10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2022; 18(12):4704-4713. doi:10.7150/ijbs.72663 This issue Cite
Review
Signaling mechanisms of SARS-CoV-2 Nucleocapsid protein in viral infection, cell death and inflammation
1. Medical Research Center and Guangdong-Hong Kong Joint Laboratory for Immunity and Genetics of Chronic Kidney Disease, Guangdong Provincial People's Hospital, Guangdong Academy of Medical Sciences, Guangzhou, China.
2. Department of Nephrology, The Third Affiliated hospital, Southern Medical University, Guangzhou, China.
3. Departments of Medicine & Therapeutics, Li Ka Shing Institute of Health Sciences, and Lui Che Woo Institute of Innovative Medicine, The Chinese University of Hong Kong, Hong Kong, China.
4. The Chinese University of Hong Kong-Guangdong Academy of Sciences/Guangdong Provincial People's Hospital Joint Research Laboratory on Immunological and Genetic Kidney Diseases, The Chinese University of Hong Kong, Hong Kong, China.
Received 2022-3-7; Accepted 2022-5-22; Published 2022-7-11
Abstract
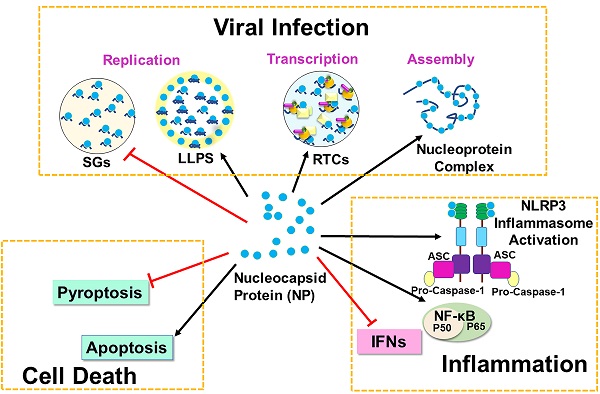
COVID-19 which is caused by severe acute respiratory syndrome coronavirus (SARS-CoV-2) has posed a worldwide pandemic and a major global public health threat. SARS-CoV-2 Nucleocapsid (N) protein plays a critical role in multiple steps of the viral life cycle and participates in viral replication, transcription, and assembly. The primary roles of N protein are to assemble with genomic RNA into the viral RNA-protein (vRNP) complex and to localize to the replication transcription complexes (RTCs) to enhance viral replication and transcription. N protein can also undergo liquid-liquid phase separation (LLPS) with viral genome RNA and inhibit stress granules to facilitate viral replication and assembly. Besides the function in viral life cycle, N protein can bind GSDMD to antagonize pyroptosis but promotes cell death via the Smad3-dependent G1 cell cycle arrest mechanism. In innate immune system, N protein inhibits IFN-β production and RNAi pathway for virus survival. However, it can induce expression of proinflammatory cytokines by activating NF-κB signaling and NLRP3 inflammasome, resulting in cytokine storms. In this review article, we are focusing on the signaling mechanisms of SARS-CoV-2 N protein in viral replication, cell death and inflammation.
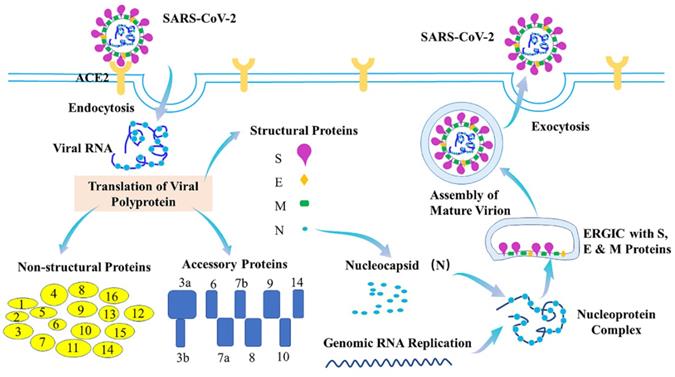
1. Life cycle of SARS-CoV-2
The coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus (SARS-CoV-2). With causing more than 518 million confirmed cases and more than 6.25 million deaths worldwide (World Health Organization (WHO)), COVID-19 has resulted in public health crises and widespread economic disruption. Typical clinical symptoms of COVID-19 are fever, dry cough, fatigue and shortness of breath, while severe patients may progress to acute respiratory distress syndrome (ARDS), acute lung injury, septic shock, or even death [1-4]. SARS-CoV-2 with ~30kb viral genome RNA is an enveloped, positive-stranded RNA virus which belongs to the β-coronaviruses. SARS-CoV-2 genome consists of 14 functional open reading frames (ORFs), including two regions (ORF1a and ORF1b) for 16 non-structural proteins (Nsp1-Nsp16), nine regions for nine putative accessory proteins, and other regions for four structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins [5-7]. Of them, SARS-CoV-2 S protein binds to its cellular receptor, angiotensin-converting enzyme 2 (ACE2) [8,9], to enter the cells. Additionally, the host serine protease TMPRSS2 is important for priming of the S protein for receptor interactions and entry [9]. Many host proteins can also function as cofactors for viral entry, such as heparin sulfate proteoglycans, C-type lectins, neuropilin-1 and furin [10-13]. After being entry, the viral and host membranes can fuse together and then release the positive sense, single-stranded RNA genome of SARS-CoV-2 that directly translates into the structural and nonstructural proteins [9]. N protein can also bind to the viral genome RNA to form the ribonucleoprotein (RNP) complex, while the M and E proteins can initiate the viral assembly [14,15]. The 16 nonstructural proteins (Nsps) can facilitate formation of the viral replication-transcription complex [16,17] and promote the traffic to the ER or Golgi membranes. In addition, they can combine with genomic RNA and N proteins to create nascent viral particles. Occurring within the ER-to-Golgi intermediate compartment (ERGIC), the assembly of mature SARS-CoV-2 virions-containing vesicles can fuse with the plasma membrane during exocytosis and release SARS-CoV-2 into the extracellular space [18,19] (Figure 1).
