10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2024; 20(7):2532-2554. doi:10.7150/ijbs.95122 This issue Cite
Review
Autophagy in Its (Proper) Context: Molecular Basis, Biological Relevance, Pharmacological Modulation, and Lifestyle Medicine
1. Department of Medicine and Medical Specialities, Faculty of Medicine and Health Sciences, University of Alcalá, 28801 Alcala de Henares, Spain.
2. Ramón y Cajal Institute of Sanitary Research (IRYCIS), 28034 Madrid, Spain.
3. Department of Surgery, Medical and Social Sciences, Faculty of Medicine and Health Sciences, University of Alcalá, 28801 Alcala de Henares, Spain.
4. Unit of Biochemistry and Molecular Biology, Department of System Biology (CIBEREHD), University of Alcalá, 28801 Alcala de Henares, Spain
5. Department of General and Digestive Surgery, Príncipe de Asturias Universitary Hospital, 28805 Alcala de Henares, Spain.
6. Pathological Anatomy Service, Central University Hospital of Defence-UAH Madrid, 28801 Alcala de Henares, Spain.
7. Immune System Diseases-Rheumatology, Oncology Service an Internal Medicine (CIBEREHD), Príncipe de Asturias University Hospital, 28806 Alcala de Henares, Spain.
* These authors contributed equally.
Received 2024-2-6; Accepted 2024-4-4; Published 2024-4-22
Abstract
Autophagy plays a critical role in maintaining cellular homeostasis and responding to various stress conditions by the degradation of intracellular components. In this narrative review, we provide a comprehensive overview of autophagy's cellular and molecular basis, biological significance, pharmacological modulation, and its relevance in lifestyle medicine. We delve into the intricate molecular mechanisms that govern autophagy, including macroautophagy, microautophagy and chaperone-mediated autophagy. Moreover, we highlight the biological significance of autophagy in aging, immunity, metabolism, apoptosis, tissue differentiation and systemic diseases, such as neurodegenerative or cardiovascular diseases and cancer. We also discuss the latest advancements in pharmacological modulation of autophagy and their potential implications in clinical settings. Finally, we explore the intimate connection between lifestyle factors and autophagy, emphasizing how nutrition, exercise, sleep patterns and environmental factors can significantly impact the autophagic process. The integration of lifestyle medicine into autophagy research opens new avenues for promoting health and longevity through personalized interventions.
Keywords: ATG proteins, autophagosome, aging, pharmacological modulation, lifestyle habits
Introduction
Autophagy is a conserved cellular self-eating process that plays a critical role in maintaining cellular homeostasis and responding to stress. It involves the degradation and recycling of damaged components, organelles, and molecules to ensure an adequate function of the cell. The process of autophagy includes three main types: macroautophagy, microautophagy, and chaperone-mediated autophagy. Macroautophagy forms autophagosomes, double-membrane vesicles that engulf cytoplasmic components and fuse with lysosomes for degradation and recycling. In microautophagy, materials are directly taken up into lysosomes by invagination. Chaperone-mediated autophagy selectively transports proteins to lysosomes for degradation with the help of chaperones. Autophagy plays a crucial role in cell survival, proliferation, metabolism, senescence, modulation of host defenses, and various other processes that contribute to maintaining cell integrity and function (1). Because of that, a growing number of studies have identified autophagy as a major hallmark of health, but also of disease (2,3). In this context, autophagy modulation arises as a promising translational intervention for sustaining health, delaying aging and also to prevent or ameliorate the initiation and progression of an array of diseases (4,5). Two major strategies directed to regulate autophagy can be remarked: pharmacological agents, frequently directed to specific activators/inhibitors of autophagy (6) and lifestyle medicine, a growing area of research that include different strategies like diet, physical activity, sleep and other modifiable habits that also plays a major role in the activation/inhibition of autophagy (7).
This integrative narrative review aims to shed light on the role of autophagy in cellular homeostasis and its broader implications in diverse biological processes. By comprehensively exploring the cellular and molecular basis of autophagy, its importance in aging and disease, the broad pharmacological modulation and the impact of lifestyle factors, we hope to improve our understanding of autophagy and stimulate innovative research and clinical applications that harness the full potential of this process for human health and well-being.
Cellular and molecular basis of autophagy
Macroautophagy
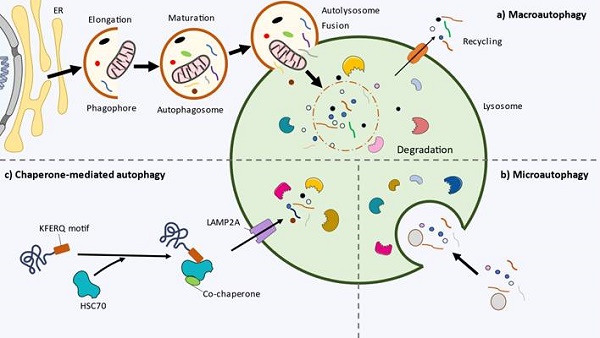
Macroautophagy, commonly referred to as autophagy, consists of the formation of an autophagosome, a double-membraned cytosolic vesicle, and the uptake of dysfunctional organelles and aggregated proteins leading to the degradation and recycling of these components after fusion with lysosomes (Figure 1a). In mammalian cells it develops in the following steps.
Initiation
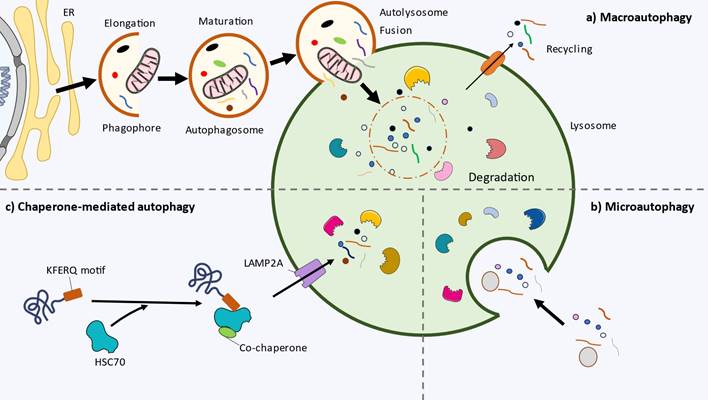
The initiation phase involves the activation of autophagy in response to various internal and external stimuli, such as nutrient deprivation (amino acid starvation or glucose depletion), hypoxia, oxidative stress, accumulation of damaged organelles, physical exercise or pathogen invasion (8,9). In these contexts, the upstream regulators of autophagy mammalian target of rapamycin complex 1 (mTORC1) and AMP-activated protein kinase (AMPK) are inhibited and activated, respectively. Thus this leads to the assembly and activation of the ULK complex, composed of ULK1/2 (unc-51 like autophagy activating kinase 1/2; yeast Atg1), ATG 13, ATG 101 and FIP200 (focal adhesion kinase (FAK)-interacting protein of 200 kDa, also known as RBCC1) (10). Once activated, ULK1/2 phosphorylates itself and the ATG13 and FIP200 proteins, leading to the assembly of the ULK1/2 complex (11). This complex then localizes to a specific site in the cell called the phagophore assembly site (PAS, also referred to as preautophagosomal structure), which serves as a platform for autophagosome biogenesis. The current consensus states that upon activation the ULK complex translocates to a specific localization of endoplasmic reticulum (ER) marked by ATG9 (12). Several mechanisms seem to be implicated in the recruitment of ULK1. They include the interaction of ULK1 with the gamma-aminobutyric acid receptor-associated protein (GABARAP), small GTPase RAB1A/ C9orf72 and ER contact proteins VAMP-associated protein A (VAPA) and VAPB (13).
Despite the similarity between ULK1 and ULK2 and the redundant or interchangeable role that had been assigned to them previously, there is growing evidence about their separate functions due to differences in autophagy‑related interactors and their post‑translational and transcriptional regulators (14). Furthermore, although it is well described the role of ULK1/2 complex in the initiation of autophagy, under certain conditions autophagy is initiated independently from it or mTOR, e.g.: acute hypoxia-induced autophagy or ammonia-induced autophagy (15,16).
mTOR, AMPK and p53 are the main upstream regulators of autophagy (17). Briefly, in basal conditions mTOR inhibits autophagy by phosphorylation of ULK1/2, ATG13 and the transcription factor EB (TFEB), which is in charge of the transcription of the autophagy machinery (18,19). During nutrient starvation, AMPK becomes activated and phosphorylates both ULK1/2, at additional sites to enhance its activation, and mTOR, specifically in the T-loop, resulting in mTOR inhibition. These events collectively trigger autophagy activation (20,21). The activation of the transcription factor p53 leads to the upregulation of ULK1/2, AMPK, or TSC2 through transcriptional processes. Conversely, p53 can also exert a negative regulatory effect on autophagy by inhibiting the expression of ULK1 and ATG7. Additionally, p53 can directly interact with BECN1, further modulating autophagy in response to various cellular conditions (22,23).
Nucleation
Activation of ULK1/2 complex leads to recruitment of class III phosphatidylinositol 3-kinase complex I (PI3KC3-C1), which consists of the lipid kinase PIK3C3/VPS34 (phosphatidylinositol 3-kinase catalytic subunit type 3), PIK3R4/VPS15/p150 (phosphoinositide-3-kinase regulatory subunit 4), BECN1/Beclin 1, ATG14/ATG14L and NRBF2 (nuclear receptor binding factor 2) (24). ULK1 phosphorylates the subunits of PI3K3C3-C1: PIK3C3 (S249), BECN1 (S15/30), ATG14 (S29) and PIK3R4 (S813/S861/S865/S879/S1039/S1289) (25-28). The phosphorylation of PIK3C3 facilitates its binding with GABARAP, GABARAPL1 (GABARAP-like 1), and LC3. Additionally, the phosphorylation of BECN1 and ATG14 enhances the lipid kinase activity of the complex. Together, these coordinated phosphorylation events enable the precise initiation of autophagy. The translocation of the PI3KC3-C1 is mediated by the subunit ATG14, essential for autophagy (29).
PIK3C3 is responsible for generating phosphatidylinositol 3-phosphate (PI3P), a key signaling lipid, at a cup-shaped ER subdomain known as the omegasome. It phosphorylates phosphatidylinositol at the 3' position on the inositol ring (30). PI3P and PIK3C3 also have significant roles in other cellular processes, including endocytic trafficking, phagocytosis, cytokinesis, and nutrient sensing (31). Inside the omegasomes, the phagophores (also known as isolation membranes) nucleate, elongate and engulf the cargo, eventually forming the autophagosome which dissociates from the ER (32,33). The curvature of the omegasome appears to facilitate the exposure of PI3P, resulting in the recruitment of PI3P-binding proteins, particularly double FYVE containing protein 1 (DFCP1) and WIPI/PROPPIN (WD-repeat protein interacting with phosphoinositides) family (34). DFCP1 is an ER-localized protein that relocates to the omegasome upon autophagy induction, but its precise function remains poorly understood. It is commonly used as a marker for the omegasome, and recent research indicates its involvement in lipid droplet formation within the ER (35). Moreover, recent studies have revealed that DFCP1 possesses ATPase activity, which mediates the release of autophagosomes specifically in selective autophagy processes, but not in non-selective autophagy (36). On the other hand, mammals possess four WIPI proteins, WIPI1-4, each characterized by seven bladed β-propellers. These proteins act as essential effectors of autophagy by binding to PI3P through the FRRG motif, inserting into the membrane through a hydrophobic loop in blade six, and subsequently recruiting downstream ATG proteins (37,38). In nucleation step, isoforms WIPI2B and WIPI2D bind and recruit the ATG5-ATG12-ATG16L1 complex by interacting with ATG16L1 (39,40). In this sense, Jensen et al. showed recently that the interaction between the ATG12-ATG5-ATG16L1 complex and the early phagophore rim may stabilize membrane curvature and promote the growth of the autophagosome (41). Interestingly, it seems that WIPI2 is recruited to the membrane by RAB11A, a protein of recycling endosomes, where PI3P is also formed upon starvation (42).
Cellular and molecular basis of autophagy. a) Macroautophagy. The initiation of autophagy and the nucleation of phagophore starts inside the endoplasmic reticulum (ER). During elongation, the phagophore engulfs the cytoplasmic materials. The phagophore closes forming the double-membrane vesicle autophagosome. Then, it fuses with the lysosome (autolysosome), where the inner autophagosomal membrane and cargo are degraded by the lysosomal hydrolases. The remaining catabolites are transported to cytoplasm and recycled by the cell. b) Microautophagy. The cytoplasmic material is captured through direct invagination of the lysosome. c) Chaperone-mediated autophagy (CMA). In multicellular organisms, proteins with the pentapeptide motif KFERQ are recognised and transported by the chaperone HSC70 and cochaperones to the lysosome. The LAMP2A receptor imports the content to the lysosome.
Elongation
During the elongation phase of autophagy, the phagophore undergoes expansion and elongation, a process facilitated by the recruitment of autophagy-related proteins and the lipidation of LC3, a ubiquitin-like protein. This crucial step enables the autophagosome to engulf cellular cargo for subsequent degradation and recycling. Key to the elongation phase is the lipidation of the microtubule-associated protein 1 light chain 3 (LC3), a member of the ATG8 family (43). LC3 is initially synthesized as an inactive precursor, which Arg C-terminus residues are removed by the cysteine protease ATG4. Then, ATG7 (E1-like activating enzyme), ATG3 (E2-like conjugating enzyme) and the supramolecular complex ATG12-ATG5-ATG16L1 (E3-like ligase complex) act subsequently to covalently link LC3 to an amino group of phosphatidylethanolamine (PE) (44). The lipidation of LC3 is crucial for both the expansion and potential closure of the phagophore and serves as a late marker of autophagosomes. Moreover, Nath et al. proposed that ATG3 structure is adapted to function at highly curved membranes (45). Furthermore, ATG3 catalytic site is rearranged, enhancing its E2 conjugase activity, by the interaction with the intermediate conjugate ATG12-ATG5 (46). The PIK3C3-C1 aids LC3 lipidation by utilizing the amphipathic lipid packing sensor (ALPS) motif of ATG14, which targets membrane curvature during the process (47). In addition, ATG8 family, both LC3 and GABARAPs, are important mediators of selective autophagy and vesicle trafficking (reviewed below).
The elongation of the phagophore requires a significant supply of lipids to sustain its growth. Although the intricate details of this process are not yet fully elucidated, it is widely acknowledged that it involves inputs from almost all intracellular compartments (34). Three potential mechanisms have been proposed to facilitate this lipid supply: vesicle-mediated delivery, direct extrusion from preexisting organelles, and direct protein-mediated lipid transport (48). These three mechanisms are likely interconnected and may act in a coordinated manner to ensure a continuous supply of lipids during autophagosome elongation.
The first mechanism involves vesicle-mediated delivery, where vesicles derived from various cellular organelles, such as the ER, Golgi apparatus, and endosomes, transport lipids to the phagophore membrane. ATG9 is the only transmembrane member of the ATG family. ATG9 vesicles are involved in transporting membrane materials for phagophore expansion, mainly in the first stages (seeds of membrane formation) (49). Notably, ATG9 harbors scramblase activity, essential for membrane growth, and it forms associations with ATG16L1 in endosomes, being mobilized from the Golgi-endosomal complex through the TRAPPIII (trafficking protein particle III) complex.(50) However, ATG9 is not found in the autophagosome and it is thought to recycle back to its original location after contributing its membrane content to the growing autophagosome (51). In the case of yeast, ATG13 HORMA domain recruits ATG9 during autophagosome formation (52). Lastly, ATG9 vesicles have been proposed to establish membrane contact sites with lipid donor compartments, further contributing to the process (53). Coat protein complex II (COPII) vesicles have ben also reported to contribute to the phagophore elongation (54-56).
