin-mediated signaling is required for organizer formation in the zebrafish. Development. 2000;127:3899-3911
3. Shimizu T, Yamanaka Y, Ryu SL, Hashimoto H, Yabe T, Hirata T, Bae YK, Hibi M, Hirano T. Cooperative roles of Bozozok/Dharma and Nodal-related proteins in the formation of the dorsal organizer in zebrafish. Mech Dev. 2000;91:293-303
4. Bellipanni G, Varga M, Maegawa S, Imai Y, Kelly C, Myers AP, Chu F, Talbot WS, Weinberg ES. Essential and opposing roles of zebrafish beta-catenins in the formation of dorsal axial structures and neurectoderm. Development. 2006;133:1299-1309
5. Liu JX, Zhang D, Xie X, Ouyang G, Liu X, Sun Y, Xiao W. Eaf1 and Eaf2 negatively regulate canonical Wnt/beta-catenin signaling. Development. 2013;140:1067-1078
6. Reim G, Brand M. Maternal control of vertebrate dorsoventral axis formation and epiboly by the POU domain protein Spg/Pou2/Oct4. Development. 2006;133:2757-2770
7. Bauer H, Lele Z, Rauch GJ, Geisler R, Hammerschmidt M. The type I serine/threonine kinase receptor Alk8/Lost-a-fin is required for Bmp2b/7 signal transduction during dorsoventral patterning of the zebrafish embryo. Development. 2001;128:849-858
8. Saneyoshi T, Kume S, Amasaki Y, Mikoshiba K. The Wnt/calcium pathway activates NF-AT and promotes ventral cell fate in Xenopus embryos. Nature. 2002;417:295-299
9. Flores MV, Lam EY, Crosier KE, Crosier PS. Osteogenic transcription factor Runx2 is a maternal determinant of dorsoventral patterning in zebrafish. Nat Cell Biol. 2008;10:346-352
10. Kofron M, Klein P, Zhang F, Houston DW, Schaible K, Wylie C, Heasman J. The role of maternal axin in patterning the Xenopus embryo. Dev Biol. 2001;237:183-201
11. Ramel MC, Lekven AC. Repression of the vertebrate organizer by Wnt8 is mediated by Vent and Vox. Development. 2001;131:3991-4000
12. Lekven AC, Thorpe CJ, Waxman JS, Moon RT. Zebrafish wnt8 encodes two wnt8 proteins on a bicistronic transcript and is required for mesoderm and neurectoderm patterning. Dev Cell. 2001;1:103-114
13. Pelegri F, Maischein HM. Function of zebrafish beta-catenin and TCF-3 in dorsoventral patterning. Mech Dev. 1998;77:63-74
14. Hadjihannas MV, Bruckner M, Behrens J. Conductin/axin2 and Wnt signalling regulates centrosome cohesion. EMBO Rep. 2010;11:317-324
15. Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172-1183
16. Behrens J, Jerchow BA, Wurtele M, Grimm J, Asbrand C, Writz R, Kuhl M, Wedlich D, Birchmeier W. Functional interaction of an axin homolog, conductin, with beta-catenin, APC, and GSK3beta. Science. 1998;280:596-599
17. Simone F, Luo RT, Polak PE, Kaberlein JJ, Thirman MJ. ELL-associated factor 2 (EAF2), a functional homolog of EAF1 with alternative ELL binding properties. Blood. 2003;101:2355-2362
18. Simone F, Polak PE, Kaberlein JJ, Luo RT, Levitan DA, Thirman MJ. EAF1, a novel ELL-associated factor that is delocalized by expression of the MLL-ELL fusion protein. Blood. 2001;98:201-209
19. Xiao W, Zhang Q, Jiang F, Pins M, Kozlowski JM, Wang Z. Suppression of prostate tumor growth by U19, a novel testosterone-regulated apoptosis inducer. Cancer Res. 2003;63:4698-4704
20. Xiao W, Zhang Q, Habermacher G, Yang X, Zhang AY, Cai X, Hahn J, Liu J, Pins M, Doglio L, Dhir R, Gingrich J, Wang Z. U19/Eaf2 knockout causes lung adenocarcinoma, B-cell lymphoma, hepatocellular carcinoma and prostatic intraepithelial neoplasia. Oncogene. 2008;27:1536-1544
21. Xiao W, Ai J, Habermacher G, Volpert O, Yang X, Zhang AY, Hahn J, Cai X, Wang Z. U19/Eaf2 binds to and stabilizes von hippel-lindau protein. Cancer Res. 2009;69:2599-2606
22. Liu JX, Hu B, Wang Y, Gui JF, Xiao W. Zebrafish eaf1 and eaf2/u19 mediate effective convergence and extension movements through the maintenance of wnt11 and wnt5 expression. J Biol Chem. 2009;284:16679-16692
23. Wan X, Ji W, Mei X, Zhou J, Liu JX, Fang C, Xiao W. Negative feedback regulation of Wnt4 signaling by EAF1 and EAF2/U19. PLoS One. 2010;5:e9118
24. Liu JX, Xu QH, Li S, Yu X, Liu W, Ouyang G, Zhang T, Chen LL. Transcriptional factors Eaf1/2 inhibit endoderm and mesoderm formation via suppressing TGF-beta signaling. Bba-Gene Regul Mech. 2017;1860:1103-1116
25. Xie XW, Liu JX, Hu B, Xiao W. Zebrafish foxo3b negatively regulates canonical Wnt signaling to affect early embryogenesis. PLoS One. 2011;6:e24469