2. SARS-CoV-2 Nucleocapsid protein
The SARS-CoV-2 nucleocapsid (N) protein is approximately 90% identical with the SARS-CoV N protein [20]. N protein is a key player in viral replication, viral genomic RNA (gRNA) packaging into new virions and modulation of host-cell response to infection. N protein has two conserved and independently folded structural domains, called N-terminal RNA binding domain (NTD) which is responsible for the RNA binding and C-terminal dimerization domain (CTD) [21]. N protein exhibits three important roles in coronavirus life cycle (Figure 2). First, the primary role is to assemble with genomic RNA into the viral RNA-protein (vRNP) complex [22]. Virion formation occurs via the accumulation of the SARS-CoV-2 structural proteins [S, E, M and N] and gRNA at the ER-Golgi intermediate compartment (ERGIC) membrane. The N protein can interact with a luminal domain of the M protein through C-terminal dimerization domain (CTD) (247-365), which may be essential for mediating the recruitment of N-containing RNPs to the ERGIC membrane [23]. Previous study also suggests that the interaction between N protein and the E protein may plays an important role in SARS-CoV-2 assembly [24]. Together with N protein, a single strand of SARS-CoV-2 gRNA forms dense, locally ordered ribonucleoprotein (RNP) regions which may be further organized into more complex arrangements [25-27]. Second, N protein can undergo liquid-liquid phase separation (LLPS) with viral genome RNA and potentially facilitates the viral assembly. Many RNA-binding proteins have been found to undergo LLPS with RNA to participate the biological and disease processes [28-32]. The LLPS is dependent on the length and concentration of ssRNA. What is more, N protein forms typical sphere-like droplets with short ssRNAs, but not solid-like structures with long ssRNAs. The free Zn2+ in cytosol is essential for N protein/RNA LLPS [21]. It is also reported that viral RNA can induce assembly of the N protein into phase-separated condensates in vitro and pinpoints a ~40 residue region in the central intrinsically disordered region (IDR) with a key role in RNA-driven phase separation [23].
Life cycle of SARS-CoV-2. During the viral infection, SARS-CoV-2 S protein binds to ACE2 to inject its genome into host cell via endocytosis. The viral genome comprises 14 ORFs, encoding 16 Nsps, 9 ORF proteins and 4 structural proteins. Nucleocapsid (N) protein binds to viral genome RNA into ribonucleoprotein (RNP) complex, assisting membrane (M) and envelope (E) proteins to initiate viral assembly. The assembly of mature SARS-CoV-2 virions-containing vesicles which occurs within the ERGIC can fuse with the plasma membrane during exocytosis and release SARS-CoV-2 into the extracellular space.
SARS-CoV-2 Nucleocapsid protein. The nucleocapsid (N) protein plays three important roles in SARS-CoV-2 life cycle. The primary role is to assemble with genomic RNA into the viral RNA-protein (vRNP) complex. Second, N protein can undergo liquid-liquid phase separation (LLPS) with viral genome RNA and potentially facilitate viral assembly. Third, N protein can localize to replication transcription complexes (RTCs) at the early stage of infection, where it enhances replication and the transcription of viral RNA by recruiting virus proteins and host factors.
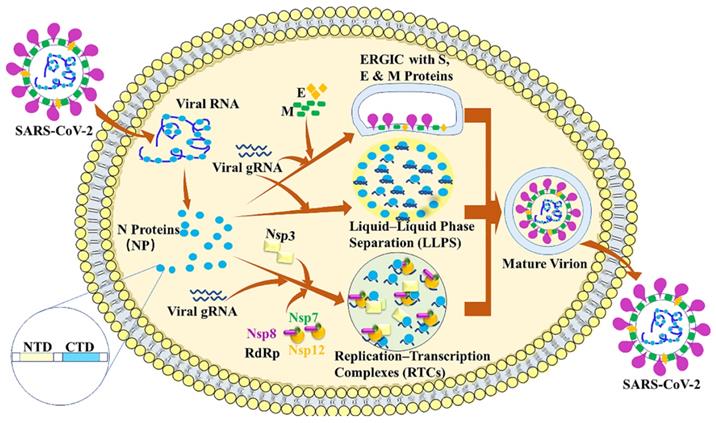
Third, N protein can localize to replication transcription complexes (RTCs) at the early stages of infection, where it enhances replication and the transcription of viral RNA by recruiting host factors [15,33-36]. Previous studies have demonstrated that N protein and RNA recruited components of the RNA polymerase (RdRp) complex (Nsp7, Nsp8 and Nsp12) are responsible for replicating viral gRNA [37,38]. The phosphorylation of the SR domain of the N protein can decrease the degree of recruitment of RdRp components to condensates [37]. The interaction between N and Nsp3 is essential for viral replication. The N-terminal of N has an RNA-binding domain (N-NTD) that can bind the RNA genome, whereas, the C-terminal domain (N-CTD) can interact with the viral Nsp3 [39].