The second mechanism suggests a direct extrusion of lipids from preexisting organelles to the phagophore. This idea is supported by observations of certain autophagosomes forming in locations other than the ER, such as mitochondria or early recycling endosomes (42,57). Another possibility is the direct extrusion of phagophore from the ER (58). This direct transfer of lipids ensures a rapid and efficient supply of essential membrane components.
Lastly, the third proposed mechanism involves direct protein-mediated lipid transport. In this case, the best characterized is the ATG2 protein, which transfers tens of lipids, such as glycerophospholipids, between the ER and the phagophore in expansion (59). WIPI4 forms a complex with ATG2 to act as a tethering factor supporting the phagophore elongation (60,61).
Closure
Once the phagophore has expanded, its edges come closer together. The initial membrane structure undergoes transformation into a complete, sealed double-membrane autophagosome. This requires the scission of the inner and outer membranes of the phagophore, that become separated entities (62). Some key regulators of this crucial step are endosomal sorting complexes required for transport (ESCRT) machinery, ATG2-5, Rab GTPases and soluble N-ethylmaleimide sensitive factor attachment protein receptors (SNAREs) (63). Lastly, the closure of the phagophore requires phospholipid metabolism, specifically the synthesis of phosphatidylcholine (64).
Maturation
The maturation of the autophagosome is a sequential process that occurs after the phagophore closure and culminates in the formation of the autolysosome. Throughout this maturation, the autophagosome undergoes various transformations and interacts with endolysosomal compartments (65). Initially, the autophagosome fuses with early endocytic vesicles, early endosomes, multivesicular bodies, and late endosomes/lysosomes, leading to the formation of amphisomes. These amphisomes contain a mixture of both autophagic and endocytic materials (66). Subsequently, the autophagosome further matures and fuses with lysosomes, transforming into autolysosomes. In autolysosomes, a distinct lysosome-specific content, including hydrolases, combines with the autophagic material, facilitating the degradation of cargo and recycling of cellular components (67).
Fusion
After the formation of the autophagosome, it undergoes fusion with lysosomes, autophagolysosome, to release its contents for degradation. First, autophagosomes must move to lysosomes, usually at the perinuclear space, in a microtubule- and dynein-dynactin motor complex-dependent manner (68). Membrane-tethering factors have to be recruited to bring into close proximity both compartments, connecting the opposing membranes and/or promoting the formation of SNARE complexes, such as the heterohexameric complex HOPS (homotypic fusion and protein sorting), PLEKHM1 or TECPR1 (67). HOPS complex is the best-studied and mediates autophagosome-lysosome fusion through interaction with syntaxin 17 (STX17) (69). Following tethering, the SNARE proteins present on the autophagosomal membrane, such as syntaxin 17, interact with SNAREs on the lysosomal membrane, including VAMP8 (vesicle-associated membrane protein 8) and SNAP29 (synaptosomal-associated protein 29) (70,71). These SNARE interactions drive the fusion of the two membranes, bringing the contents of the autophagosome into the lysosomal lumen (72).
Another critical player in the fusion process is the small GTPase protein RAB7, which regulates late endosome-lysosome and autophagosome-lysosome fusion. RAB7 promotes the movement and docking of lysosomes to autophagosomes, interacting with FYVE and coiled-coil domain-containing protein 1 (FYCO1) and Rab-interacting lysosomal protein (RILP), and the fusion of the membranes, by the collaboration with HOPS complex and SNARES (73,74). Finally, PI3KC3-C2 contains the UV-irradiation resistance-associated gene (UVRAG) subunit, instead of ATG14, and is a critical regulator of the last steps of autophagy. UVRAG recruits RAB7 and HOPS complex, mediating the fusion (75,76).
Degradation and autophagic lysosome reformation
Once the autophagolysosome has formed, it initiates the lysosomal degradation of autophagic materials. Initially, yeast Atg15 (or its unidentified mammalian counterpart) degrades the inner autophagosomal membrane, while leaving the outer membrane intact (77). This may due to the acquisition of different properties at the scission step of both membranes. Subsequently, a multitude of over 60 lysosomal hydrolases effectively digest the autophagic material to molecular level, functioning optimally under acidic pH conditions (78). It is well accepted that catabolites are transported to the cytoplasm via lysosomal transporters, decreasing autophagolysosome volume, and subsequently recycled by the cell to synthesize new molecules, structures, or organelles.
Autophagic lysosome reformation (ALR) is the final step of autophagy that restores the level of free lysosomes to maintain lysosome homeostasis (79). It initiates upon mTOR reactivation, a negative feedback to avoid excess of autophagy. During ALR, lysosomal membrane components assemble into tubular structures that protrude from autophagolysosomes. Subsequently, small vesicles, called proto-lysosomes, bud from these tubular structures (80). In their initial state, proto-lysosomes are pH-neutral and lack lysosomal luminal proteins. As they mature, they acquire acidity and incorporate lysosomal luminal proteins, transforming into nascent lysosomes (81). Overall, the main molecular players of macroautophagy are summarized in Figure 2.
Selective vs non-selective autophagy
Autophagy can be categorized into non-selective (bulk autophagy) or selective processes based on the material being degraded. Non-selective autophagy involves the random engulfment of various cytoplasmic contents, triggered by general stress conditions like nutrient deprivation, with the derived catabolites supporting cell survival (82). On the other hand, selective autophagy serves as cellular quality control, targeting and degrading specific components such as damaged organelles, protein aggregates, and invading pathogens. This process relies on specific autophagy receptors or adaptors that recognize cargo and interact with the autophagosome machinery (83,84). The cargo is then sequestered into autophagosomes for degradation in lysosomes. Selective autophagy has been found to target various cellular components, including aggregated proteins (aggrephagy), mitochondria (mitophagy), peroxisomes (pexophagy), ribosomes (ribophagy), endoplasmic reticulum (reticulophagy), lipid droplets (lipophagy), glycogen (glycophagy) and pathogens (xenophagy). Autophagy receptors, such as p62, optineurin, NDP52, or NBR1, play a crucial role in the autophagy process. These receptors have the unique ability to simultaneously bind to both the cargo that needs to be degraded and the autophagy machinery components, like LC3 or FIP200 (85,86). Defects in the PINK1 and Parkin signaling in mitophagy are involved in the development of Parkinson´s disease (87,88).
Alternative autophagy
Alternative autophagy is a recently discovered form of autophagy that exhibits distinct characteristics from the canonical pathway, with the autophagosome membranes derived from the trans-Golgi membrane and its biogenesis independent of ATG5 and ATG7 (89). Despite lacking these essential autophagy-related proteins, this alternative pathway is still capable of selectively degrading specific cellular cargo. Golgi stressors like etoposide, amphotericin B1, or genotoxic stress can induce alternative autophagy (90). Research into various diseases, including cardiovascular diseases, neurodegenerative disorders, cancer development (oncogenesis), and inflammatory bowel disease (IBD), has highlighted the significant role of alternative autophagy in their pathophysiological processes, offering new opportunities for investigating autophagy regulation complexities (91).
Microautophagy
Microautophagy is a distinct and less well-understood form of autophagy that involves the direct engulfment of cytoplasmic components by the lysosome or late endosome. This process allows for the selective degradation of specific cytoplasmic components, including organelles and protein aggregates, and plays a crucial role in cellular homeostasis and quality control (92). The lysosomal or endosomal membrane undergoes invagination, forming a tubular or vesicular structure protruding into the cytoplasm (Figure 1b).
Regarding the molecular machinery, endosomal microautophagy relies on multiple endosomal sorting complexes required for transport (ESCRT) systems (93). However, it is possibly the lest studied form of autophagy. Microautophagy can be also a selective process, e.g.: micro-ER-phagy through SEC62 receptor or proteins with the pentapeptide motif KFERQ via chaperone HSC70 (94).
Schematic representation of the key molecular players of macroautophagy, steps and role. mTORC1: mammalian target of rapamycin complex 1, AMP-activated protein kinase: AMPK, ULK1/2: unc-51 like autophagy activating kinase 1/2, FIP200: focal adhesion kinase (FAK)-interacting protein of 200 kDa, PAS: phagophore assembly site, ER: endoplasmic reticulum, GABARAP: gamma-aminobutyric acid receptor-associated protein, VAMPA/B: VAMP-associated protein A/B, PI3KC3-C1: class III phosphatidylinositol 3-kinase complex I, PI3P: phosphatidylinositol 3-phosphate, DFCP1: double FYVE containing protein 1, WIPI1-4: WD-repeat protein interacting with phosphoinositides 1-4, LC3: microtubule-associated protein 1 light chain 3, PE: phosphatidylethanolamine, COPII: coat protein complex II, ESCRT: endosomal sorting complexes required for transport, SNAREs: soluble N-ethylmaleimide sensitive factor attachment protein receptors, HOPS: homotypic fusion and protein sorting, STX17: syntaxin 17, VAMP8: vesicle-associated membrane protein 8, SNAP29: synaptosomal-associated protein 29, FYCO1: FYVE and coiled-coil domain-containing protein 1, RILP: Rab-interacting lysosomal protein, UVRAG: UV-irradiation resistance-associated gene.
Microautophagy can be further categorized into three main types based on the engulfment mechanism: type 1, microautophagy with lysosomal protrusion, type 2, with lysosomal invagination; and type 3, with endosomal invagination (95).
Chaperone-mediated autophagy (CMA)
Chaperone-mediated autophagy (CMA) is a selective form of autophagy that specifically targets and degrades individual cytosolic proteins (Figure 1C). Unlike macroautophagy, CMA directly delivers substrates to lysosomes for degradation (96). This highly regulated process plays a crucial role in maintaining cellular protein quality control, responding to cellular stress, and providing an alternative energy source during nutrient deprivation (97).
The hallmark of CMA is the recognition and targeting of substrate proteins by chaperone proteins. One of the key chaperones involved in CMA is HSC70, which selectively recognizes the pentapeptide motif KFERQ in the substrate proteins. Upon binding, HSC70 escorts the target protein to the lysosomal membrane, where it interacts with the lysosome-associated membrane protein type 2A (LAMP2A), a lysosomal membrane protein that forms a translocation complex that recognizes HSC70-substrate complexes (98,99). The substrate is then unfolded and translocated across the lysosomal membrane into the lysosomal lumen for degradation by lysosomal proteases. Overall, Figure 1 summarizes the main types of autophagy and their different steps.
Biological relevance of autophagy. An overview
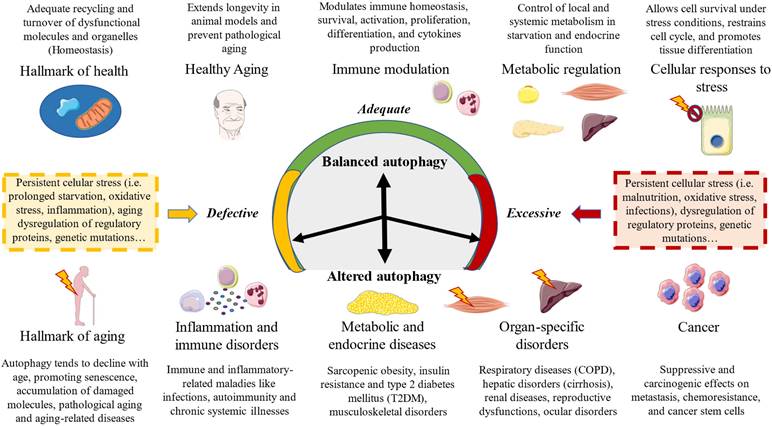
Autophagy is an essential factor related to several physiological processes, equally representing a pivotal pathophysiological mechanism of a broad spectrum of diseases (13). López-Otín & Kroemer described the hallmarks of health, a set of biological mechanisms that aid to understand health not only as the absence of pathology but also as a set of organizational and dynamic features that collectively maintain normal physiology (2). They divided the hallmarks of health into three major dimensions: spatial compartmentalization, sustainment of homeostasis over time, and adequate responses to stress. In this picture, autophagy is known to represent a central process implicated in adequate recycling and turnover of dysfunctional molecules and organelles that aid to keep homeostasis in the different cells and tissues. Thus, autophagy acts as an important cytoprotective mechanism implicated in the maintenance of cellular energy levels, components, and metabolites, as well as the elimination of cellular molecular damage (100). However, autophagy also orchestrates an array of biological processes, playing a dual and controversial role in different cellular events, aiding to understand its relevance in health and disease (13). In this section, a brief summary of the role of autophagy in different biological processes will be performed.
Autophagy in the cellular response to stress
Autophagy is mostly considered a cellular stress response. Different environmental stressors are responsible for modulating the autophagy response such as nutrient deprivation, oxidative stress, hypoxia, endoplasmic reticulum stress, inflammation, intracellular pathogens, protein aggregates, toxic molecules and certain pharmacological agents (8,101). As a part of the cellular machinery implicated in stress responses, autophagy is induced by transcriptional and posttranscriptional reprogramming that leads to the activation of the autophagy proteins. Different transcriptional factors like MIT/TFE, PPARα, TFEB, ATF4, E2F1, C/EBPβ, FOXO, NF-κB, E93, STAT, and p53 are critically involved in the activation of autophagy in response to stress, whereas a broad spectrum of non-coding RNAs involved in the regulation of the Atg gene and other epigenetic mechanisms like histone modifications and DNA methylation can also participate in the activation of autophagy (102,103).
Autophagy is also integrated and related to several cellular processes like the cell cycle, apoptosis, and other processes (8). Under stressful conditions, compelling evidence supports that autophagy is able to induce and possibly execute cell cycle arrest programs, delimiting cell division and growth (104). This relationship can have important consequences for certain pathologies like cancer. Likewise, the relationship between autophagy and apoptosis is complex and frequently controversial. Both processes can be activated simultaneously by various stress pathways (105). Despite autophagy commonly blocks the induction of apoptosis, and apoptosis prevents the autophagic process, autophagy and apoptosis can appear concomitantly (106). From an evolutive perspective, autophagy is evolutionarily a more ancient event than apoptosis, and as both processes controls cell death and survival, they need to be narrowly linked (107). As autophagy has both pro-survival and pro-death functions, the cellular fate will depend on the extent of stress or damage and what the cell is capable of tolerating, as well as the functioning of the apoptosis process and its interplay with autophagy. Indeed, loss of autophagy and apoptosis function and communication is associated with different pathologies like cancer, neurodegenerative or cardiovascular diseases amongst others (107-109).
The relevance of autophagy as a cellular stress response can be observed in different regions of the body. Among various cell types, nervous system cells, especially postmitotic neurons, are remarkably susceptible to an array of insults, making autophagy a crucial stress response mechanism that safeguards the well-being and survival of these vulnerable cells (110). Conversely, prolonged exposure to stress can lead to a disruption in the autophagy control, leading to a defective or exacerbated autophagy, which is associated with a range of human diseases (111).
Autophagy and aging
One of the most widely known mechanisms by which autophagy influences health and disease is through its tight association with aging. Several animal models have found that maintenance of proper autophagic activity is associated with extended longevity (112). Conversely, disabled autophagy is considered a primary hallmark of aging, also leading to unrestrained inflammasome activation, cellular damage, and oncogenesis, among other consequences (113). It should be noticed that both a reduction and an exacerbated autophagy are tightly associated with pathological aging and aging-related diseases. Previous works have found that autophagy is a process that seems to decrease with age, potentially contributing to the accumulation of damaged organelles and macromolecules, metabolic alterations in the cells and organelles, an increase in endoplasmic reticulum stress or decreased lysosomal proteolytic activity, among other consequences (112,114). On the other hand, uncontrolled autophagy might accelerate aging by increasing the number of senescent cells, cellular fast muscle fibers atrophy, cardiac hypertrophy, molecular and metabolic dysregulation, sarcopenia, neurodegeneration and other detrimental effects (114). Thus, balanced autophagy is essential to promote healthy aging and different strategies aimed to modulate this process are growingly being considered to ameliorate pathological aging and aging-related diseases (112).
