10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2023; 19(10):3115-3127. doi:10.7150/ijbs.85285 This issue Cite
Research Paper
FABP5 suppresses colorectal cancer progression via mTOR-mediated autophagy by decreasing FASN expression
Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, Nanjing, China.
*These authors contributed equally to this work.
Received 2023-4-14; Accepted 2023-6-3; Published 2023-6-12
Abstract
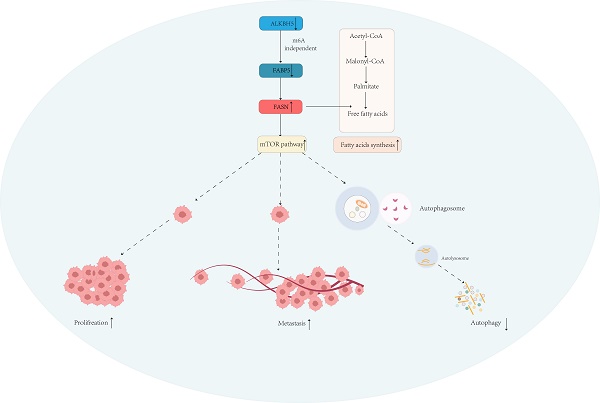
Lipid metabolism plays an important role in the occurrence and development of cancer, in particular, digestive system tumors such as colon cancer. Here, we investigated the role of the fatty acid-binding protein 5 (FABP5) in colorectal cancer (CRC). We observed marked down-regulation of FABP5 in CRC. Data from functional assays revealed inhibitory effects of FABP5 on cell proliferation, colony formation, migration, invasion as well as tumor growth in vivo. In terms of mechanistic insights, FABP5 interacted with fatty acid synthase (FASN) and activated the ubiquitin proteasome pathway, leading to a decrease in FASN expression and lipid accumulation, moreover, suppressing mTOR signaling and facilitating cell autophagy. Orlistat, a FASN inhibitor, exerted anti-cancer effects both in vivo and in vitro. Furthermore, the upstream RNA demethylase ALKBH5 positively regulated FABP5 expression via an m6A-independent mechanism. Overall, our collective findings offer valuable insights into the critical role of the ALKBH5/FABP5/FASN/mTOR axis in tumor progression and uncover a potential mechanism linking lipid metabolism to development of CRC, providing novel therapeutic targets for future interventions.
Keywords: Colorectal cancer, Lipid metabolism, FABP5, FASN, Autophagy, N6-methyladenosine, Orlistat
Introduction
Colorectal cancer (CRC) is the third most common malignancy and second most deadly cancer type worldwide, accounting for estimated 1.9 million new cases and 900,000 deaths in 2020[1, 2]. The incidence of CRC is relatively high in developed countries and continues to increase gradually in developing countries[3, 4]. Since the early symptoms of CRC are not obvious and colonoscopy is not widely used as a screening tool, timely diagnosis of CRC is a challenge and most cases are detected at the middle and late stages of disease progression. The current clinical treatments for CRC mainly include surgery, chemotherapy and radiotherapy[5]. However, surgical treatment is only suitable for patients with early diagnosis. For patients with advanced CRC, the efficacy of chemotherapy is affected by drug resistance and serious adverse reactions, and the overall treatment effect remains unsatisfactory. As a major threat to human health, CRC management is a significant challenge in the field of cancer research[6, 7]. Elucidation of the molecular mechanisms underlying its pathogenesis and identification of effective disease-targeting molecules with minimal side-effects are essential research goals for early diagnosis and treatment.
Lipids are an important component of cellular bio-membranes. In addition to energy storage and metabolism, lipids serve as critical signaling molecules for multiple cellular activities[8]. Regulation of lipid metabolism (such as lipid uptake, synthesis, and hydrolysis) is critical for maintaining cellular homeostasis[9]. Abnormal lipid metabolism is clearly associated with various diseases such as diabetes, cancer, and neurodegenerative disorders[10]. For example, intestinal tumor cells often exhibit abnormal activation of lipid metabolism[11]. Cancer cells in the tumor micro-environment can increase uptake of exogenous lipids or up-regulate endogenous lipogenesis and cholesterol synthesis in order to meet the needs of continuous proliferation during growth and metastasis[12]. Abundant lipids and lipid metabolites are utilized to provide energy and promote rapid tumor cell growth and metastasis. Accordingly, abnormal lipid metabolism is one of the hallmark features of cancer that has attracted considerable research attention in recent years[13].
Earlier studies have implicated dysregulation of lipid or lipid metabolism-related genes in the occurrence and development of CRC, supporting their value as potential biomarkers for early detection. Therefore, key genes in lipid metabolism could serve as molecular targets for CRC therapy and further elucidation of the underlying mechanisms may have clinical significance[14, 15]. Fatty acid binding proteins (FABPs) are intracellular fatty acid carriers that coordinate lipid responses, function in cellular fatty acid utilization, and are highly associated with metabolic and inflammatory pathways. Nine FABP genes have been identified in mammals to date, designated FABP1-7, FABP9 and FABP12. FABPs exist in different forms in various tissues, with unique roles and expression patterns in multiple cancer types[16].
FABP5 is a relatively low molecular weight lipid chaperone protein involved in regulation of various biological processes, such as fatty acid uptake and transport. FABP5 is highly expressed in various cancers and closely related to tumor growth, development and metastasis. In an earlier cervical cancer study, FABP5 was shown to promote epithelial-mesenchymal transition and lymph node metastasis by reprogramming fatty acid (FA) metabolism. Mechanistically, FABP5 enhanced lipolysis and FA synthesis and activated NF-κB signaling, leading to increased levels of intracellular FA, thereby inducing lymph node metastasis[17]. Another study on a mouse model of lung tumor metastasis reported that mice lacking FABP5 were more prone to metastasis. Further studies revealed that FABP5 deficiency leads to impaired NK cell maturation in the lung and FABP5 controls NK cell maturation to regulate lung tumor metastasis[18]. FABP5 may therefore be of significant clinical value in CRC, since this tumor type is closely related to lipid metabolism.
Methods
Human CRC cell lines and tissues
Human CRC cells HCT116 and SW620 were cultured in RPMI-1640 (Biological Industries, Israel) and L15 medium (Fuheng, Shanghai, China), respectively. 293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM, Biological Industries) supplemented with 10% fetal bovine serum (FBS, Yeasen Biotechnology, Shanghai, China) and 1% penicillin-streptomycin solution (New Cell & Molecular Biotech, Suzhou, China). All cells were grown in a cell incubator under 5% CO2 and 37 °C.
Quantitative real time-polymerase chain reaction (qPCR)
TRIzol (Vazyme, Nanjing, China) was used for extraction of total RNA. Relative cDNA was synthesized using a specific cDNA synthesis kit (Yeasen) under the following conditions: 42 °C for 2 min to digest genomic DNA, followed by 25 °C (5 min), 55 °C (15 min) and 85 °C (5 min) for reverse transcription. Hieff Universal Blue SYBR Green Mix (Yeasen) was used for the qPCR assay (Roche). The PCR protocol was as follows: initial denaturation at 95°C for 5 min, followed by 35 cycles at 95°C (30 s), 58°C (30 s), and 72°C (30 s). GAPDH was used as the internal control. The primers utilized are specified in Supplementary Table 1. Expression of genes was analyzed using GraphPad Prism 6 software.