3. Nucleocapsid protein undergoes Liquid-Liquid Phase Separation (LLPS) and attenuates stress granules
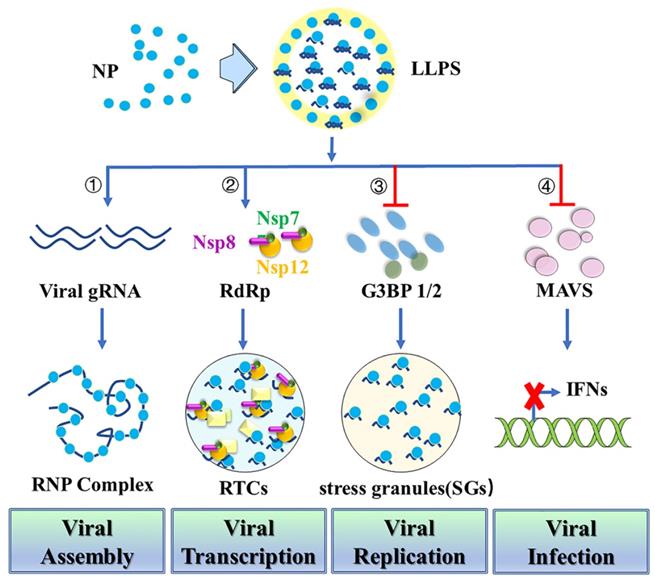
LLPS provides a highly cooperative mechanism for proteins and nucleic acids condensation into a dense phase to resemble the liquid droplets [40]. During virus infection, LLPS serves as a scaffold for virus replication and promotes the assembly of mature virions through proximity-dependent interactions [41]. N protein has sequence and structure features similar to those of other proteins that undergo LLPS with nucleic acids [42]. Thus, N protein can undergo LLPS with viral genome RNA to potentially facilitate the viral assembly (Figure 3). Negative staining electronic microscopy (EM) or cryo-EM imaging of N protein/RNA LLPS in the presence and absence of Zn2+ reveals similar loose filament-like structures as those observed in the RNP particles of another β-coronavirus MHV [43]. This suggests the potential role for N protein/RNA LLPS in viral assembly [21]. A recent study also demonstrates that N protein LLPS can promote cooperative association of the RdRp complex with polyU RNA in vitro [37]. N protein may also use LLPS-based mechanisms to enable high initiation and elongation rates during viral transcription [37]. Phosphorylation of SR-domains of N protein inhibits its RNA binding and RNA-induced LLPS [37]. Besides the function in viral assembly and transcription, the dimerization domain of N protein can also inhibit Lys63-linked poly-ubiquitination and aggregation of MAVS, thereby suppressing the innate antiviral immune response [44]. N protein acetylation at Lys375 abrogates its LLPS with RNA and the N protein-mediated suppression of MAVS signaling. Targeting the dimerization domain of N protein by a peptide can disrupt the LLPS and then inhibit SARS-CoV-2 replication in vitro and in vivo [44].
N protein can also interact with human ribonucleoproteins which are found in several LLPS-driven cytosolic protein/RNA granules [45]. This suggests that N protein may modulate protein/RNA granule formation in order to promote viral replication [46]. Stress granules (SGs) are cytoplasmic protein/RNA granules. They are formed through LLPS as an antiviral response to inhibit protein synthesis and induce innate immune signaling [47-49]. Recent studies reported that SARS-CoV-2 N protein undergoes LLPS into SGs through its N-terminal intrinsically disordered region (IDR) with SGs protein G3BP1/2 [23,50,51]. Additionally, N protein inhibits the host stress response through SGs attenuation by sequestering G3BP1/2 through its interaction with these proteins and direct interaction with host mRNAs [51]. N protein can also specifically interact with G3BP1/2, leading to a reduction in the size and/or number of stress granules [52-57]. A short sequence (residues 15-18) in the IDR NTD or three arginine residues (R92, R107, and R149) in the NTD may also play an important role in its interaction with G3BP1 and modulation of stress granules. Thus, targeting the N-G3BP1 interaction by competitive peptide can reduce viral proliferation, indicating that N-G3BP1 is important for viral replication [52,57]. Furthermore, N protein is able to impair SGs formation by inhibiting PKR autophosphorylation and activation, as well as by targeting G3BP1 [55].
4. Nucleocapsid protein suppresses host pyroptosis but promotes apoptosis
Gasdermin D (GSDMD) is cleaved by the caspase-1 dimers post-inflammasome activation, leading to cell membrane permeability, cell content leakage and finally cell death termed pyroptosis [58,59]. While SARS-CoV-2 infection promotes activation of caspase-1 and NLRP3 inflammasome, GSDMD cleavage and pyroptosis are inhibited by N protein in infected human monocytes. N protein binds GSDMD and hinders GSDMD cleavage by the activated caspase-1 dimers to antagonize pyroptosis.
Nucleocapsid protein undergoes Liquid-Liquid Phase Separation (LLPS) and attenuates stress granules. First, N protein can undergo LLPS with viral genome RNA and potentially facilitate viral assembly. Second, N protein LLPS promotes cooperative association of the RdRp complex with polyU RNA to enable high initiation and elongation rates during viral transcription. Third, N protein undergoes LLPS into SGs through its N-terminal intrinsically disordered region (IDR) with SG protein G3BP1/2 to promote viral replication. Fourth, N protein which is required for LLPS with RNA inhibits MAVS and thereby suppresses the innate antiviral immune response to promote viral infection.
Apoptosis is a major type of programmed cell death. It is triggered by mitochondrion or cell-surface death receptor mediated the cleavage of downstream caspases [60-62]. Previous studies indicated that SARS-CoV N protein can induce apoptosis in HPF cells and COS-1 cells by activating the mitochondrial pathway [63-65]. Recently study also demonstrated that SARS-CoV-2 N protein can specifically enhance the M protein-induced apoptosis by strengthening M protein-mediated attenuation of PDK1-PKB/Akt interaction [66]. Our recent study also reveals that SARS-COV-2 N protein not only promotes renal cell death in ischemic-induced AKI but inhibits renal tubular cell proliferation via Smad3-p21 dependent G1 cell cycle arrest [67]. We further uncover that genetic deletion of Smad3 or pharmacological inhibition of Smad3 can protect kidneys from SARS-CoV-2 N protein-induced renal cell death. In addition, SARS-COV-2 N can interact with α-synuclein to disturb the α-synuclein proteostasis and increase cell death in S-SY5Y [68]. This may provide molecular basis for the correlation between SARS-COV-2 infections and Parkinsonism.