Autophagy and the immune system
Simultaneously, there is also a close relationship between the immune system and autophagy. Compelling evidence has found that autophagy is directly involved in the modulation of immune responses mainly by influencing the homeostasis, survival, activation, proliferation, differentiation, and cytokines production, of innate and adaptative immune cells including T and B lymphocytes, natural killer (NK) cells, macrophages, and dendritic cells (DCs) (115). Simultaneously, immunoglobulins, immune-related cells, and cytokines are directly involved in autophagy regulation both by inducing this process through specific products such as transforming growth factor (TGF)-β, interferon (IFN)-γ, interleukin (IL)-1, IL-2, and IL-12 or by inhibiting this process (IL-4, IL-10, and IL-13).(116) Thus, autophagy is a process which influences and is regulated by acute and chronic inflammatory conditions, also collaborating with the immune responses to infections and other systemic challenges (117,118). In this context, previous works have identified perturbations in the autophagy process as a critical pathogenic mechanism of immune and inflammatory-related maladies like infections, autoimmunity, metabolic disorders, inflammatory bowel disease, neurodegeneration, cardiovascular diseases and cancer (117,119). Therefore, autophagy modulation is also a major target for prevention and therapy of immune-related diseases, although the knowledge of the mechanistic processes and role of autophagy in the context of inflammation requires additional efforts (120-122).
Autophagy, metabolic and endocrine system
Autophagy has also been linked to metabolic regulation by different studies. Previous works differentiate between a metabolic and a quality control autophagy (123). The latter selectively eliminates a diverse range of cytoplasmic targets whereas the former is a response to starvation. As aforementioned, nutrient deprivation is a pivotal inductor of autophagy through the activation of AMPK and suppression of mTOR (124). Amino acids, fatty acids and glucose narrowly influences the core components of the autophagy machinery, controlling energy metabolism by tissue-specific effects and globally (endocrine effects) (125). In case of the former, nutrient deprivation and starvation are major inductors of autophagy in liver and heart, generating fatty acids and amino acids catabolized to obtain energy (126). In the liver, autophagy is linked to the processes of ketogenesis and gluconeogenesis, leading to the release of ketone bodies and glucose to feed the brain. However, as starvation prolongs, degradation and autophagy of adipose and muscle tissue is required to supply substrates to the liver, demonstrating that autophagy is a central mechanism implicated in the modulation of the metabolism in the body (126). In the endocrine system, autophagy orchestrates intracellular hormone levels both in peptide-secreting cells (targeting the secretory granules to control the levels of stored hormone) as well as in steroid secreting cells (targeting steroid-producing organelles) (127). As starvation favorably modulates autophagy, overnutrition can drive to an important dysregulation of autophagy, leading to the onset of a broad spectrum of disorders with a metabolic and endocrine base (125). For example, different conditions like sarcopenic obesity, insulin resistance and type 2 diabetes mellitus (T2DM) are characterized by an impaired autophagy related to a metabolic derangement, enhanced intracellular stresses and accelerated ageing in specific organs like the liver, pancreas, skeletal muscle and adipose tissue (128).
Autophagy and tissue differentiation
During ontogenesis across diverse organisms, autophagy orchestrates a multitude of cellular processes, encompassing survival in nutrient-deprived conditions, programmed cell death (apoptosis), phagocytic events, organelle clearance, and miRNA regulatory functions (129). By employing animal models, pivotal mechanistic insights pertaining to autophagy regulation and its involvement in pathological states have been unveiled. Embryonic studies have unveiled specific autophagy-associated molecules responsible for the elimination of paternally inherited mitochondria and have identified autophagic components responsible for the clearance of protein aggregates during development (129). It has become evident that autophagy is a crucial process for maintaining the equilibrium and functionality of various tissues, influencing their development, differentiation, and capacity to undergo remodeling in response to stimuli or during periods of stress (130).
Autophagy, as an essential recycling mechanism, profoundly influences the destiny of various types of stem cells. Its role can be attributed to its significant contribution in maintaining the balance of a wide range of peptides and proteins, including growth factors, transcription factors, and crucial enzymes that play pivotal roles in cell functions such as proliferation, differentiation, and aging (131,132).
Moreover, autophagy exerts an impact on cell fate determinations by modulating mitochondrial content, energy generation, and epigenetic programming. Notably, in the context of aging and degenerative disorders, where the regenerative capacity of stem cells declines, autophagy assumes pivotal roles in shielding stem cells from cellular stress, emerging as a promising target in the field of regenerative medicine (133).
Autophagy in diseases
Autophagy is a relevant biological process in almost all systems and regions of the body. The relationship between autophagy and disease has been explored in previous works. Autophagy can act either as a protective or as a promoter mechanism of different maladies. A balanced autophagy is associated with protective mechanisms and health effects, whereas a disturbed autophagy either by defect or excess is a major trigger of disease (Figure 3). There are many underlying causes of an aberrant autophagy, particularly a prolonged stress exposure, dysregulation of autophagy proteins and specific genetic mutations. In case of the latter, the relationship between genes and impaired autophagy is receiving growing attention in the last years, as compelling evidence is revealing direct links between genetic defects of core autophagy genes, autophagy-associated genes, and genes encoding autophagic receptors with multiple diseases (134). In this sense, there are a group of pathologies known as congenital disorders of autophagy, which are identified by monogenic mutations and include EPG5-related Vici syndrome, beta-propeller protein-associated neurodegeneration due to mutations in WDR45, SNX14-associated autosomal-recessive cerebellar ataxia and intellectual disability syndrome, and three forms of hereditary spastic paraplegia, SPG11, SPG15 and SPG49 caused by SPG11, ZFYVE26 and TECPR2 mutations, respectively (135).
Besides these specific conditions and as aforementioned, autophagy plays a key role in the regulation of multiple processes like aging, immune modulation, metabolic and endocrine function, and an altered autophagy is associated with pathological aging, immune dysfunction and inflammation as well as several endocrine and metabolic diseases. But an impaired autophagy is observed not only in these conditions, but also in virtually all organ-specific and systemic disorders (3). For instance, autophagy is considered a critical process in heart and cardiovascular health, and an aberrant autophagy is associated with molecular, cellular, structural and functional impairments in the cardiovascular system (136,137). Because of that, autophagy has been recognized as a major therapeutic target for different cardiovascular diseases, aiming to stimulate (ischemia/reperfussion, cardiac lysosomal storage disease, diabetic cardiomyopathy) or to ameliorate this process (left ventricular hypertrophy; heart failure with reduced ejection fraction; anthracycline cardiotoxicity) (138). In the same line, neurodegenerative (Parkinson's, Alzheimer's or Huntington's disease) and psychiatric disorders (major depressive disorder, bipolar disorders or schizophrenia) are also characterized by an impaired autophagy in the nervous system, having been identified as a promising translational target for these conditions (139).
A summarized view of the biological relevance of autophagy in balanced and disrupted autophagy.
The role of autophagy in cancer has also been deeply explored by prior works. The evidence seems to support that autophagy acts as a tumor suppressor and promotor, with multiple effects on metastasis, chemoresistance and cancer stem cells (140). While certain autophagy modulators like rapamycin and chloroquine are employed in anticancer therapy to regulate autophagy, a comprehensive understanding of the precise mechanisms underlying autophagy's involvement in cancer is imperative and necessitates further investigation (141). As the complexities of autophagy in cancer biology unfold, continued research efforts are warranted to optimize its therapeutic potential and exploit its dualistic nature in a targeted manner for effective cancer treatments.
Finally, abnormalities in autophagy has been demonstrated in respiratory diseases like chronic obstructive pulmonary disease (COPD), hepatic disorders like cirrhosis, renal diseases, reproductive dysfunctions, ocular disorders, and in general, the different diseases related to aging, immune and metabolic dysfunction (3). In summary, autophagy plays a crucial role in determining human health and offers potential interventions to prevent or alleviate common illnesses. Preclinical evidence suggests that autophagy defects are particularly detrimental to post-mitotic cells, and that autophagy defects in healthy cells are typically associated with diseases due to disruptions in cellular homeostasis rather than a failure to adapt to nutrient scarcity, whereas cancer cells exploit autophagy whereas cancer cells exploit autophagy to cope with intracellular stress (142).
Modulation of autophagy
Studying the modulation of autophagy, through both pharmacological and non-pharmacological approaches, is essential for understanding its role in maintaining cellular health and its involvement in various diseases. By identifying drugs and lifestyle interventions that can enhance or inhibit autophagy, researchers can develop targeted therapies for conditions like neurodegenerative disorders, cancer, and metabolic diseases. Autophagy modulation also offers insights into cellular stress responses, aging, and personalized medicine. Ultimately, this research holds promise for improving disease treatment, drug discovery, and promoting healthier aging and longevity.
Pharmacological modulation
As discussed earlier, autophagy is a crucial process that significantly influences both health and disease. This opens up a wide range of therapeutic opportunities aimed at targeting and regulating autophagic flux within cells. Generally, promoting autophagy can lead to cell survival, while inhibiting it can result in cell death due to the buildup of toxic products (143). However, more research is needed in order to achieve better results and to know the possible side effects of the agents.
Pharmacological modulation of autophagy can involve both activation and inhibition of the autophagy process, depending on the specific aims and contexts. Activators of autophagy promote the initiation and progression of autophagosome formation, while inhibitors prevent the degradation of autophagosomes or disrupt the fusion of autophagosomes with lysosomes (144). In Table 1 we present a collection of the best-described autophagic activators and inhibitors.
Pharmacological activators and inhibitors of autophagy. (*) Nutraceuticals able to modulate autophagy
| Pharmacological agent | Chemical structure | Source | Autophagy-associated targets | Mechanism of action | References |
| Activators | |||||
| Rapamycin (Sirolimus) | Macrocyclic lactone | Streptomyces hygroscopicus | mTOR | Inhibits mTORC1 (acute and chronic exposure) Inhibits mTORC2 (chronic exposure) Rapamycin binds FKBP12 and act as an allosteric inhibitor | (148) |
| Temsirolimus | Derivative of rapamycin | Synthetic analog of rapamycin | (149) | ||
| Everolimus (RAD-001) | Derivative of rapamycin | Synthetic analog of rapamycin | (150) | ||
| Erlotinib | Quinazolinamine | Synthetic compound | EGFR | Inhibition of the EGFR signaling by interaction with ATP binding site | (151) |
| Imatinib | 2-phenyl amino pyrimidine | Synthetic drug | BCR-ABL c-Abl Abl-related gene | Inhibits selectively tyrosine kinases Sequestering of the Bcr-Abl protein into autophagosomes | (152,153) |
| Dasatinib (BMS-354825) | Derivative of aminopyrimidine | Synthetic compound | Src/Abl family kinases | Reduces the phosphorylation of AKT, mTOR, p70S6K, and S6 kinase expression Requires BECN1 and ATG12 | (154,155) |
| Vorinostat (SAHA) | Hydroxamic acid derivative | Synthetic compound | HDAC | Acetylation of ATG proteins leading to hyperactivation of PIK3C3 Upregulation of LC3 Inhibition of mTOR | (156-158) |
| Arsenic Trioxide | Amphoteric oxide | Processing of the mineral arsenic | TFEB ROS PI3K/AKT/mTOR | Nuclear translocation of TFEB Inhibition of PI3K/AKT/mTOR pathway Generation of ROS | (159) |
| Epigallocatechin-3-gallate* | Polyphenol. | Green tea leaves | mTOR-AMPK | Induces activation of AMPK and inhibition of mTOR | (160,161) |
| Polygonatum cyrtonema lectin* | Lectin | Rhizomes of Polygonatum cyrtonema | Mitochondria-mediated ROS-p38-p53 pathway | Accumulation of mitochondrial ROS, activating p38 and p53 | (162,163) |
| Spermidine* | Polyamine | Dry soy bean, chicken liver, green peas, corn, shell fish, and blue cheese | Acetylation of histones | Modifies epigenetic landscape Increases the expression of ATG | (164,165) |
| Resveratrol* | Polyphenol | Grapes, apples, plums, blueberries, and peanut | mTOR, AMPK, SIRT1 | Inhibiting mTOR via several routes, activates AMPK and SIRT1 | (166-168) |
| Allicin* | Sulfoxide | Allium sativum | p53, mTOR and AMPK | Modulation of the pathways leading to autophagy | (169,170) |
| Ginsenosides* | Saponins | Ginseng root | Autophagy-related pathways | Modulation of the pathways leading to autophagy | (171,172) |
| Curcumin* | Diarylheptanoid | Curcuma longa | ROS mTOR | Inhibiition of mTOR Overexpression of autophagy-related proteins | (173-175) |
| Pharmacological agent | Chemical structure | Source | Molecular target | Mechanism of action | References |
| Inhibitors | |||||
| Wortmannin | Furanosteroid | Penicillium funiculosum Talaromyces wortmannii | PI3KC1 (transient) PI3KC3 (persistent) | Inhibition of the production of PIP3 | (146) |
| 3-Methyladenin | Adenin | Saccharomyces cerevisiae | PI3KC1 (persistent) PI3KC3 (transient) | Inhibition of PI3K Promotion of autophagic flux under nutrient-rich conditions (dual role) | (176,177) |
| Chloroquine | Aminoquinoline | Cinchona officinalis | Lysosomal pH SNAP29? | Slows down lysosomal acidification Inhibition of the fusion of autophagosomes with lysosomes | (178,179) |
| Pepstatin-A | Hexapeptide | Streptomyces sp | Cathepsin D and E | Inhibition of lysosomal proteases Accumulation of LC3-II | (146,180) |
| Bafilomycin A1 | Macrolide | Streptomyces sp | Vacuolar-ATPase SERCA | Reduce lysosomal acidification Disrupt autophagosome-lysosome fusion | (181,182) |
| Hydroxy-chloroquine | Derivate from chloroquine | Synthetic modification of cloroquine | Lysosomal pH | Slows down lysosomal acidification Inhibition of the fusion of autophagosomes with lysosomes | (183,184) |
| Spautin-1 | 4-quinazolinamine | Synthetic compound | Ubiquitin-specific peptidase 10 and 13 | Inhibits autophagy by the degradation of PI3K3 and BECN1 | (185,186) |
| SAR405 | Pyrimidine | Small molecule inhibitor | Class III PI3K | A selective ATP-competitive inhibitor of PI3K class III | (187) |
| siRNAs | 20 to 24-bp double-stranded RNA | Synthetic compound | ATG mRNA | Knockdown the autophagy machinery, e.g.: ATG5 | (188,189) |
Autophagy activators can either promote cell survival in the case of diseases characterized by accumulation of protein aggregates, such as neurodegenerative diseases or haemochromatosis, or lead to cell death when induces excessive autophagy, such as cancer cells (145). The main targets of this group are the inhibition of mTOR and activation of AMPK, the upstream regulators of macroautophagy. It is the case of rapamycin and rapalogs, arsenic trioxide or epigallocatechin-3-gallate (EGCG). A special group are the nutraceuticals, which achieve the activation of autophagy by pleiotropic pathways, e.g.: EGCG, polygonatum cyrtonema lectin, spermidine, resveratrol, allicin, ginsenosides and curcumin.