Stable transfection of cell lines
FABP5, FASN and m6A molecules plasmids were purchased from Genomeditech (Shanghai, China) for construction in PLKO1 (knockdown) or PLVX (over-expression) vectors. The short hairpin targets used are presented in Supplementary Table 2. Lentivirus was packaged into 293T cells using PEI MAX transfection reagent (Polysciences, USA). Briefly, 50 µL PEI-MAX transfection reagent was added to 5 µg FABP5 plasmid along with equivalent amounts of two auxiliary plasmids (PAX2 and PDM2G at a 1:1 ratio), followed by the addition of 1.5 mL serum-free DMEM. After 30 min, plasmids were added to cells in serum-free DMEM. After 6 h, 10% FBS was added to cells and virus collected 48 h later. Stably transfected cells were acquired after virus infection and puromycin screening.
Cell proliferation assays
Proliferation was detected with CCK-8 (New Cell & Molecular Biotech). Briefly, 5×103 cells were cultured in 100 µL medium in 96-well plates, incubated with 10 µL reagent for 2 h, and analyzed using a microplate reader at a wavelength of 450 nm. For the colony formation assay, 1 × 104 cells were seeded in a 6-well plate and cultured for one week, followed by fixing with 4% paraformaldehyde and staining with 0.25% crystal violet. The 5-ethynyl-2′-deoxyuridine (EdU) assay was conducted as described previously. Briefly, a 1:1000 dilution of EdU was added to 96-well cell plates. After 2 h, cells were subjected to fixing, EdU staining, Hoechst 33342 staining, and washing (PBS), and images obtained under a microscope.
Cell migration and invasion assays
For the cell migration and invasion assays, 8 μm micropore inserts in 24-well cell culture plates were used. For cell migration experiments, 2 × 105 cells were seeded into upper wells without FBS. For cell invasion experiments, 4 × 105 cells were seeded into upper wells coated with 50 μL diluted matrigel (Becton, Dickinson) without FBS. In transwell assay, 30% FBS was added to lower wells. Wells were fixed with 4% paraformaldehyde for 10 min and stained with 0.25% crystal violet for 20 min.
Western blot
Cellular proteins were extracted using NP40 lysis buffer (Beyotime, Nantong, China) on ice for 30 min. After measuring protein concentrations using the Bradford method (Beyotime), samples were boiled for 10 min at 100 ℃ in 1× SDS protein loading buffer (Yeasen). Following standard electrophoresis and transfer, the membrane (Millipore, USA) was blocked with skimmed milk (8%) for 60 min. Next, primary antibodies (listed in Supplementary Table 3) were added to the membrane on a glass plate for overnight incubation at 4 °C. After washing three to four times with Tris-buffered saline with Tween 20(TBST) buffer, samples were incubated with the appropriate secondary antibodies for 1 h at room temperature. After washing with TBST for three times, signals of bands were detected using the Enhanced Chemiluminescent Reagent kit (New Cell & Molecular Biotech). Image J software was used to quantitative gray value of the bands.
Co-immunocoprecipitation
RIPA lysis (1 ml) were added to 10 cm dish with cells and proteins were extracted following western blot methods. After incubation with 2 µg antibody for 2 h at 4°C, 30 µl protein A/G magnetic beads (Beyotime) was added and inverted overnight. Next day, samples were washed thoroughly with RIPA lysis buffer (Beyotime) three times, incubated in 30 µl of 2×SDS-PAGE sample loading buffer (Beyotime) and boiled at 100°C for 10 min for subsequent western blot experiments.
Animal assays
For generation of animal models, 1 × 106 CRC cells from each group were subcutaneously injected into the flanks of 4-6 week old BALB/c nude mice in 100 µL PBS. To ascertain the effect of orlistat on tumor growth in vivo, orlistat in oil (10 mg/kg/day) was intragastrically administered following transplantation of cells into mice. After three weeks, mice were sacrificed and tumors collected to evaluate volumes and weights. Subsequently, tumor tissues were fixed and slides prepared for immunohistochemistry of FABP5 and FASN. The immunohistochemistry procedure was conducted as described previously[19]. Animal studies were approved by Institutional Animal Care and Use Committee of Nanjing Medical University.
Statistical analysis
Results are presented as mean ±SD and analyzed using GraphPad Prism 6.0 software. Student's t-test was used to assess significant differences in two-group comparisons. P values < 0.05 were considered significant. All in vitro assays were independently repeated at least three times.
Results
FABP5 is down-regulated in CRC
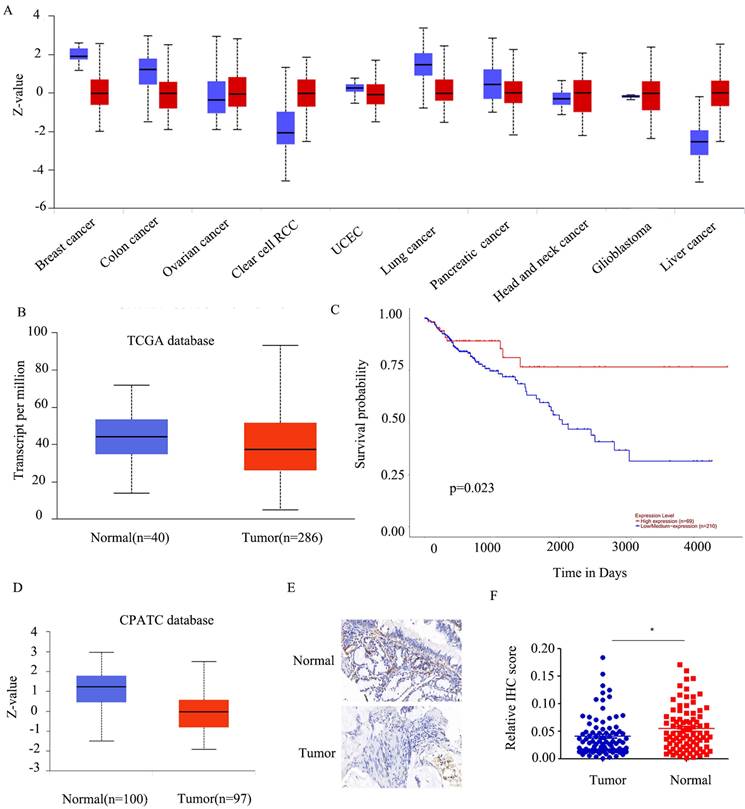
To establish the precise function of FABP5 in cancer, we investigated its expression patterns in various tumor types. FABP5 was over-expressed in renal clear cell carcinoma and liver cancer and, conversely, down-regulated in lung, breast and colon cancer (Figure 1A). Moreover, the FABP5 mRNA level was lower, but not to a significant extent, in tumor than control tissues (Figure 1B). Data from survival analyses indicated better prognosis of patients with higher FABP5 levels (Figure 1C).
Consistent with findings from the CPATC database (Figure 1D), our results showed down-regulation of FABP5 in CRC. As few reports have explored the function of FABP5 in CRC, we initially established a tumor microarray using 90 CRC and peri-tumorous tissues. And the results indicated FABP5 significantly down-regulated in CRC (Figure 1E-1F).