5. Regulation of host inflammation by Nucleocapsid protein
Host immune response including innate and adaptive immunity against SARS-CoV-2 seems crucial to control and resolve the viral infection [69-71]. The innate immune system detects viral infections through the recognition of molecular patterns. It is a primary host defense strategy to suppress viral infections, coordinate and accelerate the development of adaptive immunity [72]. Pattern recognition receptors (PRRs) respond to pathogen-associated molecular patterns (PAMPs) can trigger the activation of inflammatory responses and the release of inflammatory cytokines to limit viral infection [73]. Several families of PRRs have been described: the Toll-like receptor (TLR) [74], the RIG-I-like receptor (RLR) [75], the NOD-like receptor (NLR), and the C-type lectin receptor (CLR) [76]. Single-stranded RNA derived from genomic, subgenomic or replicative intermediates of SARS-CoV-2 can be sensed by RLRs, which include MAD5, RIG-I and LGP2 [77-80]. RIG-I and MDA5 are the most well-studied RLRs and critically regulate IFN pathways. After virus infection, RIG-I and MDA5 sense the RNA and then translocate to the mitochondria, where they interact with the adaptor protein mitochondrial antiviral signaling (MAVS) to form a MAVS signalosome. This complex formation activates downstream proteins to induce phosphorylation of IRF3, therefore facilitating its nuclear translocation and the transcription of genes encoding type I and III IFNs [81,82]. Production and subsequent release of IFNs can stimulate the downstream signals to produce hundreds of IFN-stimulated genes (ISGs) with various antiviral functions [83,84]. Previous studies demonstrated that coronaviruses (CoVs) have evolved evasion strategies to limit host control. Otherwise, they enhance replication and transmission in response to innate immune-dependent viral clearance mechanisms [69,85-87]. In SARS-COV-2 infection, a low production of type I interferons was detected in the peripheral blood or lungs of COVID-19 patients with a severe clinical picture [69,88,89]. In a recent study, SARS-COV-2 N protein can interact with TRIM25 functional domain SPRY to block the ubiquitinating activity of TRIM25 on RIG-I and then inhibit IFN-β production [90]. SARS-COV-2 N protein can also interact with G3BP1 to prevent the antiviral stress granule formation and impair the recognition of dsRNA by RIG-I [91]. It is also reported that SARS-COV-2 N protein is able to interact with RIG-I through the DExD/H domain of RIG-I and then to repress RIG-I-mediated IRF3 phosphorylation and nuclear translocation. N protein suppresses IFN-β production upon the infection of SeV or by the stimulation of poly(I:C) [92]. In addition, N protein is found to inhibit Lys63-linked poly-ubiquitination and aggregation of MAVS, thereby suppressing the innate antiviral immune response [44]. All these results indicate that SARS-COV-2 N protein can inhibit IFN-β production by targeting the RIG-I signaling pathway (Figure 4).
In most cases, innate immune responses limit viral entry, translation, replication, and assembly. However, in some individuals, the disease severity or mortality of the COVID-19 might be associated with the excessive production of proinflammatory cytokines, resulting in “cytokine storm” and acute respiratory distress syndrome [93]. Nuclear factor κB (NF-κB) is a key transcription factor of proinflammatory cytokines in immune cells [94]. Upon sensing different ligands, their receptors then recruit the adaptor proteins to promote the activation of tumor necrosis factor receptor (TNF-R)-associated factor (TRAF) signaling molecules, thus recruiting TGF-beta-activated kinase 1 (TAK1) and IκB kinase (IKK) complex [95]. Activated IKK complex can phosphorylate IκB proteins to induce ubiquitin-proteasome degradation. Degradation of IκB allows NF-κB translocation to the nucleus to initiate the transcription of downstream proinflammatory cytokines [96]. SARS-CoV-2 N protein is reported to promote activation of NF-κB signaling by enhancing the association between TAK1 and IKK complex [95]. With viral RNA, N protein undergoes LLPS to recruit TAK1 and IKK complex, and then to promote NF-κB activation. The CTD domain of SARS-CoV-2 N protein is critical for its LLPS and NF-κB activation. 1,6-hexanediol which is the inhibitor of LLPS can inhibit the phase separation of N protein and then suppress the activation of NF-κB. All these results indicate that LLPS of N protein provides a platform to induce NF-κB activation after virus infection. SARS-CoV-2 N can also function as a PAMP to directly bind to TLR2 to activate NF-κB and MAPK signaling in endothelial cells [97]. Treatment with recombinant SARS-CoV-2 N protein can induce acute lung injury via M1 macrophage polarization and NF-κB activation, which can be inhibited by N-protein denaturation, neutralizing antibody to N-protein, and NF-κB inhibitor [98]. NLRs are also reported to respond to SARS-CoV-2 infection and induce production of pro-inflammatory cytokines [99]. NLRP3, one of the best characterized inflammasome sensors, is triggered in response to virus infection and thus activates Caspase-1 with an adaptor protein ASC [100]. Active caspase-1 is formed by autocatalytic cleavage, which then catalyzes proteolytic processing of pro-interleukin (IL)-1β and pro-interleukin (IL)-18 into mature IL-1β and IL-18 [101]. IL-1β plays crucial roles in inflammatory responses and instructs adaptive immune responses by inducing expression of immunity associated genes [102]. SARS-CoV-2 N protein is reported to induce proinflammatory cytokines through promoting the assembly and activation of the NLRP3 inflammasome [100]. Indeed, N protein interacts directly with NLRP3, promotes the binding of NLRP3 with ASC, and facilitates NLRP3 inflammasome assembly. More importantly, N protein aggravates lung injury and accelerates death in acute inflammation mouse models, which can be blocked by NLRP3 inhibitor MCC950 and Caspase-1 inhibitor Ac-YVAD-cmk. Taken together, SARS-CoV-2 N protein induces proinflammatory cytokines through promoting the activation of NF-κB signaling and NLRP3 inflammasome (Figure 4).
RNAi, a post-transcriptional gene silencing mechanism, has been recognized as an antiviral immune defense after virus infection [103]. After virus infection and replication, virus-derived dsRNA is generated and can be recognized and cleaved by the host endoribonuclease Dicer. As a countermeasure, viruses such as Influenza A virus NS1 and Dengue virus 2 NS2A can encode viral protein to inhibit the RNAi pathway [103,104]. Recently study also reported that SARS-CoV-2 N protein can suppress RNAi pathway [105]. Indeed, N protein can interact with dsRNA and then sequestrates dsRNA to suppress RNAi, thereby functioning as a key immune evasion factor of SARS-CoV-2.
Regulation of host inflammation by Nucleocapsid protein. First, SARS-COV-2 N protein can inhibit IFN-β production by targeting each step of the RIG-I signaling pathway. Second, with viral RNA, N protein undergoes LLPS to recruit TAK1 and IKK complex, and then promotes NF-κB activation. Third, N protein interacts directly with NLRP3 protein to promote the assembly and activation of the NLRP3 inflammasome.