On the other hand, autophagy inhibitors are a suitable approach for cancer therapy due to the protective role of autophagy in cancer (146,147). Autophagy inhibitors can target various steps of the autophagy process. For instance, wortmannin or 3-methyladenine act on the nucleation step by targeting PI3KC3, while pepstatin-A and bafilomycin A1 affect lysosomes, either by inhibiting lysosomal acidification or by hindering the fusion with autophagosomes.
Lifestyle Medicine and Autophagy
Lifestyle medicine is a growing area of research based on the modulation of key health behaviors such as diet, physical activity, sleep, and environmental factors like tobacco, social relationships, and also stress management (190-192). Lifestyle medicine is increasingly being considered a crucial mechanism to modulate autophagy, aiding to explain the multiple benefits from this type of approaches for health and also for the management of chronic diseases (193,194). In this section we will summarize the relevance and implicated mechanisms of different lifestyle factors on autophagy, exploring the role of diet (considering nutrients, foods and dietary strategies), physical activity, sleep and circadian regulation, tobacco, alcohol, air pollution, sunlight exposure and psychosocial stress. Table 2 summarizes the main findings of this topic.
Lifestyle habits and their influence on autophagy.
| Lifestyle habit | Effectors | Influence on autophagy | Main outcomes of the autophagy | Reference |
|---|---|---|---|---|
| Mediterranean Diet | Fruits, vegetables, fish, rice, olive oil and eggs | Stimulation | Anti-aging, anti-inflammatory, improved cardiovascular health and enhanced brain function | 203-205 |
| Calorie restriction | Reduced energy intake. Adequate nutrition | Stimulation by decreasing mTOR signalling and activation of AMPK | Increase of fat mobilization, oxidation, metabolic flexibility, insulin sensitivity and redox imbalance Reduction in systemic inflammation, cardiovascular risks and body weight Anti-aging | 196, 210-212, 214-220 |
| Intermittent fasting | Periodic cycles of eating and fasting | 196, 213-220 | ||
| Physical activity | Increases FOXO, TFEB, AMPK or ROS levels and AMP/ATP ratio | Activate autophagy in skeletal muscle | Regulation of glucose, protein synthesis, muscle mass maintenance and exercise performance Attenuate aging-associated autophagic dysfunctions leading to neurodegeneration | 225-234 |
| Sleep deprivation | Autophagy machinery (BECN1, LC3 and p62) | Impairment of the levels of ATG proteins in hippocampus and striatum | Autophagy dysfunction is associated with neurodegenerative and behavioral alterations, all linked to REM sleep loss. Propofol and modafinil improve cognitive function loss by sleep deprivation in rodent models | 243-252 |
| Tobacco | Egr-1 Bicaudal D1, p62, galectin 8, NDP52 | Induction of autophagy Defective autophagosome maturation | Accumulation of bicaudal D1, p62 and autophagosomes Development of COPD | 255-258 |
| Alcohol consumption | Ethanol metabolites | Stimulation (Acute) Inhibition (Chronic) | Chronic consumption leads to ALD. Autophagy is a protective response against acute ethanol induced steatosis and liver injury. Chronic consumption inhibits autophagy and induces apoptosis | 262-266 |
| Air pollution | Particulate matter | Stimulation | PM2.5-induced oxidative stress PM2.5 leads to accumulation of LC3 and overexpression of BECN1 and ATG5 Impairment in lung function Skin aging | 267-269 |
| Sunlight exposure | UVA-induced ROS | Stimulation or impairment | Skin photoaging and cancer | 272-279 |
| Psychosocial stress | BECN1, LC3II, FKBP51 | Stimulation (acute) Impairment (chronic) | Promotes adaptive emotional outcomes (acute) Gradual decline in synaptic function and neurodegeneration (chronic) Exacerbate IBD, gut microbiota dysbiosis and inflammation Antidepressants reverse the psychological consequences of chronic stress via mTOR. | 280-286 |
Diet
Diet is a major modulator of autophagy. More specifically, available literature distinguishes between specific nutrients present in certain groups of foods or in the form of supplementation and specific dietary strategies such as caloric restriction and intermittent fasting (195,196). All these components can directly act through the biochemical regulation of different products such as AMPK, SIRT-1, eIF5A, or GCN2, ultimately leading to the modulation of the main proteins implicated in the autophagy process like ULK1, TFEB, FOXO1, ATF-4 or CHOP (195).
Firstly, there are many nutrients with significant health/translational properties (nutraceuticals) that have been demonstrated to modulate the process of autophagy. To provide some examples, amino acids (i.e., leucine), fatty acids (i.e., omega 3 polyunsaturated fatty acids), vitamins (carotenoids and retinoids, ascorbic acid, calciferol, tocopherols, and tocotrienols), coenzyme Q10, bioactive compounds (i.e., mainly polyphenols like curcumin, caffeine, EGCG resveratrol, allicin), minerals like zinc or iron, ergothioneine, lipoic acid, N acetylcysteine and spermidine.(197-201) Some of the most relevant nutraceuticals with a role in the modulation of autophagy, targets, and biological mechanisms appear summarized in Table 1. Not only isolated nutrients but also those contained in foods are important modulators of autophagy. Plant foods are perhaps the major modulators of these processes, especially regarding their content in a broad spectrum of nutrients involved in the regulation of this process, such as vitamins and several bioactive compounds (195,202). After plants, foods like fish and seafood, eggs, and in general, different groups of food commonly consumed in healthy dietary patterns such as Mediterranean Diet (Med Diet) have also significant effects on upregulating autophagy (203,204). Generally, healthy, non-processed or minimally processed foods and dietary patterns tend to stimulate the process of autophagy, thus explaining their anti-aging, anti-inflammatory, and beneficial health outcomes (205). Conversely, unhealthy foods and dietary patterns such as westernized diets, abundant in refined sugar, unhealthy fats, ultra-processed foods, salt and controversial additives are mostly related to an inhibition in the autophagy process (206,207). Thus, dietary patterns, foods, and nutrients are major modulators of autophagy, aiding not only to preserve health but also representing promising translational approaches for several diseases (136,164,197,208,209).
Additionally, not only the quality and type of nutrients and foods ingested but also the quantity and timing of intake seem to be relevant for modulating autophagy. Thus, calorie restriction (CR) and intermittent fasting are both strategies tightly linked to the activation of this process. CR is a type of nutritional intervention of reduced energy intake but supported with an adequate nutrition, representing a valuable clinical strategy in the medical management of obesity, cancer and cardiometabolic disorders (210). Besides, CR has also been related to anti-aging effects, being its stimulatory role in autophagy defined as a critical mechanism explaining this interplay (211,212). Intermittent fasting could be defined as the complete deprivation of food but not water, with intervening periods of normal food intake (213). Their stimulatory effects on autophagy are similar to those observed in CR, although in comparison it is commonly associated with greater adherence rates than CR (214). Indeed, it is common that intermittent fasting entails a reduced caloric intake by limiting the number of daily intakes, explaining the similar effects of both strategies. CR and intermittent fasting stimulates autophagy mainly by decreasing mTOR signaling by reducing insulin and IGF-1 levels and increasing the AMP/ATP ratio that leads to the activation of AMPK as well as several products involved in the stimulation of this process like ATG6, ATG7, ATG8, LC3-II, Beclin1, p62, Sirt1, LAMP2, ULK1 and ATG101 (196,214). The favorable effects on autophagy of both CR and intermittent fasting seems to be involved in the increase of fat mobilization, oxidation, metabolic flexibility, insulin sensitivity and redox imbalance together with a reduction in systemic inflammation, cardiovascular risks and body weight (215). Hence, either CR or intermittent fasting are valuable and similar strategies to preserve health and aid in the clinical management of several diseases (216). Nonetheless, despite the heterogeneity observed in the available literature, some articles reflect that intermittent fasting and CR could potentially exert different effects and applications depending not only on adherence but also on the purposes of the intervention (217,218). Also, there are different strategies of intermittent fasting that could bring different health outcomes such as alternate-day fasting, other similar full-day fasting patterns, and time-restricted feeding (219). More studies are warranted to fully understand the precise relationship between CR and intermittent fasting with autophagy, especially to compare their effects on different tissues or in various types of fasting strategies (214,220).
Overall, diet could be considered a pivotal modulator of autophagy. Including specific types of nutrients and foods, commonly present in a healthy dietary pattern like Mediterranean diet favorably regulates this process, whereas refined and ultra-processed foods usually have the opposite effect. CR and intermittent fasting are two widely studied nutritional strategies that can also stimulate autophagy and exert significant anti-aging and translational applications, although further studies are required in healthy individuals and patients with different pathologies.
Physical activity
Physical activity (PA) is another major representant of lifestyle medicine with several benefits for sustaining health and a critical support for several chronic diseases (221). Physical inactivity or sedentary behavior emerges as a consequence of multiple factors that not only depend on the individual but also on society itself. According to the World Health Organization (WHO), physical inactivity is considered the fourth leading risk factor for global mortality (222). Thus, sedentary individuals face an increased risk of developing a wide spectrum of pathologies, ranging from cardiovascular diseases, metabolic disorders, cancer, osteoporosis, and other musculoskeletal issues, ultimately leading to a diminished quality of life (223). PA and exercise have multiple systemic effects on the body, leading to important adaptations and effects in the musculoskeletal, cardiovascular, nervous, metabolic and immune system, with a crosstalk between all these systems through different products (exerkines) (224). In this context, previous studies have found that PA plays a central role in the regulation of autophagy, potentially aiding to explain its multiple benefits on health and disease (225). Compelling evidence supports that PA increases several activators of the autophagy in the skeletal muscle like FOXO, TFEB, AMPK or ROS amongst others, promoting an increase of autophagy capacity and autophagy flux, eventually driving to the elimination of damaged organelles and proteins, improving mitochondrial function/oxidative capacity, regulating glucose, protein synthesis, muscle mass maintenance and exercise performance (226). Conversely, decreased activation of autophagy in the skeletal muscle is related to significant local and systemic consequences, driving to significant concerns like myopathies, muscle atrophy, exercise intolerance and insulin resistance (227). Not only the skeletal muscle, but also other regions of the body benefits from the positive effects of PA in the autophagy. The brain is one of the regions more favored by PA, as it regulates multiple processes and products essential for brain health and cognitive functions, aiding to prevent or improve the medical management of neurodegenerative and age-related disorders in the brain (228). Among the benefits of PA in the brain, it has been shown that exercise ameliorates autophagy and apoptosis dysfunction, leading to significant improvements in the affected patients (229). Also, an increased detection of autophagy markers like Beclin-1, ATG12, ATG16 and LAMP-2 is also observed in peripheral blood mononuclear cells (PBMCs) after 8 weeks of PA, along with a decreased apoptosis and inflammasome NLRP3 activation (230). Therefore, the association between PA and autophagy is overwhelmingly important, supporting the potential of considering PA as a non-pharmacological approach to favorably modulate autophagy in health and disease conditions.
On the other hand, different studies have found that the effects of PA on autophagy seems to be tissue- and exercise dependent on different exercise regimens to compare their precise role on this process. Pinto et al.(231) compared the acute effects of endurance (END), exhaustive (ET), strength (ST), and concurrent (CC) physical exercise on various markers of autophagy in the gastrocnemius muscle, heart, and liver of mice. They observed that each type of PA presented different effects on autophagy markers in these regions, suggesting that each variant of exercise could be differentially receipted according to the patient´s features. In more detail, for gastrocnemious muscle samples the main alterations were observed after 6 hours of ST, whereas the markers of autophagy for the CC group were increased. The Beclin 1 and ATG5 levels in the heart were downregulated in the ET group. Sqstm1/p62, one of the autophagy markers, were shown to be increased in the cardiac tissue of the END and ST groups, whereas for the ET group, the levels of the liver protein ATG5 were downregulated. Simultaneously, a systematic review conducted by Chen et al.(232) observed that acute ST was acutely associated with a decreased autophagy demonstrated by reduced levels of LC3-II and increased SQSTM1, although long-term ST was oppositely associated with an increased LC3-II and autophagy with a decrease in SQSTM1. They equally observed that other markers of autophagy like ULK1, Beclin-1, ATG12, BCL2 can also differentially change according to the type of exercise and in a tissue-dependent manner, and also that moderate and vigorous END was not associated with changes in the autophagy process, although other works have found a positive effect of this type of training on the autophagy (233,234). Likewise, trained subjects with a combination of ST with sprint exercises also presented enhanced autophagy defined as an increase in LC3-II and a decrease in p62 in comparison to other trained subjects that did not follow this protocol (235). Additionally, there are studies comparing the effects of high intensity interval training (HIIT) with moderate-intensity continuous training (MICT) on autophagy (236). Interestingly, they observed that both regimens were associated with enhanced autophagy 3 h post-exercise in skeletal muscle, although in PBMCs, the autophagy increased after HIIT and MICT in men, but not in women, suggesting a differential effect of both trainings depending on the tissue and sex.
Overall, PA exerts significant improvements in the autophagy that plays a pivotal role in the body adaptations to the exercise and also in several systemic processes essential for health and alleviation of various diseases. However, the type of training program, the considered tissue, sex and other factors are important to determine the precise effects of PA on autophagy. Future works should be performed to deeply characterize this intricate relationship.
Sleep and circadian regulation
Sleep fulfills several important functions for the homeostasis of the organism: restoration of energy levels, reparation of tissues, memory consolidation, maintenance of brain function, support of immune and endocrine system and psychological well-being (237-239). Numerous studies have demonstrated that sleep deprivation negatively affects health and quality of life, including cognitive functions such as mood, cognition, and memory (240-242). It has been demonstrated that basal autophagy is crucial for preventing the buildup of aberrant cytosolic proteins in neurons, whereas disruption of basal autophagy may result in neurodegeneration, as seen by a significant loss of neurons (243). In addition, autophagic activity follows the circadian rhythm (244,245). In Drosophila, sleep decreases autophagosome levels under unperturbed conditions and a strong and sustained downregulation of autophagosomes increases sleep (246). Also, in models expressing the mutant Huntingtin protein the induction of autophagy via overexpression of Atg8a (homolog of mammal LC3s and GABARAPs) achieves the reestablishment of normal sleep habits (247).
In mice, 5 days of sleep fragmentation led to the dysregulation of autophagy in specific brain regions, with the striatum and hippocampus showing heightened sensitivity. In the striatum, levels of BECN1, LC3-II, and p62 were increased, while in the hippocampus, LC3-II was elevated, but BECN1 and p62 were decreased (248). Interestingly, these changes had no impact on autophagy in the frontal cortex, indicating a region-specific response to sleep fragmentation. Another study showed that sleep-deprivation in rats also increases the expression of Beclin1, PINK1, parkin, p62, and LC3. Then, the treatment with propofol significantly reduces the levels of these proteins in hippocampal neurons, i.e., inhibits excessive autophagy, and could improve the impairment of learning and memory caused by sleep deprivation (249). Similar results are obtained in mice treated with modafinil, which also reduces excessive autophagy in hippocampus and likely activates P13K/Akt/mTOR/P70S6K signaling pathway (250). A study in rats revealed that sleep deprivation induces oxidative stress in the thyroid, which combined with alterations in autophagy- and apoptosis-related proteins, lead to an imbalance between autophagy and apoptosis in the organ (251). Consequently, an increased number of apoptotic cells were observed, suggesting potential damage to the thyroid. Finally, Kumar et al. performed a systematic review that indicate that autophagy dysfunction is connected to both acute and chronic neurodegenerative changes, as well as pathophysiological and behavioral alterations, all of which are linked to rapid eye movement (REM) sleep loss (252). Moreover, the increased levels of noradrenaline during REM sleep disruption may potentially affect autophagy in neurons, resulting in disturbances to neuronal integrity, homeostasis, and overall brain functions. These disturbances could ultimately play a role in the development of associated neurodegenerative disorders.