FABP5 is silenced in CRC. A) FABP5 protein levels in ten cancers from the CPATC database. (B) FABP5 was down-regulated in CRC compared to normal tissues in the TCGA database(p=2.3e-1). (C) Low expression of FABP5 was associated with shorter survival probability in CRC cases from the TCGA database. (D) Protein expression of FABP5 in CRC from the CPATC database(p=7.73e-13). (E) Representative immunohistochemical staining of FABP5 in CRC tissues from patients, magnification: ×73. (F) Relative average optical density values in 90 CRC and paired normal tissues. (*P < 0.05)
Over-expression of FABP5 is associated with reduced CRC cell proliferation, migration and invasion
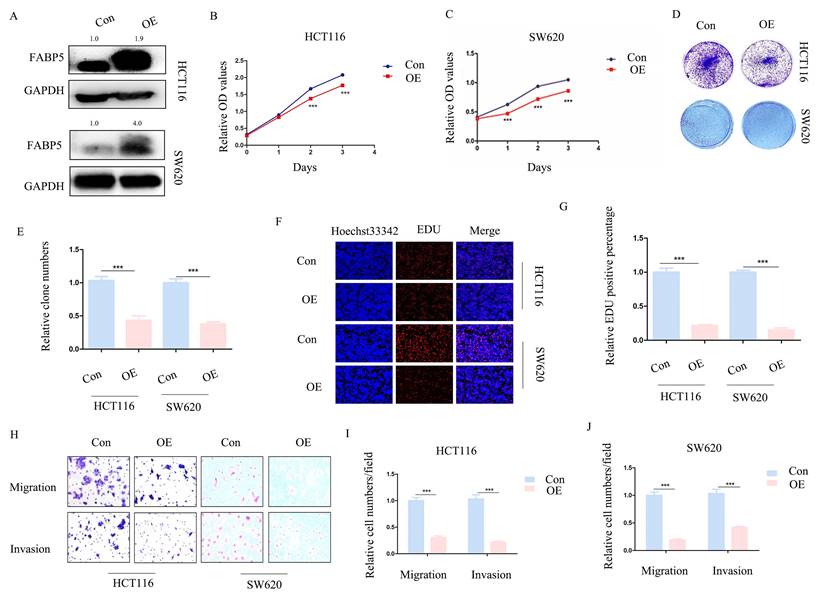
To ascertain whether FABP5 plays a tumor suppressor role in CRC, stable FABP5 over-expressing HCT116 and SW620 cell lines were generated via stable transfection. Western blot results validated the efficiency of FABP5 over-expression (Figure 2A). In CCK-8 and EdU assays, up-regulation of FABP5 was concomitant with suppression of cell proliferation (Figure 2B-2C; 2F-2G). Moreover, data from the clone formation assay showed decreased clone numbers in FABP5-expressing HCT116 and SW620 cells (Figure 2D-2E). In the transwell assay, up-regulation of FABP5 suppressed cell migration and invasion (Figure 2H-2J). Overall, FABP5 functioned as anti-cancer in CRC.
Knockdown of FABP5 promotes CRC cell proliferation, migration and invasion
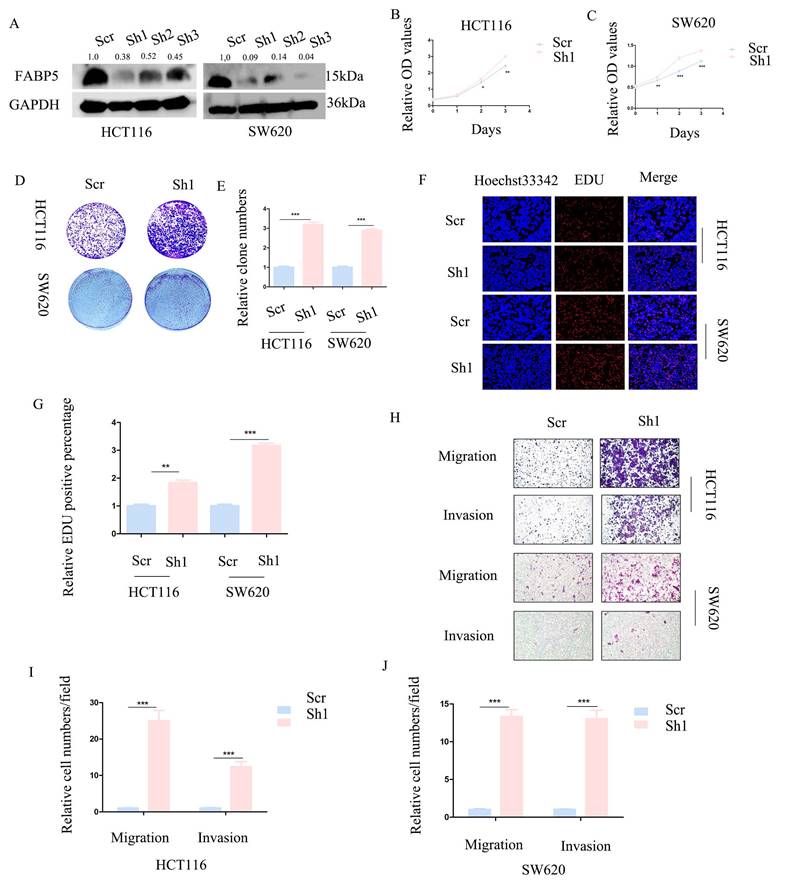
To further establish the anti-cancer function of FABP5 in CRC, HCT116 and SW620 cell lines with stable knockdown of FABP5 were generated. Western blot analysis validated the efficiency of FABP5 knockdown (Figure 3A). In CCK-8 and EdU experiments, down-regulation of FABP5 promoted cell proliferation (Figures 3B-3C, 3F-3G). Notably, suppression of FABP5 increased the clone numbers of HCT116 and SW620 in the clone formation assay (Figure 3D-3E). Data from the transwell assay showed that FABP5 silencing facilitated migration and invasion of HCT116 and SW620 cells (Figure 3H-3J), supporting a tumor suppressor role of FABP5 in CRC cells.