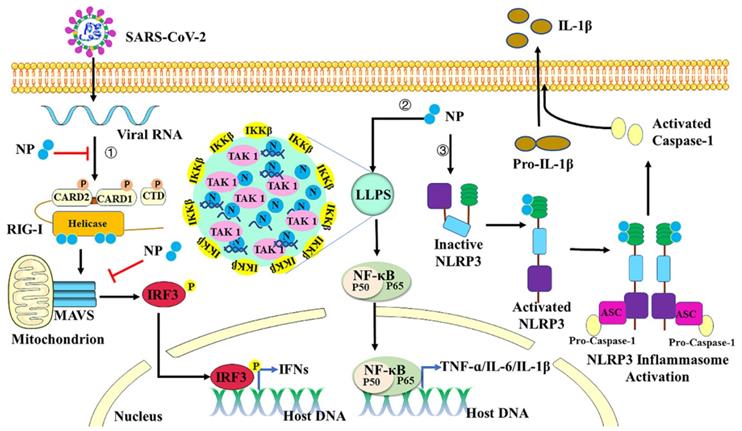
Based on the function of SARS-CoV-2 N protein, we proposed a working model for SARS-CoV-2 induced inhibition of host innate immunity. In the early stage of infection, N protein can interact with TRIM25 or G3BP1 to influence RIG-I activation. N protein can also interact with DExD/H domain of RIG-I directly and inhibit RIG-I activation. What is more, N protein is capable of interacting with an adaptor protein MAVS to inhibit the ubiquitination of MAVS. Taken together, N protein represses RIG-I-mediated phosphorylation of TBK1 and IRF3 to suppress their nuclear translocation and IFN-β expression. At the same time, N protein can also suppress RNAi to evade the innate immune system. However, at the later stage of infection, the viral replication, transcription, and assembly are actively processed with the involvement of various viral proteins and inflammatory pathways, which may trigger the overactivation of innate immunity, resulting in cytokine storm syndrome and disease progression.
6. Conclusions and perspectives
Here, we summarize the signaling mechanisms of SARS-CoV-2 N protein in viral replication, cell death, and inflammation. N protein as a structure protein plays a critical role in multiple steps of the viral life cycle. N protein assembles with genomic RNA into the viral RNA-protein (vRNP) complex to facilitate viral assembly. Moreover, it contributes to forming helical ribonucleoprotein during the packaging of the RNA genome and regulating the viral RNA synthesis during replication and transcription. Importantly, N protein has multiple functions in cell death and inflammation. N protein can inhibit pyroptosis but promotes apoptosis to induce cell death. N protein can inhibit RIG-I and RNAi signaling but promotes NF-κB signaling and NLRP3 inflammasome in the innate immune system to trigger the “cytokine storm”.
Recent phosphoproteomic analyses revealed that SARS-CoV-2 N protein is highly phosphorylated within the RS domain [106-109]. It is known that phosphorylation regulates the states and functions of the N protein. Phosphorylation of N protein by glycogen synthase kinase 3(GSK-3) is required for viral transcription and replication [110]. GSK-3 is essential for SARS-CoV-2 N phosphorylation as blockade of GSK-3 with inhibitors can block N phosphorylation and virus replication in SARS-CoV-2 infected lung epithelial cells [111]. Thus, research into a better understanding of the phosphorylation of SARS-CoV-2 N protein in viral transcription and replication is needed.
Based on the high homology (90%) of the amino acid sequences and fewer mutations over time among coronavirus N proteins [112], SARS-CoV-2 N protein may function similarly to SARS-CoV N or MERS-CoV N protein. The SARS-CoV N protein has been reported to interact with numerous host cell proteins, such as TRIM25 [113], Smad3 [114], the chemokine Cxcl16 [115], translation elongation factor-1 alpha [116], pyruvate kinase [117], and 14-3-3 [118]. Although SARS-CoV-2 N can also interact with TRIM25 [90], Smad3 [67,119], 14-3-3 [120], and others [121], further research into the interaction between SARS-CoV 2 N and host cell proteins under disease conditions may provide valuable information for potential druggable targets. Thus, understanding of the roles and mechanisms of N protein in the pathogenesis of diseases may be the first step towards the development of anti-SARS-CoV-2 drugs and vaccines to prevent and control the SARS-CoV-2 pandemics.
Abbreviations
ARDS: acute respiratory distress syndrome; ASC: apoptosis-associated speck-like protein with the CARD domain; COVID-19: coronavirus disease 2019; CLR: C-type lectin receptor; CTD: C-terminal dimerization domain; ERGIC: ER-to-Golgi intermediate compartment; IDR: intrinsically disordered region; RIG-I: retinoic-acid inducible gene-I; MDA5: melanoma differentiation-associated protein 5; IKK: IκB kinase; LLPS: liquid-liquid phase separation; MAVS: mitochondrial antiviral signaling; N: Nucleocapsid; NF-κB: Nuclear factor κB; NLR: NOD-like receptor; Nsps: nonstructural proteins; NTD: N-terminal RNA binding domain; ORFs: open reading frames; PAMPs: pathogen-associated molecular patterns; RdRp: RNA dependent RNA polymerase; RLR: RIG-I-like receptor; PRRs: Pattern recognition receptors; RTCs: replication transcription complexes; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; SGs: Stress granules; TAK1: TGF-beta-activated kinase 1; TLR: Toll-like receptor; TRAF: tumor necrosis factor receptor (TNF-R)-associated factor; vRNP: viral RNA-protein; WHO: World Health Organization.
Acknowledgements
This study was supported by the Grants from Research Council of Hong Kong (14117418, 14104019, and 14101121), the Guangdong-Hong Kong-Macao-Joint Labs Program from Guangdong Science and Technology Department (2019B121205005), the High-level Hospital Construction Project from Guangdong Provincial People's Hospital, Guangdong Academy of Medical Science (KJ012019108), and National Natural Science Foundation of China (81902053,82100723).