Tobacco
The tobacco epidemic is a significant global public health challenge. It is one of the most notoriously overused drugs among rural and urban populations in underdeveloped countries (253). The rate of tobacco product consumption and the number of smokers have both been steadily rising globally over the past ten years. As a result, the World Health Organization (WHO) estimates that almost 7,000,000 deaths are directly related to tobacco use (254).
In vitro exposure of bronchial epithelial cells and in vivo exposure of mice to cigarette smoke extracts (CSEs) and cigarette smoke, respectively, lead to defective autophagosome maturation, impairing the autophagic flux (255). These may occurr due to the observed accumulation of bicaudal D1, p62 and ubiquitin-associated p62 oligomers. Another study reveals a connection between tobacco smoking and impaired autophagy in alveolar macrophages, the cells responsible for clearing bacteria from the lungs. Smokers have an increased risk of lung infections, and the research shows that alveolar macrophages in smokers accumulate autophagosomes and p62, indicative of a dysfunctional autophagy process (256). This defect hinders the clearance of protein aggregates, impairs mitochondria function, and disrupts the delivery of bacteria to lysosomes. In addition, autophagy is increased in lung tissue from COPD patients. In vitro experiments using human pulmonary epithelial cells exposed to CSE showed a rapid induction of autophagy. The study identified the transcription factor Egr-1 as a critical factor promoting autophagy and apoptosis in response to cigarette smoke exposure (257). Lastly, CSE was found to impair autophagy in U937 macrophage-like cells, leading to the accumulation of galectin-8 and the autophagic adaptor protein NDP52, which is also observable in lung tissue and blood circulation of COPD patients. Soluble galectin-8 induced the release of interleukin-6 (IL-6) in bronchial epithelial cells via PI3Kα signaling (258). The study suggests that increased galectin-8 due to impaired autophagy induced by CSE may contribute to the development of COPD and could serve as a potential biomarker for the disease.
Alcohol
Drinking alcohol is linked to a variety of harmful medical effects that essentially affect every system in the body (259). Additionally, drinking alcohol has been linked to a number of cancers, which may be explained by the genotoxic effects of acetaldehyde, the generation of reactive oxygen and nitrogen species (ROS and RNS), modifications to the metabolism and DNA methylation of folate, impaired immune surveillance, nutritional deficiencies, and elevated estrogen levels (260).
In this sense, chronic alcohol consumption is a common risk factor for the onset of alcoholic liver disease (ALD). ROS and local inflammation are the main causes of alcohol-induced hepatocellular injury, which harms the structure of the liver and ultimately results in increased cell loss from the liver (261). There is not fully comprehension of how drinking alcohol affects the autophagic system of the liver. The studies suggest that alcohol can induce autophagy in the liver, acting as a cellular protective mechanism against acute ethanol-induced steatosis and liver injury (262,263). Ethanol metabolism through alcohol dehydrogenase (ADH) and CYP2E1 generates reactive metabolites that are required for autophagy induction. Ethanol-induced autophagy selectively removes damaged mitochondria and lipid droplets accumulated in liver cells with ethanol treatment, contributing to its beneficial effects in alcohol-induced liver disease (264). Pharmacological induction of autophagy by rapamycin or carbamazepine suppresses acute alcohol-induced steatosis and liver injury in mice (265). However, the effects of chronic ethanol exposure on autophagy are the inhibition of hepatocellular autophagy and induction of apoptotic cell death in Wistar rats (266). Nonetheless, enhancing hepatic autophagy seems to be beneficial in ALD, and specific enhancers of autophagy may hold promise as potential therapeutic interventions.
Air pollution
Air pollution primarily consists of particulate matter (PM) containing hazardous airborne particles suspended in the atmosphere. Studies have shown that air pollution can trigger autophagy in human lung epithelial cell lines, potentially leading to impairment in pulmonary function. The underlying mechanism involves oxidative stress induced by PM2.5 (particles with a diameter of less than 2.5 µm), which leads to accumulation of LC3 and elevated expression of BECN1 and ATG5, key components involved in autophagy (267). Conversely, inhibiting autophagy or depleting its resources can result in varied toxic effects, depending on the specific context (268). Mammalian cells rely on selective autophagy to effectively eliminate particulate matter, nanoparticles, toxic metals, and exposure to smoke without harming cytosolic components (269). One of the first organs exposed to PM is the skin. Results from Park et al. showed that PM contributes to skin aging by impairing collagen synthesis and triggering inflammation, along with an increase in autophagy which may indicate a potential reparative role in response to PM-induced stress (270).
Sunlight exposure
The primary risk factor for the development of skin cancer and skin photoaging is exposure to ultraviolet (UV) radiation from sunshine and indoor tanning beds. The most prevalent is UVA, which makes up around 95% of solar UV radiation and the remaining 5% of solar UV light is made up of UVB, and UVC is blocked by ozone. UV light is the primary source of vitamin D for humans (271).
UVA, UVB, and UVC stimulate the creation of autophagosomes and the overexpression of autophagy markers. It is believed to be a protective response against skin from ultraviolet radiation-driven photoaging or skin cancer (272,273). Autophagy degrades oxidized phospholipids and protein aggregates generated by UVA-induced ROS production, which are the best-known mechanism that activate autophagy.(274) Deeply, oxidative stress seems to activate AMPK autophagy signaling (275).
On the one hand, UVA exposure leads to the up-regulation of p62, triggering a positive feedback loop with Nrf2 (nuclear factor (erythroid-derived 2)-like 2) to counteract oxidative stress. Moreover, p62 also forms a negative feedback loop with PTEN (phosphatase and tensin homologue deleted on chromosome 10), a tumor suppressor in melanoma cells, suggesting that p62 acts as an oncogene in UVA-associated melanoma development and progression (276). PTEN stimulates the autophagic flux by the inhibition of PI3K/AKT/mTOR signaling (277,278). On the other hand, the alterations in autophagy induced by UVA are a result of impaired autophagic flux caused by the inactivation of cathepsin B, which may play a role in UVA-induced skin photodamage (279).
Psychosocial stress
Alterations in autophagy have been linked to typical stress response pathways, and abnormal autophagy appears to be related to neuropsychiatric diseases (280). Studies conducted after death on people with major depressive disorder revealed deficiencies in the autophagy regulating mechanism mTOR. Additionally, it was discovered that antidepressants work through the mTOR pathway and reverse the psychological consequences of chronic stress (281).
Findings from the impact of abused substances indicate the vital role of autophagy in regulating emotional and cognitive behavior in response to psychological stress, particularly in the catecholamine synapse and neuronal protection (282). Autophagy plays a significant role in promoting adaptive emotional outcomes that contribute to overall wellbeing. However, in situations of chronic or persistent stress, where changes in neurotransmitter activity lead to maladaptive neuronal alterations, autophagy may be continuously impaired, leading to a gradual decline in synaptic function and ultimately to neurodegeneration (282). Indeed, chronic nicotine administration in mice exposed to mild chronic unpredictable stress, improved cognitive impairments and neuropathological alteration of dentate gyrus neurons caused by chronic stress in mice (283). The neuroprotective effects of nicotine were associated with an enhancement of the autophagy signaling pathway, involving the upregulation of autophagy markers (BECN1 and LC3 II). Paradoxically, these effects were also linked to the activation of the PI3K/Akt/mTOR signaling pathway.
On the other hand, epigenetic modifications play a crucial role in regulating autophagy and are linked to stress response and neuropsychiatric disorders. The HSP90 co-chaperone FKBP51 activates autophagy genes in response to antidepressants by reducing DNA methylation and increasing gene expression, including BDNF involved in neurogenesis (284). Stress conditions are associated with epigenetic changes that influence autophagic clearance of RNA from mobile genetic elements, while chronic stress may impact autophagy-related genes, potentially affecting neurogenesis (285). Autophagy's compensatory role in clearing aberrant transcripts may be crucial during chronic stress. Lastly, an interesting studio analyzed the relationship between psychosocial stress and intestinal autophagy in both intestinal bowel disease (IBD) patients and animal model. The findings suggest that psychosocial stress may exacerbate IBD by enhancing intestinal autophagy through gut microbiota modulation and inflammation, highlighting autophagy as a potential therapeutic target for psychosocial stress-related IBD (286).
Conclusions
Autophagy is a growing area of research that has been related to several physiological and pathological processes, although their biological significance remains to be further explored. Compelling evidence define autophagy as a pivotal mechanism in the context of cellular stress response, aging, immunity, metabolism, tissue differentiation, and systemic illnesses like cancer, cardiovascular disease, or neurological disorders. Besides, different types of autophagy like macroautophagy, microautophagy, and chaperone-mediated autophagy are currently recognized by the available literature, each of them entailing a complex molecular background. Strategies involved in autophagy modulation include pharmacological interventions with potential clinical applications (Table 1). Besides, lifestyle medicine has also a notable role in the modulation of autophagy, as shown in Table 2. Diet, exercise, sleep habits, and environmental factors can all have a relevant influence on the process. The inclusion of lifestyle medicine in autophagy research creates new opportunities for individualized therapies that enhance health and longevity, although further efforts in this field are still warranted.
Acknowledgements
Funding
The study was supported by the Comunidad de Madrid (P2022/BMD-7321) and ProACapital, Halekulani S.L. and MJR.
Data availability statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Author contributions
All authors have read and agreed to the published version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nature Cell Biology. 2013;15(7):713-20
2. López-Otín C, Kroemer G. Hallmarks of Health. Cell. 2021;184(1):33-63
3. Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P. et al. Autophagy in major human diseases. The EMBO Journal. 2021;40(19):1-64
4. Maiuri MC, Kroemer G. Therapeutic modulation of autophagy: which disease comes first? Cell Death and Differentiation. 2019;26(4):680-9
5. Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature Reviews Drug Discovery. 2012;11(9):709-30
6. Vakifahmetoglu-Norberg H, Xia HG, Yuan J. Pharmacologic agents targeting autophagy. Journal of Clinical Investigation. 2015;125(1):5-13
7. Khandia R, Dadar M, Munjal A, Dhama K, Karthik K, Tiwari R. et al. A comprehensive review of autophagy and its various roles in infectious, non-infectious, and lifestyle diseases: Current knowledge and prospects for disease prevention, novel drug design, and therapy. Cells. 2019 8(7)
8. Kroemer G, Mariño G, Levine B. Autophagy and the Integrated Stress Response. Molecular Cell. 2010;40(2):280-93
9. Pierzynowska K, Gaffke L, Cyske Z, Puchalski M, Rintz E, Bartkowski M. et al. Autophagy stimulation as a promising approach in treatment of neurodegenerative diseases. Metabolic Brain Disease. 2018;33(4):989-1008
10. Grasso D, Renna FJ, Vaccaro MI. Initial steps in Mammalian autophagosome biogenesis. Frontiers in Cell and Developmental Biology. 2018;6(OCT):1-10
11. Hurley JH, Young LN. Mechanisms of autophagy initiation. Annual Review of Biochemistry. 2017;86:225-44
12. Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H. et al. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nature Communications. 2016 7(12420)
13. Lu G, Wang Y, Shi Y, Zhang Z, Huang C, He W. et al. Autophagy in health and disease: From molecular mechanisms to therapeutic target. MedComm. 2022;3(3):1-52
14. Demeter A, Romero-Mulero MC, Csabai L, Ölbei M, Sudhakar P, Haerty W. et al. ULK1 and ULK2 are less redundant than previously thought: computational analysis uncovers distinct regulation and functions of these autophagy induction proteins. Scientific Reports. 2020;10(1):1-17
15. Feng Y, Kang HH, Wong PM, Gao M, Wang P, Jiang X. Unc-51-like kinase (ULK) complex-independent autophagy induced by hypoxia. Protein and Cell. 2019;10(5):376-81
16. Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(27):11121-6
17. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annual Review of Genetics. 2009;43:67-93
18. Wong PM, Feng Y, Wang J, Shi R, Jiang X. Regulation of autophagy by coordinated action of mTORC1 and protein phosphatase 2A. Nature Communications. 2015;6:1-11
19. Al-Bari MAA, Xu P. Molecular regulation of autophagy machinery by mTOR-dependent and -independent pathways. Annals of the New York Academy of Sciences. 2020;1467(1):3-20
20. Ren Y, Xie H, Shen HM. Regulation of Autophagy by AMPK. In: Autophagy and Signaling. CRC Press. 2017 p. 3-24
21. Wang S, Li H, Yuan M, Fan H, Cai Z. Role of AMPK in autophagy. Frontiers in Physiology. 2022;13(November):1-11
22. Mrakovcic M, Fröhlich LF. P53-mediated molecular control of autophagy in tumor cells. Biomolecules. 2018 8(2)
23. Rahman MA, Park MN, Rahman MH, Rashid MM, Islam R, Uddin MJ. et al. p53 Modulation of Autophagy Signaling in Cancer Therapies: Perspectives Mechanism and Therapeutic Targets. Frontiers in Cell and Developmental Biology. 2022;10(January):1-14
24. Birgisdottir ÅB, Mouilleron S, Bhujabal Z, Wirth M, Sjøttem E, Evjen G. et al. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy. 2019;15(8):1333-55
25. Egan DF, Chun MGH, Vamos M, Zou H, Rong J, Miller CJ. et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Molecular Cell. 2015;59(2):285-97
26. Park JM, Seo M, Jung CH, Grunwald D, Stone M, Otto NM. et al. ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy. 2018;14(4):584-97
27. Wold MS, Lim J, Lachance V, Deng Z, Yue Z. ULK1-mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington's disease models. Molecular Neurodegeneration. 2016;11(1):1-13
28. Mercer TJ, Ohashi Y, Boeing S, Jefferies HBJ, De Tito S, Flynn H. et al. Phosphoproteomic identification of ULK substrates reveals VPS15-dependent ULK/VPS34 interplay in the regulation of autophagy. The EMBO Journal. 2021;40(14):1-25
29. Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T. et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. The Journal of cell biology. 2010Aug23;190(4):511-21
30. Nascimbeni AC, Codogno P, Morel E. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS Journal. 2017;284(9):1267-78
31. Backer JM. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochemical Journal. 2016;473(15):2251-71
32. Uemura T, Yamamoto M, Kametaka A, Sou Y shin, Yabashi A, Yamada A. et al. A Cluster of Thin Tubular Structures Mediates Transformation of the Endoplasmic Reticulum to Autophagic Isolation Membrane. Molecular and Cellular Biology. 2014;34(9):1695-706
33. Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A. et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. Journal of Cell Biology. 2008;182(4):685-701
34. Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 2018;14(2):207-15
35. Li D, Zhao YG, Li D, Zhao H, Huang J, Miao G. et al. The ER-Localized Protein DFCP1 Modulates ER-Lipid Droplet Contact Formation. Cell Reports. 2019;27(2):343-358.e5
36. Nähse V, Raiborg C, Wee Tan K, Mørk S, Lyngaas Torgersen M, Wenzel M. et al. ATPase activity of DFCP1 controls selective autophagy. Nature Communications. 2023 14(4052)
37. Almannai M, Marafi D, El-Hattab AW. WIPI proteins: Biological functions and related syndromes. Frontiers in Molecular Neuroscience. 2022 15
38. Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-Site Recognition of Phosphatidylinositol 3-Phosphate by PROPPINs in Autophagy. Molecular Cell. 2012;47(3):339-48
39. Gong X, Wang Y, Tang Y, Wang Y, Zhang M, Li M. et al. ATG16L1 adopts a dual-binding site mode to interact with WIPI2b in autophagy. Science Advances. 2023 9(9)