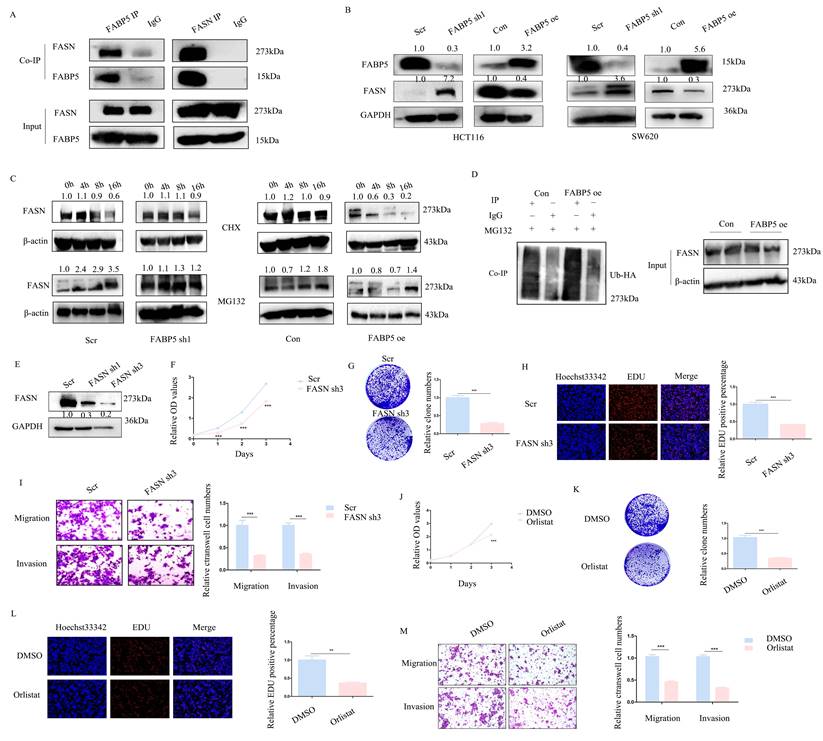
FABP5 interacts with FASN and promotes its ubiquitin proteasome pathway
With the aid of combined immunoprecipitation and mass spectrometry analyses, 395 proteins interacting with FABP5 were identified. Among these proteins, FASN attracted us attention, which was a fatty acid synthase. Subsequent Co-IP experiments confirmed interactions of FABP5 with FASN (Figure 4A). Notably, knockdown of FABP5 led to an increase in FASN expression and, conversely, over-expression of FABP5 induced a decrease in FASN (Figure 4B). After treatment with CHX, the stability of FASN was increased in FABP5 knockdown and decreased in FABP5 over-expression groups. Moreover, the FASN level was markedly increased upon MG132 treatment (Figure 4C). Co-IP assay of FASN and ubiquitin consistently showed that over-expression of FABP5 led to an increase in ubiquitin combined with FASN (Figure 4D). To further resolve its function in CRC, FASN was inhibited via shRNA or treatment with orlistat. Under conditions of knockdown of FASN (Figure 4E, Supplementary Figure 1A), cell proliferation (Figure 4F, 4H; Supplementary Figure 1B, 1E-1F), clone formation (Figure 4G, Supplementary Figure 1C-1D), cell migration and invasion (Figure 4I; Supplementary Figure 1G-1H) were inhibited. Similar phenomena were observed in the orlistat treatment group (Figure 4J-4M; Supplementary Figure 1I-1Q). The collective results clearly indicate that FABP5 regulates FASN via stimulation of its ubiquitin proteasome pathway.
Over-expression of FABP5 suppresses cell proliferation, migration and invasion. Western blot analysis of alterations in protein levels of FABP5 in over-expressing and control CRC cells. (B, C) Over-expression of FABP5 inhibited proliferation of HCT116 (B) and SW620 (C) cells, determined via the CCK8 assay. (D, E) Up-regulation of FABP5 induced a decrease in the number of colonies. (F, G) EdU assay showed suppression of proliferation in both cell types under conditions of FABP5 over-expression, magnification: ×200. (H-J) FABP5 suppressed migration and invasion of HCT116 and SW620 cells, magnification: ×100. (**P < 0.01, ***P < 0.001)
Knockdown of FABP5 promotes malignant biological behaviors of CRC cells. (A) Western blot analysis of the protein levels of FABP5 in knockdown and control CRC cells. (B, C) CCK-8 data showing that silencing of FABP5 promoted proliferation of HCT116 (B) and SW620 (C) cells. (D, E) Down-regulation of FABP5 induced an increase in the number of colonies. (F, G) Down-regulation of FABP5 accelerated proliferation of HCT116 and SW620 cells, as observed with the EdU assay, magnification: ×200. (H-J) Silence of FABP5 facilitated cell migration and invasion of both cell types, magnification: × 100. (*P < 0.05, **P < 0.01, ***P < 0.001).
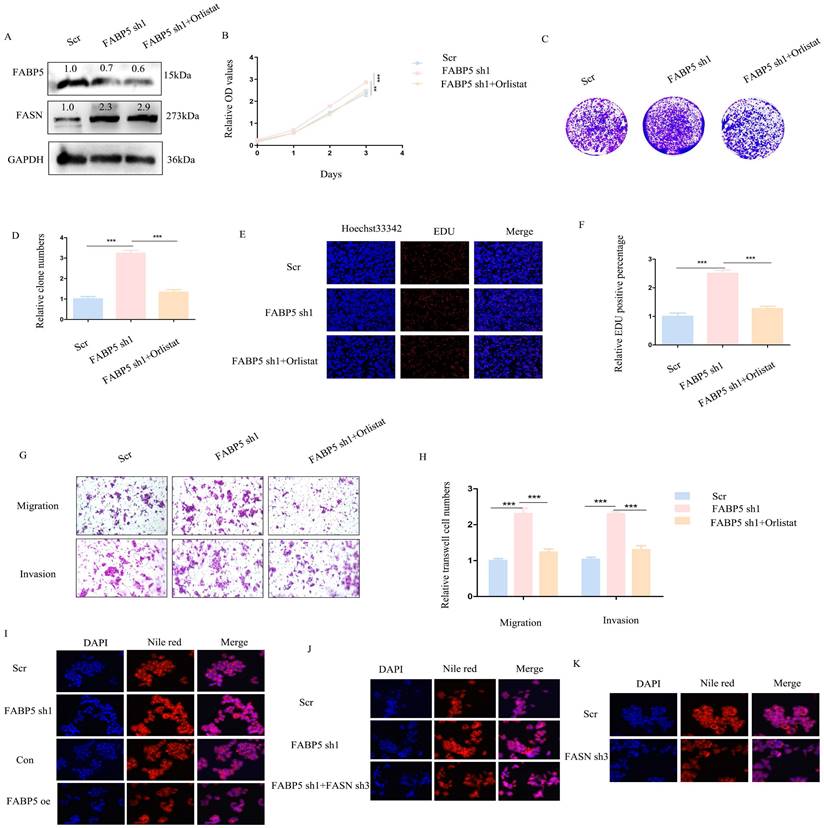
Knockdown or inhibition of FASN suppresses malignant biological behaviors activated by down-regulation of FABP5
Above results indicated down-regulation of FABP5 promotes malignant biological behaviors in CRC. To explore whether oncogenic activity is mediated by up-regulation of FASN, knockdown of FASN or orlistat treatment in FABP5 down-regulation stably transfected cells was performed. Firstly, efficiency of knockdown was detected via western blot (Figure 5A; Supplementary Figure 2A). CCK-8, colony formation and EdU experiments revealed that down-regulation of FASN or inhibition of its activity led to suppression of cell proliferation promoted by knockdown of FABP5 (Figure 5B-5F; Supplementary Figure 2B-2F). In the transwell assay, silencing or inhibition of FASN reversed the increase in migration and invasion induced by down-regulation of FABP5 (Figure 5G-5H; Supplementary Figure 2G-2H). Nile red staining demonstrated that over-expression of FABP5 decreased while its knockdown increased lipid accumulation (Figure 5I). FASN depletion resulted in attenuation of lipid accumulation induced by FABP5 silencing, highlighting a key role of FASN in FABP5-regulated malignant behaviors (Figure 5J). Moreover, sole knockdown of FASN in wild-type CRC cells consistently led to a decrease in lipid accumulation (Figure 5K). These results suggest that oncogenic progression initiated by down-regulation of FABP5 is restored by FASN inhibition in CRC cells.