Author Contributions
W.W. and J.C. wrote the manuscript. X.Y. and H.Y.L. designed and revised the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395:497-506
2. Chu DK, Akl EA, Duda S, Solo K, Yaacoub S, Schünemann HJ. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973-87
3. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J. et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. Jama. 2020;323:1061-9
4. Zhang B, Zhou X, Qiu Y, Song Y, Feng F, Feng J. et al. Clinical characteristics of 82 cases of death from COVID-19. PLoS One. 2020;15:e0235458
5. V'Kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155-70
6. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565-574
7. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG. et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265-9
8. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-3
9. Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S. et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271-80 e8
10. Ludovico Cantuti-Castelvetri, RaviOjha, Liliana D. Pedro, MinouDjannatian, Jonas Franz, Suvi Kuivanen, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856-860
11. Bailey AL, Diamond MS. A Crisp(r) New Perspective on SARS-CoV-2 Biology. Cell. 2021;184:15-7
12. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L. et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620
13. Lempp FA, Soriaga LB, Montiel-Ruiz M, Benigni F, Noack J, Park YJ. et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature. 2021;598:342-7
14. Masters PS. Coronavirus genomic RNA packaging. Virology. 2019;537:198-207
15. McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991-3018
16. Sawicki SG, Sawicki DL, Siddell SG. A contemporary view of coronavirus transcription. J Virol. 2007;81:20-9
17. Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S. et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305-15
18. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nature Reviews Microbiology. 2016;14:523-34
19. Lee S, Channappanavar R, Kanneganti TD. Coronaviruses: Innate Immunity, Inflammasome Activation, Inflammatory Cell Death, and Cytokines. Trends Immunol. 2020;41:1083-99
20. Kang S, Yang M, Hong Z, Zhang L, Huang Z, Chen X. et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020;10:1228-38
21. Chen H, Cui Y, Han X, Hu W, Sun M, Zhang Y. et al. Liquid-liquid phase separation by SARS-CoV-2 nucleocapsid protein and RNA. Cell Research. 2020;30:1143-5
22. Chang CK, Hou MH, Chang CF, Hsiao CD, Huang TH. The SARS coronavirus nucleocapsid protein-forms and functions. Antiviral Res. 2014;103:39-50
23. Lu S, Ye Q, Singh D, Cao Y, Diedrich JK, Yates JR 3rd. et al. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nature Communications. 2021;12:502
24. Li J, Guo M, Tian X, Wang X, Yang X, Wu P. et al. Virus-Host Interactome and Proteomic Survey Reveal Potential Virulence Factors Influencing SARS-CoV-2 Pathogenesis. Med (N Y). 2021;2:99-112 e7
25. Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B. et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nature Communications. 2020;11:5885
26. Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C. et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell. 2020;183:730-8.e13
27. Cao C, Cai Z, Xiao X, Rao J, Chen J, Hu N. et al. The architecture of the SARS-CoV-2 RNA genome inside virion. Nat Commun. 2021;12:3917
28. Lin Y, David S. W. Protter, Rosen MK, and Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell. 2015;60:208-19
29. Amandine Molliex, Jamshid Temirov, Jihun Lee, Maura Coughlin, Anderson P. Kanagaraj, Hong Joo Kim, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123-33
30. Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY. et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066-77
31. Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39-58
32. Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149:1188-91
33. Verheije MH, Hagemeijer MC, Ulasli M, Reggiori F, Rottier PJ, Masters PS. et al. The coronavirus nucleocapsid protein is dynamically associated with the replication-transcription complexes. J Virol. 2010;84:11575-9
34. Zuniga S, Sola I, Moreno JL, Sabella P, Plana-Duran J, Enjuanes L. Coronavirus nucleocapsid protein is an RNA chaperone. Virology. 2007;357:215-27
35. Zuniga S, Cruz JL, Sola I, Mateos-Gomez PA, Palacio L, Enjuanes L. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol. 2010;84:2169-75
36. Cong Y, Ulasli M, Hein Schepers, Mauthe M, Philip V'kovski, Franziska Kriegenburg. et al. Nucleocapsid Protein Recruitment to Replication-Transcription Complexes Plays a Crucial Role in Coronaviral Life Cycle. Journal of Virology. 2020;94:e01925-19
37. Savastano A, Ibanez de Opakua A, Rankovic M, Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nature Communications. 2020;11:6041
38. Zhao D, Xu W, Zhang X, Wang X, Ge Y, Yuan E. et al. Understanding the phase separation characteristics of nucleocapsid protein provides a new therapeutic opportunity against SARS-CoV-2. Protein Cell. 2021;12:734-40
39. Khan MT, Zeb MT, Ahsan H, Ahmed A, Ali A, Akhtar K. et al. SARS-CoV-2 nucleocapsid and Nsp3 binding: an in silico study. Archives of Microbiology. 2021;203:59-66
40. Alberti S, Gladfelter A, Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell. 2019;176:419-34
41. Poblete-Duran N, Prades-Perez Y, Vera-Otarola J, Soto-Rifo R, Valiente-Echeverria F. Who Regulates Whom? An Overview of RNA Granules and Viral Infections. Viruses. 2016;8:180
42. Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA. et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell. 2020;181:306-24 e28
43. Gui M, Liu X, Guo D, Zhang Z, Yin CC, Chen Y. et al. Electron microscopy studies of the coronavirus ribonucleoprotein complex. Protein Cell. 2017;8:219-24
44. Wang S, Dai T, Qin Z, Pan T, Chu F, Lou L. et al. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nature Cell Biology. 2021;23:718-32
45. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM. et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459-68
46. Cascarina SM, Ross ED. A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates. FASEB J. 2020;34:9832-42
47. White JP, Lloyd RE. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175-83
48. McCormick C, Khaperskyy DA. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol. 2017;17:647-60
49. Eiermann N, Haneke K, Sun Z, Stoecklin G, Ruggieri A. Dance with the Devil: Stress Granules and Signaling in Antiviral Responses. Viruses. 2020;12:984
50. Wang J, Shi C, Xu Q, Yin H. SARS-CoV-2 nucleocapsid protein undergoes liquid-liquid phase separation into stress granules through its N-terminal intrinsically disordered region. Cell Discovery. 2021;7:5