40. Gong X, Pan L. ATG16L1 is equipped with two distinct WIPI2-binding sites to drive autophagy. Autophagy. 2023.
41. Jensen LE, Rao S, Schuschnig M, Cada AK, Martens S, Hummer G. et al. Membrane curvature sensing and stabilization by the autophagic LC3 lipidation machinery. Science Advances. 2022;8(50):1-12
42. Puri C, Vicinanza M, Ashkenazi A, Gratian MJ, Zhang Q, Bento CF. et al. The RAB11A-Positive Compartment Is a Primary Platform for Autophagosome Assembly Mediated by WIPI2 Recognition of PI3P-RAB11A. Developmental Cell. 2018;45(1):114-131.e8
43. Bento CF, Renna M, Ghislat G, Puri C, Ashkenazi A, Vicinanza M. et al. Mammalian Autophagy: How Does It Work? Annual Review of Biochemistry. 2016;85:685-713
44. Nishimura T, Tooze SA. Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discovery. 2020 6(1)
45. Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S. et al. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nature Cell Biology. 2014;16(5):415-24
46. Sakoh-Nakatogawa M, Matoba K, Asai E, Kirisako H, Ishii J, Noda NN. et al. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nature Structural and Molecular Biology. 2013;20(4):433-9
47. Brier LW, Ge L, Stjepanovic G, Thelen AM, Hurley JH, Schekman R. Regulation of LC3 lipidation by the autophagyspecific class III phosphatidylinositol-3 kinase complex. Molecular Biology of the Cell. 2019;30(9):1098-107
48. Melia TJ, Lystad AH, Simonsen A. Autophagosome biogenesis: From membrane growth to closure. Journal of Cell Biology. 2020;219(6):1-18
49. Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-kakuta C. et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. Journal of Cell Biology. 2012;198(2):219-33
50. Olivas TJ, Wu Y, Yu S, Luan L, Choi P, Nag S. ATG9 vesicles comprise the seed membrane of mammalian autophagosomes. The Journal of cell biology. 2023 222(7)
51. Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM. et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Molecular Biology of the Cell. 2012;23(10):1860-73
52. Suzuki SW, Yamamoto H, Oikawa Y, Kondo-kakuta C, Kimura Y, Hirano H. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(11):3350-5
53. Sawa-makarska J, Baumann V, Coudevylle N, Bülow S Von, Nogellova V, Abert C. et al. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science. 2020;369(1206):1-10
54. Bhatti JS, Bhatti GK, Reddy PH. Mitochondrial dysfunction and oxidative stress in metabolic disorders - A Step towards mitochondria based therapeutic strategies. Biochimica et biophysica acta. 2017May1;1863(5):1066
55. Goder V. Specific COPII vesicles transport ER membranes to sites of autophagosome formation. Molecular & Cellular Oncology. 2017;4(1):1-3
56. Shima T, Kirisako H, Nakatogawa H. COPII vesicles contribute to autophagosomal membranes. Journal of Cell Biology. 2019;218(5):1503-10
57. Hailey DW, Rambold AS, Satpute-krishnan P, Mitra K, Sougrat R, Kim PK. Mitochondria Supply Membranes for Autophagosome Biogenesis during Starvation. Cell. 2010;141(4):656-67
58. Biazik J, Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy. 2015;11(3):439-51
59. Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T. et al. ATG2 transports lipids to promote autophagosome biogenesis. Journal of Cell Biology. 2019;218(6):1787-98
60. Chowdhury S, Otomo C, Leitner A, Ohashi K, Aebersold R, Lander GC. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(42):9792-801
61. Zheng JX, Li Y, Ding YH, Liu JJ, Zhang MJ, Dong MQ. et al. Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy. 2017;13(11):1870-83
62. Knorr RL, Lipowsky R, Dimova R. Autophagosome closure requires membrane scission. Autophagy. 2015;11(11):2134-7
63. Jiang W, Chen X, Ji C, Zhang W, Song J, Li J. et al. Key regulators of autophagosome closure. Cells. 2021;10(11):1-13
64. Polyansky A, Shatz O, Fraiberg M, Shimoni E, Dadosh T, Mari M. et al. Phospholipid imbalance impairs autophagosome completion. The EMBO Journal. 2022;41(23):1-21
65. Zhao YG, Codogno P, Zhang H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nature Reviews Molecular Cell Biology. 2021;22(11):733-50
66. Zhao YG, Zhang H. Autophagosome maturation: An epic journey from the ER to lysosomes. Journal of Cell Biology. 2019;218(3):757-70
67. Reggiori F, Ungermann C. Autophagosome Maturation and Fusion. Journal of Molecular Biology. 2017;429(4):486-96
68. Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell structure and function. 2008;33(1):109-22
69. Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T. et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Molecular Biology of the Cell. 2014;25(8):1327-37
70. Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256-69
71. Li Y, Cheng X, Li M, Wang Y, Fu T, Zhou Z. et al. Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(35):21391-402
72. Tian X, Teng J, Chen J. New insights regarding SNARE proteins in autophagosome-lysosome fusion. Autophagy. 2021;17(10):2680-8
73. Hyttinen JMT, Niittykoski M, Salminen A, Kaarniranta K. Maturation of autophagosomes and endosomes: A key role for Rab7. Biochimica et Biophysica Acta - Molecular Cell Research. 2013;1833(3):503-10
74. Kuchitsu Y, Fukuda M. Revisiting Rab7 functions in mammalian autophagy: Rab7 knockout studies. Cells. 2018 7(11)
75. Yu X, Long YC, Shen HM. Differential regulatory functions of three classes of phosphatidylinositol and phosphoinositide 3-kinases in autophagy. Autophagy. 2015;11(10):1711-28
76. Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA. et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nature Cell Biology. 2008;10(7):776-87
77. Tsuboyama K, Koyama-Honda I, Sakamaki Y, Koike M, Morishita H, Mizushima N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354(6315):1036-41
78. Schröder BA, Wrocklage C, Hasilik A, Saftig P. The proteome of lysosomes. Proteomics. 2010;10(22):4053-76
79. Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J. et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465(7300):942-6
80. Chen Y, Yu L. Recent progress in autophagic lysosome reformation. Traffic. 2017;18(6):358-61
81. Yim WWY, Mizushima N. Lysosome biology in autophagy. Cell Discovery. 2020 6(1)
82. Sica V, Galluzzi L, Bravo-San Pedro JM, Izzo V, Maiuri MC, Kroemer G. Organelle-Specific Initiation of Autophagy. Molecular Cell. 2015;59(4):522-39
83. Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nature Cell Biology. 2014;16(6):495-501
84. Vargas JNS, Hamasaki M, Kawabata T, Youle RJ, Yoshimori T. The mechanisms and roles of selective autophagy in mammals. Nature Reviews Molecular Cell Biology 2022 24:3. 2022Oct27;24(3):167-85
85. Kirkin V, Rogov V V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Molecular Cell. 2019;76(2):268-85
86. Zaffagnini G, Martens S. Mechanisms of Selective Autophagy. Journal of Molecular Biology. 2016;428(9):1714-24
87. Imai Y. PINK1-Parkin signaling in Parkinson's disease: Lessons from Drosophila. Neuroscience Research. 2020;159:40-6
88. Ge P, Dawson VL, Dawson TM. PINK1 and Parkin mitochondrial quality control: A source of regional vulnerability in Parkinson's disease. Molecular Neurodegeneration. 2020;15(1):1-18
89. Shimizu S. Molecules and Cells Biological Roles of Alternative Autophagy. Molecules and Cells. 2018;41(1):50-4
90. Urbanska K, Orzechowski A. The Secrets of Alternative Autophagy. Cells. 2021;10(3241):1-15
91. Feng H, Wang N, Zhang N, Liao H han. Alternative autophagy: mechanisms and roles in different diseases. Cell Communication and Signaling. 2022;20(1):1-13
92. Seki T, Katsuki H. Mammalian microautophagy: mechanism and roles in disease. In: Autophagy in Health and Disease. Academic Press. 2022 p. 385-97
93. Wang L, Klionsky DJ, Shen HM. The emerging mechanisms and functions of microautophagy. Nature Reviews Molecular Cell Biology 2022 24:3. 2022Sep12;24(3):186-203
94. Li WW, Li J, Bao JK. Microautophagy: Lesser-known self-eating. Cellular and Molecular Life Sciences. 2012;69(7):1125-36
95. Oku M, Sakai Y. Three Distinct Types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. BioEssays. 2018;40(6):1-6
96. Cuervo AM, Wong E. Chaperone-mediated autophagy: Roles in disease and aging. Cell Research. 2014;24(1):92-104
97. Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nature Reviews Molecular Cell Biology. 2018;19(6):365-81
98. Losmanová T, Janser FA, Humbert M, Tokarchuk I, Schläfli AM, Neppl C, et al. Erratum: Chaperone-Mediated Autophagy Markers LAMP2A and HSC70 Are Independent Adverse Prognostic Markers in Primary Resected Squamous Cell Carcinomas of the Lung (Oxidative Medicine and Cellular Longevity (2020) 2020 (8506572) DOI: 10.1155/2020/8506572). Oxidative Medicine and Cellular Longevity. 2021;2021
99. Robert G, Jacquel A, Auberger P. Chaperone-mediated autophagy and its emerging role in hematological malignancies. Cells. 2019;8(10):1-19
100. Stead ER, Castillo-Quan JI, Miguel VEM, Lujan C, Ketteler R, Kinghorn KJ. et al. Agephagy - Adapting Autophagy for Health During Aging. Frontiers in Cell and Developmental Biology. 2019Nov28;7:308
101. Saha S, Panigrahi DP, Patil S, Bhutia SK. Autophagy in health and disease: A comprehensive review. Biomedicine and Pharmacotherapy. 2018;104(February):485-95
102. Ma Q, Long S, Gan Z, Tettamanti G, Li K, Tian L. Transcriptional and Post-Transcriptional Regulation of Autophagy. Cells. 2022;11(3):1-12
103. Lei Y, Klionsky DJ. Transcriptional regulation of autophagy and its implications in human disease. Cell Death & Differentiation. 2023;30(6):1416-29
104. Mathiassen SG, De Zio D, Cecconi F. Autophagy and the cell cycle: A complex landscape. Frontiers in Oncology. 2017;7(MAR):1-16
105. Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: The interplay of autophagy and apoptosis. Nature Reviews Molecular Cell Biology. 2014;15(2):81-94
106. Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nature reviews Molecular cell biology. 2008Dec;9(12):1004
107. Fan YJ, Zong WX. The cellular decision between apoptosis and autophagy. Chinese Journal of Cancer. 2013;32(3):121
108. Ghavami S, Shojaei S, Yeganeh B, Ande SR, Jangamreddy JR, Mehrpour M. et al. Autophagy and apoptosis dysfunction in neurodegenerative disorders. Progress in Neurobiology. 2014Jan1;112:24-49
109. Cong L, Bai Y, Guo Z. The crosstalk among autophagy, apoptosis, and pyroptosis in cardiovascular disease. Frontiers in Cardiovascular Medicine. 2022Oct28;9:997469
110. Peker N, Gozuacik D. Autophagy as a Cellular Stress Response Mechanism in the Nervous System. Journal of Molecular Biology. 2020;432(8):2560-88
111. Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: Implications for cell injury and human disease. Annual Review of Pathology: Mechanisms of Disease. 2013;8(October 2012):105-37
112. Barbosa MC, Grosso RA, Fader CM. Hallmarks of Aging: An Autophagic Perspective. Frontiers in Endocrinology. 2018 9(JAN)
113. López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194-217
114. Tabibzadeh S. Role of autophagy in aging: The good, the bad, and the ugly. Aging Cell. 2023;22(1):1-15
115. Jiang GM, Tan Y, Wang H, Peng L, Chen HT, Meng XJ. et al. The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Molecular Cancer 2019 18:1. 2019Jan24;18(1):1-22
116. Harris J. Autophagy and cytokines. Cytokine. 2011;56(2):140-4
117. Qian M, Fang X, Wang X. Autophagy and inflammation. Clinical and Translational Medicine. 2017 6(1)
118. Pang Y, Wu L, Tang C, Wang H, Wei Y. Autophagy-Inflammation Interplay During Infection: Balancing Pathogen Clearance and Host Inflammation. Frontiers in Pharmacology. 2022;13(February):1-12
119. Deretic V. Autophagy in inflammation, infection, and immunometabolism. Immunity. 2021;54(3):437-53
120. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469(7330):323-35
121. Zuo H, Chen C, Sa Y. Therapeutic potential of autophagy in immunity and inflammation: current and future perspectives. Pharmacological Reports. 2023;75(3):499-510
122. Ortega MA, Fraile-Martinez O, Garcia-Montero C, Alvarez-Mon MA, Gomez-Lahoz AM, Albillos A. et al. An Updated View of the Importance of Vesicular Trafficking and Transport and Their Role in Immune-Mediated Diseases: Potential Therapeutic Interventions. Membranes. 2022;12(6):1-20
123. Deretic V, Kroemer G. Autophagy in metabolism and quality control: opposing, complementary or interlinked functions? Autophagy. 2022;18(2):283-92
124. Riffelmacher T, Clemens F, Katharina A. Autophagy dictates metabolism and differentiation of in fl ammatory immune cells. Autophagy. 2018;14(2):199-206
125. Kim KH, Lee M shik. Autophagy—a key player in cellular and body metabolism. Nature Reviews Endocrinology. 2014;10:322-37
126. Rabinowitz JD, White E. Autophagy and Metabolism. Science. 2010;330:1344-8
127. Weckman A, Ieva A Di, Rotondo F, Syro L V, Ortiz LD, Kovacs K. et al. Autophagy in the endocrine glands. Journal of Molecular Endocrinology. 2014;52(2):151-63
128. Kitada M, Koya D. Autophagy in metabolic disease and ageing. Nature Reviews Endocrinology. 2021;17(11):647-61
129. Allen EA, Baehrecke EH. Autophagy in animal development. Cell Death & Differentiation. 2020;27:903-18
130. Perrotta C, Cattaneo MG, Molteni R, Palma C De, Sano F Di. Autophagy in the Regulation of Tissue Differentiation and Homeostasis. Frontiers in Cell and Developmental Biology. 2020;8(December):1-20
131. Adelipour M, Regi L, Ghavami S, Narayan K, Dhingra S, Allameh A. The role of autophagy in the metabolism and differentiation of stem cells. BBA - Molecular Basis of Disease. 2022;1868:166412
132. Trejo-Iriarte CG, Ortega MA, Asúnsolo Á, Gómez-Clavel JF, Muñoz AG, Mon MÁ. et al. Mesenchymal adipose stem cells maintain the capacity for differentiation and survival in culture beyond the long term. Journal of Histotechnology. 2021;44(4):217-33
133. Chang NC. Autophagy and Stem Cells: Self-Eating for Self-Renewal. Frontiers in Cell and Developmental Biology. 2020;8(March):1-11
134. Fraiberg M, Elazar Z. Genetic defects of autophagy linked to disease. In: Progress in Molecular Biology and Translational Science. Academic Press. 2020 p. 293-323
135. Ebrahimi-Fakhari D, Saffari A, Wahlster L, Lu J, Byrne S, Hoffmann GF. et al. Congenital disorders of autophagy: An emerging novel class of inborn errors of neuro-metabolism. Brain. 2016;139(2):317-37
136. Abdellatif M, Sedej S, Carmona-Gutierrez D, Madeo F, Kroemer G. Autophagy in cardiovascular aging. Circulation Research. 2018;123(7):803-24
137. Koutouroushis C, Sarkar O. Role of Autophagy in Cardiovascular Disease and Aging. Cureus. 2021 13(11)
138. Schiattarella GG, Hill JA. Therapeutic targeting of autophagy in cardiovascular disease. Journal of Molecular and Cellular Cardiology. 2016Jun1;95:86-93
139. Bar-Yosef T, Damri O, Agam G. Dual role of autophagy in diseases of the central nervous system. Frontiers in Cellular Neuroscience. 2019;13(May):1-14
140. Yun CW, Jeon J, Go G, Lee JH, Lee SH. The dual role of autophagy in cancer development and a therapeutic strategy for cancer by targeting autophagy. International Journal of Molecular Sciences. 2021;22(1):1-22
141. Yun CW, Lee SH. The roles of autophagy in cancer. International Journal of Molecular Sciences. 2018;19(11):1-18
