10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(12):1709-1714. doi:10.7150/ijbs.27168 This issue Cite
Review
Alternative polyadenylation analysis in animals and plants: newly developed strategies for profiling, processing and validation
Department of Animal Sciences and Center for Reproductive Biology, Washington State University, Pullman, WA 99164-7620
Abstract
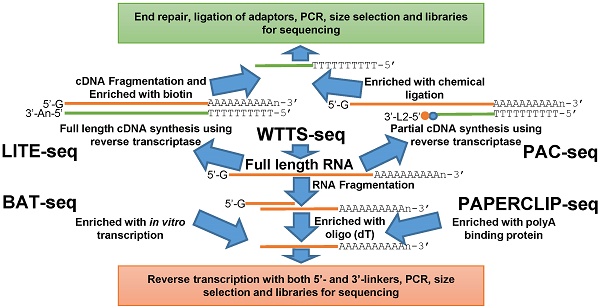
Alternative polyadenylation is an essential RNA processing event that contributes significantly to regulation of transcriptome diversity and functional dynamics in both animals and plants. Here we review newly developed next generation sequencing methods for genome-wide profiling of alternative polyadenylation (APA) sites, bioinformatics pipelines for data processing and both wet and dry laboratory approaches for APA validation. The library construction methods LITE-Seq (Low-Input 3'-Terminal sequencing) and PAC-seq (PolyA Click sequencing) tag polyA+ cDNA, while BAT-seq (BArcoded, three-prime specific sequencing) and PAPERCLIP (Poly(A) binding Protein-mediated mRNA 3′End Retrieval by CrossLinking ImmunoPrecipitation) enrich polyA+ RNA. Interestingly, only WTTS-seq (Whole Transcriptome Termini Site sequencing) targets both polyA+ RNA and polyA+ cDNA. Varieties of bioinformatics pipelines are well established to pursue read quality control, mapping, clustering, characterization and pathway analysis. The RHAPA (RNase H alternative polyadenylation assay) and 3'RACE-seq (3' rapid amplification of cDNA end sequencing) methods directly validate APA sites, while WTSS-seq (whole transcriptome start site sequencing), RNA-seq (RNA sequencing) and public APA databases can serve as indirect validation methods. We hope that these tools, pipelines and resources trigger huge waves of interest in the research community to investigate APA events underlying physiological, pathological and psychological changes and thus understand the information transfer events from genome to phenome relevant to economically important traits in both animals and plants.
Keywords: alternative polyadenylation, profiling tools, processing pipelines, validation approaches, genome function.

 Global reach, higher impact
Global reach, higher impact