51. Nabeel-Shah S, Lee H, Ahmed N, Burke GL, Farhangmehr S, Ashraf K. et al. SARS-CoV-2 nucleocapsid protein binds host mRNAs and attenuates stress granules to impair host stress response. iScience. 2022;25:103562
52. Huang W, Ju X, Tian M, Li X, Yu Y, Sun Q. et al. Molecular determinants for regulation of G3BP1/2 phase separation by the SARS-CoV-2 nucleocapsid protein. Cell Discovery. 2021;7:69
53. Somasekharan SP, Gleave M. SARS-CoV-2 nucleocapsid protein interacts with immunoregulators and stress granules and phase separates to form liquid droplets. FEBS Lett. 2021;595:2872-96
54. Luo L, Li Z, Zhao T, Ju X, Ma P, Jin B. et al. SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production. Sci Bull (Beijing). 2021;66:1194-204
55. Zheng ZQ, Wang SY, Xu ZS, Fu YZ, Wang YY. SARS-CoV-2 nucleocapsid protein impairs stress granule formation to promote viral replication. Cell Discovery. 2021;7:38
56. Cai T, Yu Z, Wang Z, Liang C, Richard S. Arginine methylation of SARS-CoV-2 nucleocapsid protein regulates rna binding, its ability to suppress stress granule formation, and viral replication. the Journal of Biological Chemistry. 2021;297:100821
57. Kruse T, Benz C, Garvanska DH, Lindqvist R, Mihalic F, Coscia F. et al. Large scale discovery of coronavirus-host factor protein interaction motifs reveals SARS-CoV-2 specific mechanisms and vulnerabilities. Nat Commun. 2021;12:6761
58. Kayagaki N, Stowe IB, Lee BL, O'Rourke K, Anderson K, Warming S. et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666-71
59. Boucher D, Monteleone M, Coll RC, Chen KW, Ross CM, Teo JL. et al. Caspase-1 self-cleavage is an intrinsic mechanism to terminate inflammasome activity. J Exp Med. 2018;215:827-40
60. Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770-6
61. Schultz DR, Harrington WJ. Apoptosis: programmed cell death at a molecular level. Semin Arthritis Rheum. 2003;32:345-69
62. Bagci EZ, Sen SM, Camurdan MC. Analysis of a mathematical model of apoptosis: individual differences and malfunction in programmed cell death. J Clin Monit Comput. 2013;27:465-79
63. Zhao G, Shi SQ, Yang Y, Peng JP. M and N proteins of SARS coronavirus induce apoptosis in HPF cells. Cell Biol Toxicol. 2006;22:313-22
64. Milan SURJIT, Boping LIU, Shahid JAMEEL, CHOW VTK, LAL SK. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 2004;383:13-8
65. Zhang L, Wei L, Jiang D, Wang J, Cong X, Fei R. SARS-CoV nucleocapsid protein induced apoptosis of COS-1 mediated by the mitochondrial pathway. Artif Cells Blood Substit Immobil Biotechnol. 2007;35:237-53
66. Ren Y, Wang A, Fang Y, Shu T, Wu D, Wang C. et al. SARS-CoV-2 Membrane Glycoprotein M Triggers Apoptosis With the Assistance of Nucleocapsid Protein N in Cells. Front Cell Infect Microbiol. 2021;11:706252
67. Wang W, Chen J, Hu D, Pan P, Liang L, Wu W. et al. SARS-CoV-2 N Protein Induces Acute Kidney Injury via Smad3-Dependent G1 Cell Cycle Arrest Mechanism. Adv Sci (Weinh). 2022;9:e2103248
68. Semerdzhiev SA, Fakhree MAA, Segers-Nolten I, Blum C, Claessens M. Interactions between SARS-CoV-2 N-Protein and α-Synuclein Accelerate Amyloid Formation. ACS Chem Neurosci. 2022;13:143-50
69. Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R. et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036-45 e9
70. Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z. et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27:883-90 e2
71. Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M. et al. Immunology of COVID-19: Current State of the Science. Immunity. 2020;52:910-41
72. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783-801
73. Kanneganti TD. Intracellular innate immune receptors: Life inside the cell. Immunol Rev. 2020;297:5-12
74. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805-20
75. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M. et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730-7
76. Hardison SE, Brown GD. C-type lectin receptors orchestrate antifungal immunity. Nat Immunol. 2012;13:817-22
77. Yin X, Riva L, Pu Y, Martin-Sancho L, Kanamune J, Yamamoto Y. et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021;34:108628
78. Yang DM, Geng TT, Harrison AG, Wang PH. Differential roles of RIG-I like receptors in SARS-CoV-2 infection. Mil Med Res. 2021;8:49
79. Rebendenne A, Valadao ALC, Tauziet M, Maarifi G, Bonaventure B, McKellar J. et al. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J Virol. 2021;95:e02415-20
80. Thorne LG, Reuschl AK, Zuliani-Alvarez L, Whelan MVX, Turner J, Noursadeghi M. et al. SARS-CoV-2 sensing by RIG-I and MDA5 links epithelial infection to macrophage inflammation. EMBO J. 2021;40:e107826
81. Loo YM, Gale M Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680-92
82. Horner SM, Liu HM, Park HS, Briley J, Gale M Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590-5
83. Stark GR, Darnell JE Jr. The JAK-STAT pathway at twenty. Immunity. 2012;36:503-14
84. Wack A, Terczynska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol. 2015;16:802-9
85. Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y. et al. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 2020;32:108185
86. Li JY, Liao CH, Wang Q, Tan YJ, Luo R, Qiu Y. et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286:198074
87. Burke JM. et al. SARS-CoV-2 infection triggers widespread host mRNA decay leading to an mRNA export block. RNA. 2021;27:1318-29
88. Hadjadj J. et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718-24
89. Acharya D, Liu G, Gack MU. Dysregulation of type I interferon responses in COVID-19. Nat Rev Immunol. 2020;20:397-8
90. Gori Savellini G, Anichini G, Gandolfo C, Cusi MG. SARS-CoV-2 N Protein Targets TRIM25-Mediated RIG-I Activation to Suppress Innate Immunity. Viruses. 2021;13:1439
91. Zheng Y, Deng J, Han L, Zhuang MW, Xu Y, Zhang J. et al. SARS-CoV-2 NSP5 and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. Signal Transduct Target Ther. 2022;7:22
92. Chen K, Xiao F, Hu D, Ge W, Tian M, Wang W. et al. SARS-CoV-2 Nucleocapsid Protein Interacts with RIG-I and Represses RIG-Mediated IFN-β Production. Viruses. 2020;13:47
93. Tufan A, Avanoglu Guler A, Matucci-Cerinic M. COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs. Turkish Journal of Medical Science. 2020;50:620-32
94. Hayden MS, Ghosh S. NF-kappaB in immunobiology. Cell Res. 2011;21:223-44
95. Chen ZJ. Ubiquitin Signaling in the NF-κB Pathway. Nature Cell Biology. 2005;7:758-65
96. Iwai K. Diverse ubiquitin signaling in NF-κB activation. Trends Cell Biol. 2012;22:355-64
97. Qian Y, Lei T, Patel PS, Lee CH, Monaghan-Nichols P, Xin HB. et al. Direct activation of endothelial cells by SARS-CoV-2 nucleocapsid protein is blocked by Simvastatin. bioRxiv. 2021.