FABP5 functions as a tumor suppressor via interacting with FASN. (A) Co-IP of FABP5 and FASN in HCT116 cells. (B) Western blot analysis of FASN levels in FABP5 over-expressing and silenced HCT116 and SW620 cells. (C) Western blot analysis of FASN in FABP5 over-expressing and depleted HCT116 cells subjected to CHX (10 µmol/L) and MG132 (10 μmol/L) treatment. (D) Co-IP analysis of FASN and ubiquitin in FABP5 over-expression HCT116 cells treated with MG132 (10 μmol/L) for 6 h. (E) Western blot showing FASN knockdown efficiency in HCT116 cells. (F) Silencing of FASN inhibited proliferation of HCT116 cells. (G, H) Down-regulation of FASN induced a decrease in colony number and proliferation of cells. (I) Silencing of FASN suppressed cell migration and invasion, magnification: ×100. (J-L) Cell proliferation was significantly decreased upon treatment with orlistat (50 µmol/L for 24 h), as observed with CCK-8 (J), colony formation (K) and EdU assays (L), magnification: ×200. (M) Orlistat inhibited migration and invasion of HCT116 cells, magnification: ×100. (**P < 0.01, ***P < 0.001)
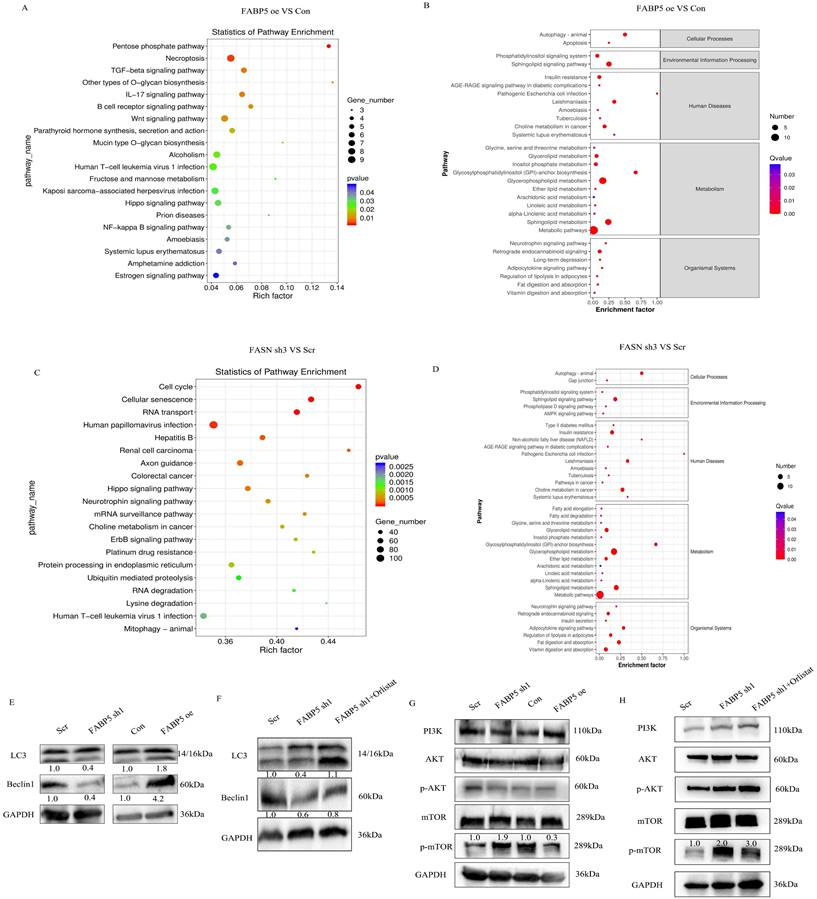
FABP5 promotes autophagy via inactivation of the mTOR pathway mediated by FASN
To explore the mechanisms underlying the tumor suppressor role of FABP5 in CRC, RNA-seq and lipid-omics were performed in FABP5 over-expression and FASN knockdown along with the respective control groups. Multiple genes associated with numerous biological processes and signaling pathways were dysregulated, including TGF-β, Hippo, Wnt, NF-κB and mTOR, as observed via RNA-seq (Figure 6A, 6C; Supplementary Figure 3A). Lipid-omic analyses further revealed that FABP5 and FASN regulated autophagy through effects on lipid metabolism (Figure 6B, 6D; Supplementary Figure 3B). Western blot results confirmed that up-regulation of FABP5 promoted while knockdown of FABP5 inhibited autophagy (Figure 6E). Moreover, silence of FASN or treatment with orlistat rescued inhibition of autophagy induced by FABP5 silencing (Figure 6F, Supplementary Figure 2J, Supplementary Figure 3C, 3E). Similar to this finding, over-expression of FABP5 suppressed, while knockdown of FABP5 activated, the mTOR pathway (Figure 6G). Both silencing of FASN and treatment with FASN inhibitor inactivated the mTOR pathway (Figure 6H; Supplementary Figure 2I; Supplementary Figure 3D, 3F), supporting a tumor suppressor role of FABP5 via inhibition of mTOR through FASN.
Orlistat restores CRC cell malignant behaviors induced by FABP5 down-regulation. (A) Western blot showing FABP5 and FASN protein levels under conditions of FABP5 silencing and 50 μmol/L orlistat treatment for 24 h. (B-F) CCK-8, colony formation and EdU assays (Magnification: ×200) showing cell proliferation under conditions of FABP5 knockdown and orlistat treatment. (G, H) Orlistat reverses CRC cell migration and invasion (Magnification: ×100) induced by FABP5 silencing. (I-K) Nile red staining (Magnification: ×200) in FABP5/FASN altered cells. (***P < 0.001)
FABP5 interacts with FASN to promote cell autophagy via mTOR. (A-D) Enrichment analysis of transcriptome and lipid metabolomes. (E, F) Western blot analysis of LC3 and Beclin1 in FABP5 over-expression or down-regulation and FABP5 down-regulation+orlistat treatment groups. (G, H) Western blot analysis of PI3K/AKT/mTOR in FABP5 over-expression or down-regulation and FABP5 +orlistat treatment groups.
