10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(12):1724-1731. doi:10.7150/ijbs.28850 This issue Cite
Research Paper
TFmapper: A Tool for Searching Putative Factors Regulating Gene Expression Using ChIP-seq Data
Faculty of Health Sciences, University of Macau, Taipa, Macau, China
Abstract
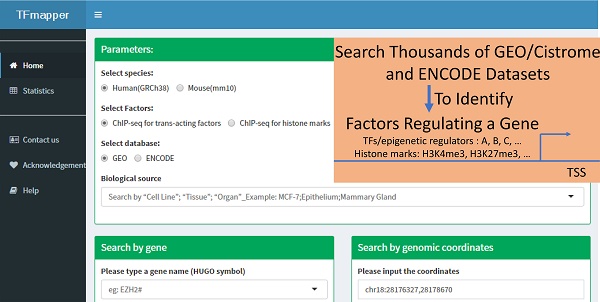
Background: Next-generation sequencing coupled to chromatin immunoprecipitation (ChIP-seq), DNase I hypersensitivity (DNase-seq) and the transposase-accessible chromatin assay (ATAC-seq) has generated enormous amounts of data, markedly improved our understanding of the transcriptional and epigenetic control of gene expression. To take advantage of the availability of such datasets and provide clues on what factors, including transcription factors, epigenetic regulators and histone modifications, potentially regulates the expression of a gene of interest, a tool for simultaneous queries of multiple datasets using symbols or genomic coordinates as search terms is needed.
Results: In this study, we annotated the peaks of thousands of ChIP-seq datasets generated by ENCODE project, or ChIP-seq/DNase-seq/ATAC-seq datasets deposited in Gene Expression Omnibus (GEO) and curated by Cistrome project; We built a MySQL database called TFmapper containing the annotations and associated metadata, allowing users without bioinformatics expertise to search across thousands of datasets to identify factors targeting a genomic region/gene of interest in a specified sample through a web interface. Users can also visualize multiple peaks in genome browsers and download the corresponding sequences.
Conclusion: TFmapper will help users explore the vast amount of publicly available ChIP-seq/DNase-seq/ATAC-seq data and perform integrative analyses to understand the regulation of a gene of interest. The web server is freely accessible at
Keywords: Chromatin immunoprecipitation, Next-generation sequencing, Regulation of gene expression

 Global reach, higher impact
Global reach, higher impact