98. Xia J, Tang W, Wang J, Lai D, Xu Q, Huang R. et al. SARS-CoV-2 N Protein Induces Acute Lung Injury in Mice via NF-ĸB Activation. Front Immunol. 2021;12:791753
99. Diamond MS, Kanneganti TD. Innate immunity: The first line of defense against SARS-CoV-2. Nature Immunology. 2022;23:165-76
100. Kelley N, Jeltema D, Duan Y, He Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int J Mol Sci. 2019;20:3328
101. Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397-411
102. Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519-50
103. Ding SW, Han Q, Wang J, Li WX. Antiviral RNA interference in mammals. Curr Opin Immunol. 2018;54:109-14
104. Qiu Y, Xu YP, Wang M, Miao M, Zhou H, Xu J. et al. Flavivirus induces and antagonizes antiviral RNA interference in both mammals and mosquitoes. Sci Adv. 2020;6:eaax7989
105. Mu J, Xu J, Zhang L, Shu T, Wu D, Huang M. et al. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci China Life Sci. 2020;63:1413-6
106. Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M. et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182:685-712.e19
107. Klann K, Bojkova D, Tascher G, Ciesek S, Münch C, Cinatl J. Growth Factor Receptor Signaling Inhibition Prevents SARS-CoV-2 Replication. Mol Cell. 2020;80:164-74.e4
108. Yaron TM, Heaton BE, Levy TM, Johnson JL, Jordan TX, Cohen BM. et al. The FDA-approved drug Alectinib compromises SARS-CoV-2 nucleocapsid phosphorylation and inhibits viral infection in vitro. bioRxiv. 2020.
109. Davidson AD, Williamson MK, Lewis S, Shoemark D, Carroll MW, Heesom KJ. et al. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68
110. Wu CH, Yeh SH, Tsay YG, Shieh YH, Kao CL, Chen YS. et al. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J Biol Chem. 2009;284:5229-39
111. Liu X, Verma A, Garcia G Jr, Ramage H, Lucas A, Myers RL. et al. Targeting the coronavirus nucleocapsid protein through GSK-3 inhibition. Proc Natl Acad Sci U S A. 2021;118:e2113401118
112. Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671-80.e2
113. Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P. et al. The Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Inhibits Type I Interferon Production by Interfering with TRIM25-Mediated RIG-I Ubiquitination. J Virol. 2017;91:e02143-16
114. Zhao X, Nicholls JM, Chen YG. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-beta signaling. J Biol Chem. 2008;283:3272-80
115. Zhang YP, Zhang RW, Chang WS, Wang YY. Cxcl16 interact with SARS-CoV N protein in and out cell. Virol Sin. 2010;25:369-74
116. Zhou B, Liu J, Wang Q, Liu X, Li X, Li P. et al. The nucleocapsid protein of severe acute respiratory syndrome coronavirus inhibits cell cytokinesis and proliferation by interacting with translation elongation factor 1alpha. J Virol. 2008;82:6962-71
117. Wei WY, Li HC, Chen CY, Yang CH, Lee SK, Wang CW. et al. SARS-CoV nucleocapsid protein interacts with cellular pyruvate kinase protein and inhibits its activity. Arch Virol. 2012;157:635-45
118. Surjit M, Kumar R, Mishra RN, Reddy MK, Chow VT, Lal SK. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J Virol. 2005;79:11476-86
119. Chen J, Wu W, Wang W, Tang Y, Lan HY. Role of TGF-β sinagling in CIVID-19. Integr Med Nephrol Androl. 2022 (in press)
120. Tugaeva KV, Hawkins D, Smith JLR, Bayfield OW, Ker DS, Sysoev AA. et al. The Mechanism of SARS-CoV-2 Nucleocapsid Protein Recognition by the Human 14-3-3 Proteins. J Mol Biol. 2021;433:166875
121. Zheng X, Sun Z, Yu L, Shi D, Zhu M, Yao H. et al. Interactome Analysis of the Nucleocapsid Protein of SARS-CoV-2 Virus. Pathogens. 2021;10:1155
Author contact
![]() Corresponding authors: Hui-Yao Lan, Departments of Medicine & Therapeutics, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong, China. E-mail: hylanedu.hk; and Xueqing Yu, Guangdong Provincial People's Hospital, Guangdong Academy of Medical Sciences, Guangzhou, China. E-mail: yuxqsysu.edu.cn
Corresponding authors: Hui-Yao Lan, Departments of Medicine & Therapeutics, Li Ka Shing Institute of Health Sciences, The Chinese University of Hong Kong, Hong Kong, China. E-mail: hylanedu.hk; and Xueqing Yu, Guangdong Provincial People's Hospital, Guangdong Academy of Medical Sciences, Guangzhou, China. E-mail: yuxqsysu.edu.cn

 Global reach, higher impact
Global reach, higher impact