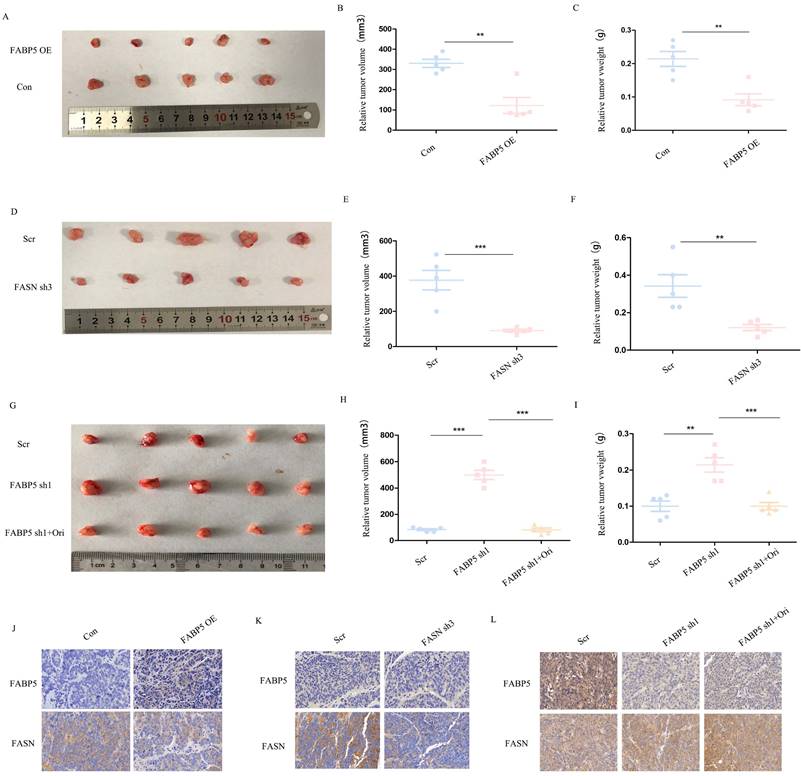
Over-expression of FABP5 and knockdown of FASN inhibits tumor growth in vivo
We further examined the roles of FABP5 and FASN in vivo with the aid of tumor xenograft models. To this end, SW620 cells transfected with FABP5 over-expression or FASN knockdown vector, FABP5 knockdown vector plus orlistat and control vector were implanted in nude mice. Notably, the weights and volumes of tumors were decreased in mouse xenografts injected with SW620 cells bearing FABP5 over-expression and FASN knockdown vectors (Figure 7A-7F). Conversely, SW620 cells with stable FABP5 knockdown showed accelerated tumor growth compared with the control group, which was suppressed by orlistat (Figure 7G-7I). Immunohistochemical staining of FABP5 and FASN revealed FABP5 negatively regulated FASN expression in vivo (Figure 7J-7L). In summary, our collective results support an essential role of FABP5 in FASN-mediated CRC progression, both in vitro and in vivo.
ALKBH5 regulates FABP5 in an m6A-independent manner
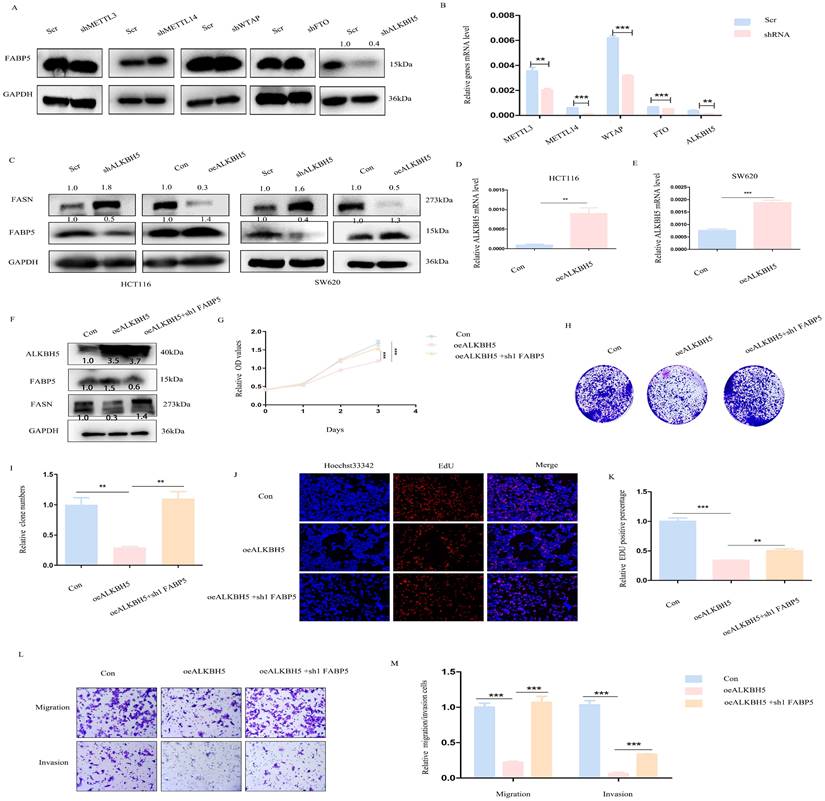
Next, we focused on the mechanisms underlying down-regulation of FABP5 in CRC. The m6A prediction server, SRAMP, revealed abundant m6A modification sites in FABP5. Upon knockdown of m6A writers (METTL3, METTL14, WTAP) and erasers (FTO and ALKBH5), positive regulation of FABP5 was observed in the absence of ALKBH5 (Figure 8A, 8B). Furthermore, ALKBH5 negatively modulated the FASN level (Figure 8C-8E). To explore whether ALKBH5 inhibited CRC via FABP5, we additionally depleted FABP5 in ALKBH5 over-expressing cells (Figure 8F). The anti-cancer effects of ALKBH5 (including suppression of cell proliferation, migration and invasion) were partly restored by silencing of FABP5 which indicated FABP5 could be downstream gene of ALKBH5(Figure 8G-8M).
FABP5 suppresses tumor growth via regulation of FASN in vivo. (A-C) Primary tumor samples obtained from mice subcutaneously injected with HCT116 cells transfected with FABP5 over-expression and control cell groups (A). Relative tumor volumes (B) and weights (C) at the endpoint (n = 5). (D-F) Primary tumor samples were obtained from mice subcutaneously injected with HCT116 cells transfected with FASN knockdown and control group (D). Relative tumor volumes (E) and weights (F) measured at the endpoint (n = 5). (G-H) Primary tumor samples obtained from mice subcutaneously injected with HCT116 cells with FABP5 silencing, FABP5 silencing plus orlistat treatment, and control groups (G). Relative tumor volumes (H) and weights (I) measured at the endpoint (n = 5). (J-L) Representative immunohistochemistry images showing expression of FABP5 and FASN in xenograft tumor tissues, magnification: ×73. (**P < 0.01, ***P < 0.001)
ALKBH5 positively regulates FABP5 to exert anti-cancer effects. (A) Western blots showing positive regulation of FABP5 by ALKBH5 via down-regulation of m6A writers (METTL3, METTL14, WTAP) and erasers (FTO, ALKBH5). (B) Knockdown efficiency of m6A molecules assessed via RT-PCR. (C) Western blot showing FABP5 and FASN expression under conditions of ALKBH5 up-regulation and down-regulation. (D, E) Efficiency of ALKBH5 over-expression assessed via RT-PCR. (F) ALKBH5, FABP5 and FASN protein levels evaluated under conditions of ALKBH5 over-expression, ALKBH5 over-expression with FABP5 knockdown and control in HCT116 cells. (G-M) FABP5 knockdown reversed the decrease in cell proliferation, colony formation, migration and invasion induced by ALKBH5 in CCK-8 (G), colony formation (H, I), EdU (J, K; Magnification: ×200) and transwell assays (L, M; Magnification: ×100). (**P < 0.01, ***P < 0.001)
Discussion
Lipid metabolism plays an important role in tumorigenesis and development[20, 21]. During malignant progression, the availability of nutrients in the tumor micro-environment constantly changes and tumor cells utilize the lipid metabolism to sustain rapid proliferation and metastasis[22, 23]. Disruption of lipid metabolism is an upcoming novel therapeutic strategy. Therefore, clarification of the molecular mechanisms underlying the association of CRC with lipid metabolism may provide effective therapeutic targets. In this study, we comprehensively explored the roles of FABP5 and FASN in CRC. Our data showed that FABP5 is down-regulated in CRC tissues and functions as a tumor suppressor via interactions with FASN. Furthermore, FABP5 is regulated upstream by ALKBH5, a m6A demethylase. Importantly, knockdown or inhibition of FASN dramatically suppressed tumor progression, both in vivo and in vitro.