142. Jiang P, Mizushima N. Autophagy and human diseases. Cell Research. 2014;24(1):69-79
143. Jung S, Jeong H, Yu SW. Autophagy as a decisive process for cell death. Experimental and Molecular Medicine. 2020;52(6):921-30
144. Bischoff P, Josset E, Dumont FJ. Novel pharmacological modulators of autophagy and therapeutic prospects. Expert Opinion on Therapeutic Patents. 2012;22(9):1053-79
145. Thellung S, Corsaro A, Nizzari M, Barbieri F, Florio T. Autophagy activator drugs: A new opportunity in neuroprotection from misfolded protein toxicity. International Journal of Molecular Sciences. 2019 20(4)
146. Yang YP, Hu LF, Zheng HF, Mao CJ, Hu WD, Xiong KP. et al. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacologica Sinica. 2013;34(5):625-35
147. Pasquier B. Autophagy inhibitors. Cellular and Molecular Life Sciences. 2016;73(5):985-1001
148. Li J, Kim SG, Blenis J. Rapamycin: One drug, many effects. Cell Metabolism. 2014;19(3):373-9
149. Liu W, Huang S, Chen Z, Wang H, Wu H, Zhang D. Temsirolimus, the mTOR inhibitor, induces autophagy in adenoid cystic carcinoma: In vitro and in vivo. Pathology Research and Practice. 2014;210(11):764-9
150. Lui A, New J, Ogony J, Thomas S, Lewis-Wambi J. Everolimus downregulates estrogen receptor and induces autophagy in aromatase inhibitor-resistant breast cancer cells. BMC Cancer. 2016;16(1):1-15
151. Li YY, Sze-Kwan L, Choi-Wo Mak, Judith, Zheng CY, Chung-Man Ho J. Erlotinib-induced autophagy in epidermal growth factor receptor mutated non-small cell lung cancer. Lung Cancer. 2013;81(3):354-61
152. Ertmer A, Huber V, Gilch S, Yoshimori T, Erfle V, Duyster J. et al. The anticancer drug imatinib induces cellular autophagy. Leukemia. 2007;21(5):936-42
153. Elzinga BM, Nyhan MJ, Crowley LC, O'Donovan TR, Cahill MR, McKenna SL. Induction of autophagy by Imatinib sequesters Bcr-Abl in autophagosomes and down-regulates Bcr-Abl protein. American Journal of Hematology. 2013;88(6):455-62
154. Le XF, Mao W, Lu Z, Carter BZ, Bast RC. Dasatinib induces autophagic cell death in human ovarian cancer. Cancer. 2010;116(21):4980-90
155. Morita M, Nishinaka Y, Kato I, Saida S, Hiramatsu H, Kamikubo Y. et al. Dasatinib induces autophagy in mice with Bcr-Abl-positive leukemia. International Journal of Hematology. 2017;105(3):335-40
156. Gammoh N, Lam D, Puente C, Ganley I, Marks PA, Jiang X. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(17):6561-5
157. Dupéré-Richer D, Kinal M, Ménasché V, Nielsen TH, Del Rincon S, Pettersson F. et al. Vorinostat-induced autophagy switches from a death-promoting to a cytoprotective signal to drive acquired resistance. Cell Death and Disease. 2013;4(2):1-11
158. Patra S, Praharaj PP, Klionsky DJ, Bhutia SK. Vorinostat in autophagic cell death: A critical insight into autophagy-mediated, -associated and -dependent cell death for cancer prevention. Drug Discovery Today. 2022;27(1):269-79
159. Fang S, Wan X, Zou X, Sun S, Hao X, Liang C. et al. Arsenic trioxide induces macrophage autophagy and atheroprotection by regulating ROS-dependent TFEB nuclear translocation and AKT/mTOR pathway. Cell Death and Disease. 2021;12(1):1-18
160. Holczer M, Besze B, Zámbó V, Csala M, Bánhegyi G, Kapuy O. Epigallocatechin-3-Gallate (EGCG) promotes autophagy-dependent survival via influencing the balance of mTOR-AMPK pathways upon endoplasmic reticulum stress. Oxidative Medicine and Cellular Longevity. 2018. 2018
161. Yin Z, Li J, Kang L, Liu X, Luo J, Zhang L. et al. Epigallocatechin-3-gallate induces autophagy-related apoptosis associated with LC3B II and Beclin expression of bladder cancer cells. Journal of Food Biochemistry. 2021;45(6):1-8
162. Liu B, Cheng Y, Bian HJ, Bao JK. Molecular mechanisms of Polygonatum cyrtonema lectin-induced apoptosis and autophagy in cancer cells. Autophagy. 2009;5(2):253-5
163. Liu T, Wu L, Wang D, Wang H, Chen J, Yang C. et al. Role of reactive oxygen species-mediated MAPK and NF-κB activation in polygonatum cyrtonema lectin-induced apoptosis and autophagy in human lung adenocarcinoma A549 cells. Journal of Biochemistry. 2016;160(6):315-24
164. Hofer SJ, Kroemer G, Kepp O. Autophagy-inducing nutritional interventions in experimental and clinical oncology. International Review of Cell and Molecular Biology. 2022Jan1;373:125-58
165. Madeo F, Bauer MA, Carmona-Gutierrez D, Kroemer G. Spermidine: a physiological autophagy inducer acting as an anti-aging vitamin in humans? Autophagy. 2019;15(1):165-8
166. Park D, Jeong H, Lee MN, Koh A, Kwon O, Yang YR. et al. Resveratrol induces autophagy by directly inhibiting mTOR through ATP competition. Scientific Reports. 2016;6(May 2015):1-11
167. Tian Y, Song W, Li D, Cai L, Zhao Y. Resveratrol as a natural regulator of autophagy for prevention and treatment of cancer. OncoTargets and Therapy. 2019;12:8601-9
168. Pineda-Ramírez N, Aguilera P. Resveratrol as an inductor of autophagy: Is there a unique pathway of activation? Neural Regeneration Research. 2021;16(1):101-3
169. Chu YL, Ho CT, Chung JG, Rajasekaran R, Sheen LY. Allicin induces p53-mediated autophagy in Hep G2 human liver cancer cells. Journal of Agricultural and Food Chemistry. 2012;60(34):8363-71
170. Xiang Y, Zhao J, Zhao M, Wang K. Allicin activates autophagic cell death to alleviate the malignant development of thyroid cancer. Experimental and Therapeutic Medicine. 2018;15(4):3537-43
171. Qomaladewi NP, Kim MY, Cho JY. Autophagy and its regulation by ginseng components. Journal of Ginseng Research. 2019;43(3):349-53
172. Ren H, Dai R, Chen Y, Xi Z, Xu H. How ginseng regulates autophagy: Insights from multistep process. Biomedicine and Pharmacotherapy. 2023;158(December 2022):114139
173. Zhu Y, Bu S. Curcumin Induces Autophagy, Apoptosis, and Cell Cycle Arrest in Human Pancreatic Cancer Cells. Evidence-based Complementary and Alternative Medicine. 2017;2017:1-13
174. Araveti PB, Srivastava A. Curcumin induced oxidative stress causes autophagy and apoptosis in bovine leucocytes transformed by Theileria annulata. Cell Death Discovery. 2019 5(1)
175. Moustapha A, Pérétout P, Rainey N, Sureau F, Geze M, Petit JM. et al. Curcumin induces crosstalk between autophagy and apoptosis mediated by calcium release from the endoplasmic reticulum, lysosomal destabilization and mitochondrial events. Cell Death Discovery. 2015 1(1)
176. Seglen PO, Gordon PB. 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proceedings of the National Academy of Sciences of the United States of America. 1982;79:1889-92
177. Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR. et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. Journal of Biological Chemistry. 2010;285(14):10850-61
178. Mauthe M, Orhon I, Rocchi C, Zhou X, Luhr M, Hijlkema KJ. et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435-55
179. Javaid HMA, Lim H, Shin S, Huh JY. Inhibition of autophagy with chloroquine dysregulates mitochondrial quality control and energetics in adipocytes. Archives of Pharmacal Research. 2022;45(10):731-42
180. Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1(2):84-91
181. Li L qing, Xie W jun, Pan D, Chen H, Zhang L. Inhibition of autophagy by bafilomycin A1 promotes chemosensitivity of gastric cancer cells. Tumor Biology. 2016;37(1):653-9
182. Seite S, Pioche T, Ory N, Plagnes-Juan E, Panserat S, Seiliez I. The autophagic flux inhibitor bafilomycine A1 affects the expression of intermediary metabolism-related genes in trout hepatocytes. Frontiers in Physiology. 2019;10(MAR):1-11
183. Nordstrøm LU, Sironi J, Aranda E, Maisonet J, Perez-Soler R, Wu P. et al. Discovery of autophagy inhibitors with antiproliferative activity in lung and pancreatic cancer cells. ACS Medicinal Chemistry Letters. 2015;6(2):134-9
184. Cook KL, Wärri A, Soto-Pantoja DR, Clarke PAG, Cruz MI, Zwart A. et al. Hydroxychloroquine inhibits autophagy to potentiate antiestrogen responsiveness in ER+ breast cancer. Clinical Cancer Research. 2014;20(12):3222-32
185. Schott CR, Ludwig L, Mutsaers AJ, Foster RA, Wood GA. The autophagy inhibitor spautin-1, either alone or combined with doxorubicin, decreases cell survival and colony formation in canine appendicular osteosarcoma cells. PLoS ONE. 2018;13(10):7-10
186. Kunimasa K, Ikeda-Ishikawa C, Tani Y, Tsukahara S, Sakurai J, Okamoto Y. et al. Spautin-1 inhibits mitochondrial complex I and leads to suppression of the unfolded protein response and cell survival during glucose starvation. Scientific Reports. 2022;12(1):1-12
187. Pasquier B. SAR405, a PIK3C3/VPS34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells. Autophagy. 2015;11(4):725-6
188. Staskiewicz L, Thorburn J, Morgan MJ, Thorburn A. Inhibiting autophagy by shRNA knockdown Cautions and recommendations. Autophagy. 2013;9(10):1449-50
189. Xiong X, Wu M, Zhang H, Li J, Lu B, Guo Y. et al. Atg5 siRNA inhibits autophagy and enhances norcantharidin-induced apoptosis in hepatocellular carcinoma. International Journal of Oncology. 2015;47(4):1321-8
190. Phillips EM, Frates EP, Park DJ. Lifestyle Medicine. Physical Medicine and Rehabilitation Clinics of North America. 2020Nov1;31(4):515-26
191. Rippe JM. Lifestyle Medicine: The Health Promoting Power of Daily Habits and Practices. American Journal of Lifestyle Medicine. 2018;12(6):499-512
192. Bodai B, Nakata T, Wong W, Clark D, Lawenda S, Tsou C. et al. Lifestyle Medicine: A Brief Review of Its Dramatic Impact on Health and Survival. Perm J. [revista en Internet] 2018 [acceso 8 de noviembre de 2021]; 22(1): 17-25. The Permanente Journal. 2018;22:17-25
193. Aman Y, Schmauck-Medina T, Hansen M, Morimoto RI, Simon AK, Bjedov I. et al. Autophagy in healthy aging and disease. Nature Aging. 2021;1(8):634-50
194. Min S, Masanovic B, Bu T, Matic RM, Vasiljevic I, Vukotic M. et al. The Association Between Regular Physical Exercise, Sleep Patterns, Fasting, and Autophagy for Healthy Longevity and Well-Being: A Narrative Review. Frontiers in Psychology. 2021 12(December)
195. McCarty MF. Nutraceutical and Dietary Strategies for Up-Regulating Macroautophagy. International Journal of Molecular Sciences. 2022 23(4)
196. Bagherniya M, Butler AE, Barreto GE, Sahebkar A. The effect of fasting or calorie restriction on autophagy induction: A review of the literature. Ageing Research Reviews. 2018;47(June):183-97
197. Rakowski M, Porębski S, Grzelak A. Nutraceuticals as Modulators of Autophagy: Relevance in Parkinson's Disease. International Journal of Molecular Sciences. 2022 23(7)
198. Son SM, Park SJ, Stamatakou E, Vicinanza M, Menzies FM, Rubinsztein DC. Leucine regulates autophagy via acetylation of the mTORC1 component raptor. Nature Communications. 2020;11(1):1-13
199. Stacchiotti A, Corsetti G. Natural Compounds and Autophagy: Allies Against Neurodegeneration. Frontiers in Cell and Developmental Biology. 2020;8(September):1-16
200. Liuzzi JP, Pazos R. Interplay Between Autophagy and Zinc. Journal of Trace Elements in Medicine and Biology. 2020;62(July):126636
201. Krishan S, Jansson PJ, Gutierrez E, Lane DJR, Richardson D, Sahni S. Iron metabolism and autophagy: A poorly explored relationship that has important consequences for health and disease. Nagoya Journal of Medical Science. 2015;77(1-2):1-6
202. Luo M, Mai M, Song W, Yuan Q, Feng X, Xia E. et al. The Antiaging Activities of Phytochemicals in Dark-Colored Plant Foods: Involvement of the Autophagy- and Apoptosis-Associated Pathways. International Journal of Molecular Sciences. 2022 23(19)
203. García-Montero C, Fraile-Martínez O, Gómez-Lahoz AM, Pekarek L, Castellanos AJ, Noguerales-Fraguas F. et al. Nutritional components in western diet versus mediterranean diet at the gut microbiota-immune system interplay. implications for health and disease. Nutrients. 2021;13(2):1-53
204. Schwingshackl L, Morze J, Hoffmann G. Mediterranean diet and health status: Active ingredients and pharmacological mechanisms. British Journal of Pharmacology. 2020;177(6):1241-57
205. Kocot AM, Wróblewska B. Nutritional strategies for autophagy activation and health consequences of autophagy impairment. Nutrition. 2022:103-104
206. Wu X, Nagy LE. MLKL contributes to Western diet-induced liver injury through inhibiting autophagy. Autophagy. 2020;16(7):1351-2
207. Budi YP, Li YH, Huang C, Wang ME, Lin YC, Jong DS. et al. The role of autophagy in high-fat diet-induced insulin resistance of adipose tissues in mice. PeerJ. 2022;10:1-19
208. Alvarez-Mon MÁ, Ortega MÁ, Garcia-Montero C, Fraile-Martínez Ó, Montserrat J, Lahera G. et al. Exploring the Role of Nutraceuticals in Major Depressive Disorder (MDD): Rationale, State of the Art and Future Prospects. Pharmaceuticals. 2021;14(821):1-32
209. Ortega MA, Fraile-Martínez Ó, García-Montero C, Alvarez-Mon MA, Lahera G, Monserrat J. et al. Biological Role of Nutrients, Food and Dietary Patterns in the Prevention and Clinical Management of Major Depressive Disorder. Nutrients. 2022 14(15)
210. Most J, Tosti V, Redman LM, Fontana L. Calorie restriction in humans: An update. Ageing Research Reviews. 2017;39:36-45
211. Xiang L, He G. Caloric restriction and antiaging effects. Annals of Nutrition and Metabolism. 2011;58(1):42-8
212. Liang Y, Gao Y, Hua R, Lu M, Chen H, Wang Z. et al. Calorie intake rather than food quantity consumed is the key factor for the anti-aging effect of calorie restriction. Aging. 2021;13(17):21526-46
213. Mattson MP, Longo VD, Harvie M. Impact of intermittent fasting on health and disease processes. Ageing Research Reviews. 2017;39:46-58
214. Chung KW, Chung HY. The effects of calorie restriction on autophagy: Role on aging intervention. Nutrients. 2019;11(12):1-18
215. Song DK, Kim YW. Beneficial effects of intermittent fasting: a narrative review. Journal of Yeungnam Medical Science. 2023;40(1):4-11
216. Cioffi I, Evangelista A, Ponzo V, Ciccone G, Soldati L, Santarpia L. et al. Intermittent versus continuous energy restriction on weight loss and cardiometabolic outcomes: A systematic review and meta-analysis of randomized controlled trials. Journal of Translational Medicine. 2018;16(1):1-15
217. Aksungar FB, Sarikaya M, Coskun A, Serteser M, Unsal I. Date of Sampling Study Model Number of Sampling. J Nutr Health Aging. 2017;21(6):681-5
218. Teong XT, Liu K, Vincent AD, Bensalem J, Liu B, Hattersley KJ. et al. Intermittent fasting plus early time-restricted eating versus calorie restriction and standard care in adults at risk of type 2 diabetes: a randomized controlled trial. Nature Medicine. 2023;29(4):963-72
219. Vasim I, Majeed CN, DeBoer MD. Intermittent Fasting and Metabolic Health. Nutrients. 2022;14(3):1-15
220. Chaudhary R, Liu B, Bensalem J, Sargeant TJ, Page AJ, Wittert GA. et al. Intermittent fasting activates markers of autophagy in mouse liver, but not muscle from mouse or humans. Nutrition. 2022;101:111662
221. Ruegsegger GN, Booth FW. Health benefits of exercise. Cold Spring Harbor perspectives in medicine. 2018;8(7):143-5
222. WHO. Physical inactivity [Internet]. Available from: https://www.who.int/data/gho/indicator-metadata-registry/imr-details/3416.