26. Rosenbluh J, Xu H, Harrington W, Gill S, Wang X, Vazquez F, Root DE, Tsherniak A, Hahn WC. Complementary information derived from CRISPR Cas9 mediated gene deletion and suppression. Nature communications. 2017;8:15403
27. Varshney GK, Carrington B, Pei W, Bishop K, Chen Z, Fan C, Xu L, Jones M, LaFave MC, Ledin J, Sood R, Burgess SM. A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nature protocols. 2016;11:2357-2375
28. Zhang T, Xu L, Wu JJ, Wang WM, Mei J, Ma XF, Liu JX. Transcriptional Responses and Mechanisms of Copper-Induced Dysfunctional Locomotor Behavior in Zebrafish Embryos. Toxicological sciences. 2015;148:299-310
29. Cui B, Ren L, Xu QH, Yin LY, Zhou XY, Liu JX. Silver_ nanoparticles inhibited erythrogenesis during zebrafish embryogenesis. Aquatic toxicology. 2016;177:295-305
30. Furthauer M, Van Celst J, Thisse C, Thisse B. Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development. 2004;131:2853-2864
31. Schier AF, Talbot WS. Molecular genetics of axis formation in zebrafish. Annu Rev Genet. 2005;39:561-613
32. Onichtchouk D, Glinka A, Niehrs C. Requirement for Xvent-1 and Xvent-2 gene function in dorsoventral patterning of Xenopus mesoderm. Development. 1998;125:1447-1456
33. Imai Y, Gates MA, Melby AE, Kimelman D, Schier AF, Talbot WS. The homeobox genes vox and vent are redundant repressors of dorsal fates in zebrafish. Development. 2001;128:2407-2420
34. Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I. et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32:97-108
35. Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230-233
36. Liu Z, Ning G, Xu R, Cao Y, Meng A, Wang Q. Fscn1 is required for the traficking of TGF-beta fanily type 1 receptors during endoderm formation. Nature communication. 2016;7:12603
37. Wilm TP, Solnica-Krezel L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development. 2005;132:393-404
38. Dick A, Hild M, Bauer H, Imai Y, Maifeld H, Schier AF, Talbot WS, Bouwmeester T, Hammerschmidt M. Essential role of Bmp7 (snailhouse) and its prodomain in dorsoventral patterning of the zebrafish embryo. Development. 2000;127:343-354
39. Dosch R, Gawantka V, Delius H, Blumenstock C, Niehrs C. Bmp-4 acts as a morphogen in dorsoventral mesoderm patterning in Xenopus. Development. 1997;124:2325-2334
40. Liu F, van den Broek O, Destree O, Hoppler S. Distinct roles for Xenopus Tcf/Lef genes in mediating specific responses to Wnt/beta-catenin signalling in mesoderm development. Development. 2005;132:5375-5385
41. Houston DW, Kofron M, Resnik E, Langland R, Destree O, Wylie C, Heasman J. Repression of organizer genes in dorsal and ventral Xenopus cells mediated by maternal XTcf3. Development. 2002;129:4015-4025
42. Ramel MC, Buckles GR, Lekven AC. Conservation of structure and functional divergence of duplicated Wnt8s in pufferfish. Dev Dyn. 2004;231:441-448
43. Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, Wu M, Kimelman D, Moon RT. Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in beta-catenin that are modulated by the Wnt signaling pathway. J Cell Biol. 1997;136:1123-1136
44. Xiong B, Rui Y, Zhang M, Shi K, Jia S, Tian T, Yin K, Huang H, Lin S, Zhao X. et al. Tob1 controls dorsal development of zebrafish embryos by antagonizing maternal beta-catenin transcriptional activity. Dev Cell. 2006;11:225-238
45. Li Y, Li Q, Long Y, Cui Z. Lzts2 regulates embryonic cell movements and dorsoventral patterning through interaction with and export of nuclear beta-catenin in zebrafish. J Biol Chem. 2011;286:45116-45130
46. Van Raay TJ, Coffey RJ, Solnica-Krezel L. Zebrafish Naked1 and Naked2 antagonize both canonical and non-canonical Wnt signaling. Dev Biol. 2007;309:151-168
Author contact
![]() Corresponding author: Jing-Xia Liu. Ph.D., College of Fisheries, Key Laboratory of Freshwater Animal Breeding, Ministry of Agriculture, Huazhong Agricultural University, Wuhan, 430070, China. E-mail: ichliuhzau.edu.cn
Corresponding author: Jing-Xia Liu. Ph.D., College of Fisheries, Key Laboratory of Freshwater Animal Breeding, Ministry of Agriculture, Huazhong Agricultural University, Wuhan, 430070, China. E-mail: ichliuhzau.edu.cn

 Global reach, higher impact
Global reach, higher impact