As fatty acids play a critical role in lipid metabolism, FABP5 and FASN have been widely studied in numerous diseases, particularly cancer[24-26]. FABP5 is up-regulated and promotes tumor development in gastric cancer, breast cancer, cervical cancer, prostate cancer and hepatocellular carcinoma[25]. FABP5 silencing has been shown to decrease cell proliferation and lead to cell cycle arrest in gastric cancer.[27] In breast cancer, FABP5 activates VEGF and up-regulates EGFR expression, in turn, increasing PPAR to promote cancer progression[28]. Similar phenomena are reported in prostate cancer, showing that FABP5 activates PPARγ and up-regulates VEGF[29]. In cervical cancer, FABP5 reprograms lipid metabolism and facilitates epithelial-mesenchymal transition and lymph node metastasis via activating the NF-κB pathway[17]. Moreover, FABP5 is reported to accelerate tumor metastasis via regulation of MMP9 and MMP2[30]. However, opposite patterns are observed in skin tumors, whereby FABP5 suppresses skin tumorigenesis through regulation of the IFN/p53/SOX2 pathway[31]. These conflicting findings suggest complex roles of FABP5 in cancer development. The function of FABP5 in CRC is not well understood at present.
Based on immunoprecipitation and mass spectrometry analyses, FASN was identified as an interacting protein with FABP5. Furthermore, our results suggested that FABP5 destabilizes FASN via activating its ubiquitin-proteasome pathway. Orlistat, a potent and long-acting specific gastrointestinal fatty acid synthase inhibitor, is the only FDA-approved over-the-counter (OTC) diet medication available worldwide. Jin and co-workers reported that orlistat could alleviate colon cancer induced by western diet-associated colitis via suppression of STAT3 and NF-κB signaling pathways[32]. The group of Zhou and colleagues demonstrated that orlistat decreases GPX4 and increases lipid peroxidation, promoting ferroptosis in lung cancer cells[33]. In the current investigation, knockdown or inhibition of FASN significantly inhibited proliferation, migration and invasion of CRC. Moreover, orlistat reversed cell malignant behaviors induced by FABP5 knockdown, clearly indicating that cancer progression is induced by FABP5 down-regulation through effects on FASN. Autophagy is considered an important biological process in tumor development. Classical autophagy is mediated by mTOR signaling[34, 35]. Data from the present study showed that over-expression of FABP5 and concomitant down-regulation of FASN result in inactivation of the mTOR pathway, and consequently, increased autophagy. However, the effect of FABP5/FASN on mTOR is not mediated by the known upstream PI3K/AKT pathway.
ALKBH5 is a type of RNA demethylase (also known as eraser), which is down-regulated in CRC. We detected numerous m6A modification sites in FABP5 mRNA in CRC. Data from knockdown studies on m6A methylase and demethylase showed positive regulation of FABP5 expression by ALKBH5. Moreover, FABP5 silencing reversed the anti-cancer activity of ALKBH5. However, a separate RNA sequencing study (MeRIP-seq) by our group showed no regulatory effects of ALKBH5 on FABP5. Accordingly, we assume that ALKBH5 regulates FABP5 in a non-m6A-dependent manner.
In summary, the present study shows that FABP5 is down-regulated and FASN is induced via suppression of the ubiquitin-proteasome pathway, leading to regulation of autophagy via mTOR signaling in CRC. Under conditions of FABP5 deficiency, lipid accumulation is elevated, ultimately accelerating CRC progression. Our study supports a crucial role of the ALKBH5/FABP5/FASN/mTOR axis in regulation of CRC progression and offer promising therapeutic targets for management of the disease.
Abbreviations
CRC: Colorectal cancer
FABP5: Fatty acid-binding protein 5
FASN: Fatty acid synthase
Co-IP: Co-Immunoprecipitation
CCK8: Cell counting kit-8
DMEM: Dulbecco's modified eagle medium
FBS: Fetal bovine serum
RT-qPCR: Quantitative real-time polymerase chain reaction
GAPDH: Glyceraldehyde-3-phosphate dehydrogenase
EdU: 5-Ethynyl-2'-deoxyuridine
Supplementary Material
Supplementary figures and tables.
Acknowledgements
Funding
This work was supported by Science Foundation Project of Ili & Jiangsu Joint Institute of Health (grant number: yl2020lhms05) and and Wuxi "Taihu talent plan" for the excellent medical expert team (Grant No. 2021-9).
Ethics approval and consent to participate
Animal study was approved by Institutional Animal Care and Use Committee (IACUC)of Nanjing Medical University.
Author contributions
TQ, TY and YM designed the study; YM, HC, CT, YP and BJ performed experiments and collected all data; YP, ZY, YL, CJ, XL and LF analyzed and dealt with the data; YM, HC, CT, YP and BJ participanted writing and figures making. TQ, KJ, TY and LX involved in critical reviewing of the manuscript. All authors read and approved the manuscript.
Availability of data and material
All of the data of this study are available from the corresponding author.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. TRANSL ONCOL. 2021 14; 101174
2. Ballester V, Rashtak S, Boardman L. Clinical and molecular features of young-onset colorectal cancer. World J Gastroenterol. 2016;22:1736-1744
3. Amintas S, Dupin C, Boutin J, Beaumont P, Moreau-Gaudry F, Bedel A. et al. Bioactive food components for colorectal cancer prevention and treatment: A good match. Crit Rev Food Sci Nutr. 2022:1-15
4. Wekha G, Ssewante N, Iradukunda A, Jurua M, Nalwoga S, Lanyero S. et al. Colorectal Cancer in Uganda: A 10-Year, Facility-Based, Retrospective Study. CANCER MANAG RES. 2021;13:7697-7707
5. Zhang F, Su T, Xiao M. RUNX3-regulated circRNA METTL3 inhibits colorectal cancer proliferation and metastasis via miR-107/PER3 axis. CELL DEATH DIS. 2022;13:550
6. Li D, Jiang S, Zhou X, Si C, Shao P, Jiang Q. et al. FBXW7 and Its Downstream NOTCH Pathway Could be Potential Indicators of Organ-Free Metastasis in Colorectal Cancer. FRONT ONCOL. 2021;11:783564
7. Jing Z, Liu Q, He X, Jia Z, Xu Z, Yang B. et al. NCAPD3 enhances Warburg effect through c-myc and E2F1 and promotes the occurrence and progression of colorectal cancer. J Exp Clin Cancer Res. 2022;41:198
8. Choi S, Snider AJ. Diet, lipids and colon cancer. Int Rev Cell Mol Biol. 2019;347:105-144