223. Park JH, Moon JH, Kim HJ, Kong MH, Oh YH. Sedentary Lifestyle: Overview of Updated Evidence of Potential Health Risks. Korean Journal of Family Medicine. 2020;41(6):365-73
224. Chow LS, Gerszten RE, Taylor JM, Pedersen BK, van Praag H, Trappe S. et al. Exerkines in health, resilience and disease. Nature Reviews Endocrinology. 2022;18(5):273-89
225. Wu NN, Tian H, Chen P, Wang D, Ren J, Zhang Y. Physical exercise and selective autophagy: Benefit and risk on cardiovascular health. Cells. 2019;8(11):3-5
226. Halling JF, Pilegaard H. Autophagy-Dependent Beneficial Effects of Exercise. Cold Spring Harbor perspectives in medicine. 2017;7(8):1-13
227. Sebastián D, Zorzano A. Self-Eating for Muscle Fitness: Autophagy in the Control of Energy Metabolism. Developmental Cell. 2020;54(2):268-81
228. Di Liegro CM, Schiera G, Proia P, Di Liegro I. Physical activity and brain health. Genes. 2019 10(9)
229. Andreotti DZ, Silva J do N, Matumoto AM, Orellana AM, de Mello PS, Kawamoto EM. Effects of Physical Exercise on Autophagy and Apoptosis in Aged Brain: Human and Animal Studies. Frontiers in Nutrition. 2020;7(July):1-16
230. Mejías-Peña Y, Estébanez B, Rodriguez-Miguelez P, Fernandez-Gonzalo R, Almar M, de Paz JA. et al. Impact of resistance training on the autophagy-inflammation-apoptosis crosstalk in elderly subjects. Aging. 2017;9(2):408-18
231. Pinto AP, da Rocha AL, Marafon BB, Rovina RL, Muñoz VR, da Silva LECM. et al. Impact of different physical exercises on the expression of autophagy markersin mice. International Journal of Molecular Sciences. 2021;22(5):1-20
232. Chen XK, Zheng C, Siu PMF, Sun FH, Wong SHS, Ma ACH. Does Exercise Regulate Autophagy in Humans? A Systematic Review and Meta-Analysis. Autophagy Reports. 2023 2(1)
233. Lira VA, Okutsu M, Zhang M, Greene NP, Laker RC, Breen DS. et al. Autophagy is required for exercise training-induced skeletal muscle adaptation and improvement of physical performance. The FASEB Journal. 2013Oct1;27(10):4184-93
234. Sanchez AMJ, Bernardi H, Py G, Candau RB. Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. American Journal of Physiology - Regulatory Integrative and Comparative Physiology. 2014;307(8):R956-69
235. Hentilä J, Hulmi JJ, Laakkonen EK, Ahtiainen JP, Suominen H, Korhonen MT. Sprint and Strength Training Modulates Autophagy and Proteostasis in Aging Sprinters. Medicine and Science in Sports and Exercise. 2020;52(9):1948-59
236. Escobar KA, Welch AM, Wells A, Fennel Z, Nava R, Li Z. et al. Autophagy response to acute high-intensity interval training and moderate-intensity continuous training is dissimilar in skeletal muscle and peripheral blood mononuclear cells and is influenced by sex. Human Nutrition and Metabolism. 2021;23:200118
237. Brinkman JE, Reddy V, Sharma S. Physiology of Sleep. In: StatPearls. StatPearls Publishing. 2023
238. Zielinski MR, McKenna JT, McCarley RW. Functions and mechanisms of sleep. AIMS Neuroscience. 2016;3(1):67-104
239. Simon KC, Nadel L, Payne JD. The functions of sleep: A cognitive neuroscience perspective. Proceedings of the National Academy of Sciences of the United States of America. 2022;119(44):24-7
240. Hanson JA, Huecker MR. Sleep Deprivation. In: StatPearls. StatPearls Publishing. 2023
241. During EH, Kawai M. The Functions of Sleep and the Effects of Sleep Deprivation. In: Sleep and Neurologic Disease. Academic Press. 2017 p. 55-72
242. Medic G, Wille M, Hemels MEH. Short- and long-term health consequences of sleep disruption. Nature and Science of Sleep. 2017;9:151-61
243. Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885-9
244. Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO Journal. 2011;30(22):4642-51
245. Ryzhikov M, Ehlers A, Steinberg D, Xie W, Oberlander E, Brown S. et al. Diurnal Rhythms Spatially and Temporally Organize Autophagy. Cell Reports. 2019;26(7):1880-1892.e6
246. Bedont JL, Toda H, Shi M, Park CH, Quake C, Stein C. et al. Short and long sleeping mutants reveal links between sleep and macroautophagy. eLife. 2021;10:1-27
247. Sharma A, Narasimha K, Manjithaya R, Sheeba V. Restoration of Sleep and Circadian Behavior by Autophagy Modulation in Huntington's Disease. Journal of Neuroscience. 2023Jun28;43(26):4907-25
248. Cheng Y, Kim WK, Wellman LL, Sanford LD, Guo ML. Short-term sleep fragmentation dysregulates autophagy in a brain region-specific manner. Life. 2021;11(10):1-15
249. Dai W, Xiao Y, Tu Y, Xiao F, Lu Y, Qin Y. et al. Propofol protects hippocampal neurons in sleep-deprived rats by inhibiting mitophagy and autophagy. Annals of Translational Medicine. 2021;9(18):1427-1427
250. Cao Y, Liu L, Wang C, Li Q, Zhang B, Wang Z. et al. Modafinil protects hippocampal neurons by suppressing excessive autophagy and apoptosis in mice with sleep deprivation. British Journal of Pharmacology. 2019;176(January):1282-97
251. Li Y, Zhang W, Liu M, Zhang Q, Lin Z, Jia M. et al. Imbalance of Autophagy and Apoptosis Induced by Oxidative Stress May Be Involved in Thyroid Damage Caused by Sleep Deprivation in Rats. Oxidative Medicine and Cellular Longevity. 2021. 2021
252. Chauhan AK, Mallick BN. Association between autophagy and rapid eye movement sleep loss-associated neurodegenerative and patho-physio-behavioral changes. Sleep Medicine. 2019;63:29-37
253. Vellios N, Ross H, Perucic AM. Trends in cigarette demand and supply in Africa. PLoS ONE. 2018;13(8):1-13
254. WHO. Tobacco [Internet]. [cited. 2023 Aug 1]. Available from: https://www.who.int/news-room/fact-sheets/detail/tobacco
255. Mercado N, Colley T, Baker JR, Vuppussetty C, Kono Y, Clarke C. et al. Bicaudal D1 impairs autophagosome maturation in chronic obstructive pulmonary disease. FASEB BioAdvances. 2019;1(11):688-705
256. Monick MM, Powers LS, Walters K, Lovan N, Zhang M, Gerke A. et al. Identification of an Autophagy Defect in Smokers' Alveolar Macrophages. The Journal of Immunology. 2010;185(9):5425-35
257. Chen ZH, Kim HP, Sciurba FC, Lee SJ, Feghali-Bostwick C, Stolz DB. et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS ONE. 2008 3(10)
258. Kono Y, Colley T, To M, Papaioannou AI, Mercado N, Baker JR. et al. Cigarette smoke-induced impairment of autophagy in macrophages increases galectin-8 and inflammation. Scientific Reports. 2021;11(1):1-13
259. Baliunas D, Rehm J, Irving H, Shuper P. Alcohol consumption and risk of incident human immunodeficiency virus infection: A meta-analysis. International Journal of Public Health. 2010;55(3):159-66
260. Boffetta P, Hashibe M. Alcohol and cancer. Lancet Oncology. 2006;7(2):149-56
261. Louvet A, Mathurin P. Alcoholic liver disease: Mechanisms of injury and targeted treatment. Nature Reviews Gastroenterology and Hepatology. 2015;12(4):231-42
262. Czaja MJ, Ding WX, Donohue TM, Friedman SL, Kim JS, Komatsu M. et al. Functions of autophagy in normal and diseased liver. Autophagy. 2013;9(8):1131-58
263. Wang L, Khambu B, Zhang H, Yin XM. Autophagy in alcoholic liver disease, self-eating triggered by drinking. Clinics and Research in Hepatology and Gastroenterology. 2015;39:S2-6
264. Ding WX, Li M, Chen X, Ni H, Lin C, Gao W. et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139(5):1740-52
265. Lin CW, Zhang H, Li M, Xiong X, Chen X, Chen X. et al. Pharmacological Promotion of Autophagy Alleviates Steatosis and Injury in Alcoholic and Non-alcoholic Fatty Liver Conditions in Mice. Journal of Hepatology. 2013;58(5):993-9
266. Menk M, Graw JA, Poyraz D, Möbius N, Spies CD, von Haefen C. Chronic alcohol consumption inhibits autophagy and promotes apoptosis in the liver. International Journal of Medical Sciences. 2018;15(7):682-8
267. Deng X, Zhang F, Rui W, Long F, Wang L, Feng Z. et al. PM2.5-induced oxidative stress triggers autophagy in human lung epithelial A549 cells. Toxicology in Vitro. 2013;27(6):1762-70
268. Martínez-García GG, Mariño G. Autophagy role in environmental pollutants exposure. In: Progress in Molecular Biology and Translational Science. Academic Press. 2020 p. 257-91
269. Rahman MA, Rahman MS, Parvez MAK, Kim B. The Emerging Role of Autophagy as a Target of Environmental Pollutants: An Update on Mechanisms. Toxics. 2023;11(2):1-15
270. Park SY, Byun EJ, Lee JD, Kim S, Kim HS. Air pollution, autophagy, and skin aging: Impact of particulate matter (PM10) on human dermal fibroblasts. International Journal of Molecular Sciences. 2018 19(9)
271. Spiro A, Buttriss JL. Vitamin D: An overview of vitamin D status and intake in Europe. Nutrition Bulletin. 2014;39(4):322-50
272. Wang M, Charareh P, Lei X, Zhong JL. Autophagy: Multiple Mechanisms to Protect Skin from Ultraviolet Radiation-Driven Photoaging. Oxidative Medicine and Cellular Longevity. 2019. 2019
273. Ma J, Teng Y, Huang Y, Tao X, Fan Y. Autophagy plays an essential role in ultraviolet radiation-driven skin photoaging. Frontiers in Pharmacology. 2022;13(October):1-11
274. Zhao Y, Zhang CF, Rossiter H, Eckhart L, König U, Karner S. et al. Autophagy is induced by UVA and promotes removal of oxidized phospholipids and protein aggregates in epidermal keratinocytes. Journal of Investigative Dermatology. 2013;133(6):1629-37
275. Sample A, He YY. Autophagy in UV Damage Response. Photochemistry and Photobiology. 2017;93(4):943-55
276. Sample A, Zhao B, Wu C, Qian S, Shi X, Aplin A. et al. The Autophagy Receptor Adaptor p62 is Up-regulated by UVA Radiation in Melanocytes and in Melanoma Cells. Photochemistry and Photobiology. 2018;94(3):432-7
277. Errafiy R, Aguado C, Ghislat G, Esteve JM, Gil A, Loutfi M. et al. PTEN increases autophagy and inhibits the ubiquitin-proteasome pathway in glioma cells independently of its lipid phosphatase activity. PLoS ONE. 2013;8(12):1-15
278. Aquila S, Santoro M, Caputo A, Panno ML, Pezzi V, Francesca DAmicis. The Tumor Suppressor PTEN as Molecular Switch Node Regulating Cell Metabolism and Autophagy: Implications in Immune System and Tumor Microenvironment. Cells. 2020;9(1725):1-19
279. Lamore SD, Wondrak GT. Autophagic-lysosomal dysregulation downstream of cathepsin B inactivation in human skin fibroblasts exposed to UVA. Photochemical and Photobiological Sciences. 2012;11(1):163-72
280. Jia J, Le W. Molecular network of neuronal autophagy in the pathophysiology and treatment of depression. Neuroscience Bulletin. 2015;31(4):427-34
281. Zschocke J, Rein T. Antidepressants encounter autophagy in neural cells. Autophagy. 2011;7(10):1247-8
282. Limanaqi F, Busceti CL, Biagioni F, Fornai F, Puglisi-Allegra S. Autophagy-Based Hypothesis on the Role of Brain Catecholamine Response During Stress. Frontiers in Psychiatry. 2020;11(September):1-12
283. Xiao X, Shang X, Zhai B, Zhang H, Zhang T. Nicotine alleviates chronic stress-induced anxiety and depressive-like behavior and hippocampal neuropathology via regulating autophagy signaling. Neurochemistry International. 2018;114:58-70
284. Chen A, Xiong LJ, Tong Y, Mao M. Neuroprotective effect of brain-derived neurotrophic factor mediated by autophagy through the PI3K/Akt/mTOR pathway. Molecular Medicine Reports. 2013;8(4):1011-6
285. Puri D, Subramanyam D. Stress - (self) eating: Epigenetic regulation of autophagy in response to psychological stress. FEBS Journal. 2019;286(13):2447-60
286. Wang SL, Shao BZ, Zhao SB, Chang X, Wang P, Miao CY. et al. Intestinal autophagy links psychosocial stress with gut microbiota to promote inflammatory bowel disease. Cell Death and Disease. 2019 10(6)
Author contact
![]() Corresponding author: Miguel A Ortega (Email: miguelangel.ortegaes).
Corresponding author: Miguel A Ortega (Email: miguelangel.ortegaes).

 Global reach, higher impact
Global reach, higher impact