9. Wang Y, Hinz S, Uckermann O, Honscheid P, von Schonfels W, Burmeister G. et al. Shotgun lipidomics-based characterization of the landscape of lipid metabolism in colorectal cancer. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865:158579
10. Yang Y, He J, Zhang B, Zhang Z, Jia G, Liu S. et al. SLC25A1 promotes tumor growth and survival by reprogramming energy metabolism in colorectal cancer. CELL DEATH DIS. 2021;12:1108
11. Piccinin E, Cariello M, Moschetta A. Lipid metabolism in colon cancer: Role of Liver X Receptor (LXR) and Stearoyl-CoA Desaturase 1 (SCD1). MOL ASPECTS MED. 2021;78:100933
12. Grube S, Dunisch P, Freitag D, Klausnitzer M, Sakr Y, Walter J. et al. Overexpression of fatty acid synthase in human gliomas correlates with the WHO tumor grade and inhibition with Orlistat reduces cell viability and triggers apoptosis. J Neurooncol. 2014;118:277-287
13. Pakiet A, Kobiela J, Stepnowski P, Sledzinski T, Mika A. Changes in lipids composition and metabolism in colorectal cancer: a review. LIPIDS HEALTH DIS. 2019;18:29
14. Yang L, Zheng L, Xie X, Luo J, Yu J, Zhang L. et al. Targeting PLA2G16, a lipid metabolism gene, by Ginsenoside Compound K to suppress the malignant progression of colorectal cancer. J ADV RES. 2022;36:265-276
15. Gong J, Lin Y, Zhang H, Liu C, Cheng Z, Yang X. et al. Reprogramming of lipid metabolism in cancer-associated fibroblasts potentiates migration of colorectal cancer cells. CELL DEATH DIS. 2020;11:267
16. Prayugo FB, Kao TJ, Anuraga G, Ta H, Chuang JY, Lin LC. et al. Expression Profiles and Prognostic Value of FABPs in Colorectal Adenocarcinomas. Biomedicines. 2021 9
17. Zhang C, Liao Y, Liu P, Du Q, Liang Y, Ooi S. et al. FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. THERANOSTICS. 2020;10:6561-6580
18. Yang S, Kobayashi S, Sekino K, Kagawa Y, Miyazaki H, Kumar SS. et al. Fatty acid-binding protein 5 controls lung tumor metastasis by regulating the maturation of natural killer cells in the lung. FEBS LETT. 2021;595:1797-1805
19. Ye M, He J, Zhang J, Liu B, Liu X, Xie L. et al. USP7 promotes hepatoblastoma progression through activation of PI3K/AKT signaling pathway. CANCER BIOMARK. 2021;31:107-117
20. Li S, Fang Y. MS4A1 as a Potential Independent Prognostic Factor of Breast Cancer Related to Lipid Metabolism and Immune Microenvironment Based on TCGA Database Analysis. Med Sci Monit. 2022;28:e934597
21. Huang J, Wang J, He H, Huang Z, Wu S, Chen C. et al. Close interactions between lncRNAs, lipid metabolism and ferroptosis in cancer. INT J BIOL SCI. 2021;17:4493-4513
22. Zhang C, Zhu N, Li H, Gong Y, Gu J, Shi Y. et al. New dawn for cancer cell death: emerging role of lipid metabolism. MOL METAB. 2022 101529
23. Mece O, Houbaert D, Sassano ML, Durre T, Maes H, Schaaf M. et al. Lipid droplet degradation by autophagy connects mitochondria metabolism to Prox1-driven expression of lymphatic genes and lymphangiogenesis. NAT COMMUN. 2022;13:2760
24. Garcia KA, Costa ML, Lacunza E, Martinez ME, Corsico B, Scaglia N. Fatty acid binding protein 5 regulates lipogenesis and tumor growth in lung adenocarcinoma. LIFE SCI. 2022;301:120621
25. Xu B, Chen L, Zhan Y, Marquez K, Zhuo L, Qi S. et al. The Biological Functions and Regulatory Mechanisms of Fatty Acid Binding Protein 5 in Various Diseases. Front Cell Dev Biol. 2022;10:857919
26. Du Q, Liu P, Zhang C, Liu T, Wang W, Shang C. et al. FASN promotes lymph node metastasis in cervical cancer via cholesterol reprogramming and lymphangiogenesis. CELL DEATH DIS. 2022;13:488
27. Wang W, Liu Z, Chen X, Lu Y, Wang B, Li F. et al. Downregulation of FABP5 Suppresses the Proliferation and Induces the Apoptosis of Gastric Cancer Cells Through the Hippo Signaling Pathway. DNA CELL BIOL. 2021;40:1076-1086
28. Levi L, Lobo G, Doud MK, von Lintig J, Seachrist D, Tochtrop GP. et al. Genetic ablation of the fatty acid-binding protein FABP5 suppresses HER2-induced mammary tumorigenesis. CANCER RES. 2013;73:4770-4780
29. Forootan FS, Forootan SS, Gou X, Yang J, Liu B, Chen D. et al. Fatty acid activated PPARgamma promotes tumorigenicity of prostate cancer cells by up regulating VEGF via PPAR responsive elements of the promoter. Oncotarget. 2016;7:9322-9339
30. Hsiao YH, Chen NC, Koh YC, Nagabhushanam K, Ho CT, Pan MH. Pterostilbene Inhibits Adipocyte Conditioned-Medium-Induced Colorectal Cancer Cell Migration through Targeting FABP5-Related Signaling Pathway. J Agric Food Chem. 2019;67:10321-10329
31. Zhang Y, Hao J, Zeng J, Li Q, Rao E, Sun Y. et al. Epidermal FABP Prevents Chemical-Induced Skin Tumorigenesis by Regulation of TPA-Induced IFN/p53/SOX2 Pathway in Keratinocytes. J INVEST DERMATOL. 2018;138:1925-1934
32. Jin BR, Kim HJ, Sim SA, Lee M, An HJ. Anti-Obesity Drug Orlistat Alleviates Western-Diet-Driven Colitis-Associated Colon Cancer via Inhibition of STAT3 and NF-kappaB-Mediated Signaling. CELLS-BASEL. 2021 10
33. Zhou W, Zhang J, Yan M, Wu J, Lian S, Sun K. et al. Orlistat induces ferroptosis-like cell death of lung cancer cells. Front Med. 2021;15:922-932
34. Ye M, Lu H, Tang W, Jing T, Chen S, Wei M. et al. Downregulation of MEG3 promotes neuroblastoma development through FOXO1-mediated autophagy and mTOR-mediated epithelial-mesenchymal transition. INT J BIOL SCI. 2020;16:3050-3061
35. Ye M, Gao R, Chen S, Wei M, Wang J, Zhang B. et al. Downregulation of MEG3 and upregulation of EZH2 cooperatively promote neuroblastoma progression. J CELL MOL MED. 2022;26:2377-2391
Author contact
![]() Corresponding authors: Qiyun Tang, Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, NO.300 Guangzhou Road, Nanjing, China. Email: tqy831com. Ye Tian, Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, NO.300 Guangzhou Road, Nanjing, China. Email: tianye6626com. Mujie Ye, Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, NO.300 Guangzhou Road, Nanjing, China. Email: mujiey0629com
Corresponding authors: Qiyun Tang, Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, NO.300 Guangzhou Road, Nanjing, China. Email: tqy831com. Ye Tian, Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, NO.300 Guangzhou Road, Nanjing, China. Email: tianye6626com. Mujie Ye, Department of Geriatric Gastroenterology, Neuroendocrine Tumor Center, Jiangsu Province Hospital, The First Affiliated Hospital of Nanjing Medical University, Institute of Neuroendocrine Tumor, Nanjing Medical University, NO.300 Guangzhou Road, Nanjing, China. Email: mujiey0629com

 Global reach, higher impact
Global reach, higher impact