10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2024; 20(7):2698-2726. doi:10.7150/ijbs.91832 This issue Cite
Review
LncRNAs as nodes for the cross-talk between autophagy and Wnt signaling in pancreatic cancer drug resistance
1. National "111" Center for Cellular Regulation and Molecular Pharmaceutics, Key Laboratory of Fermentation Engineering (Ministry of Education), Cooperative Innovation Center of Industrial Fermentation (Ministry of Education & Hubei Province), Hubei Key Laboratory of Industrial Microbiology, Hubei University of Technology, Wuhan, China, 430068.
2. Membrane Protein Disease Research Group, Department of Physiology, Faculty of Medicine and Dentistry, University of Alberta, Edmonton, AB, Canada, T6G2R3.
Received 2023-11-2; Accepted 2024-2-6; Published 2024-4-29
Abstract
Pancreatic cancer is a malignancy with high mortality. In addition to the few symptoms until the disease reaches an advanced stage, the high fatality rate is attributed to its rapid development, drug resistance and lack of appropriate treatment. In the selection and research of therapeutic drugs, gemcitabine is the first-line drug for pancreatic cancer. Solving the problem of gemcitabine resistance in pancreatic cancer will contribute to the progress of pancreatic cancer treatment. Long non coding RNAs (lncRNAs), which are RNA transcripts longer than 200 nucleotides, play vital roles in cellular physiological metabolic activities. Currently, our group and others have found that some lncRNAs are aberrantly expressed in pancreatic cancer cells, which can regulate the process of cancer through autophagy and Wnt/β-catenin pathways simultaneously and affect the sensitivity of cancer cells to therapeutic drugs. This review presents an overview of the recent evidence concerning the node of lncRNA for the cross-talk between autophagy and Wnt/β-catenin signaling in pancreatic cancer, together with the practicability of lncRNAs and the core regulatory factors as targets in therapeutic resistance.
Keywords: autophagy, drug resistance, lncRNA, pancreatic cancer, Wnt/β-catenin signaling
Introduction
Pancreatic cancer (PC) is one of the deadliest cancers. Ductal differentiation is a common phenomenon in PC (>90%) [1], and invasive ductal adenocarcinoma is a cardinal type, accounting for the majority of pancreatic tumors (>85%) [2]. Neuroendocrine tumors and acinar carcinomas are infrequent while more infrequent tumors include colloid carcinomas, pancreatoblastomas and solid-pseudopapillary neoplasms [3]. Now, pancreatic ductal adenocarcinoma (PDAC) has become a synonym for PC and can directly refer to PC [3]. According to the American Cancer Society, in 2023, there will be 64,050 new cancer cases of PC in the United States, accounting for 3.27% of the total cases (1,958,310), and 50,550 deaths, accounting for 8.29% of the total number (609,820) [4]. Meanwhile, PC currently has the lowest 5-year relative survival rate of all cancers (12%) [4]. This is associated with its poor prognosis due to factors such as low rate of early detection, rapid progression, development of drug resistance and lack of appropriate treatment. After long-term clinical treatment research, it has been established that the treatment of PC is mainly based on surgical resection, supplemented by chemotherapy [5]. Gemcitabine is the first-line drug for the treatment of advanced PC [6], and it has shown better efficacy in combination with capecitabine [7] and albumin-bound paclitaxel [8]. In addition, combination chemotherapy with fluorouracil, leucovorin, irinotecan, and oxaliplatin (FOLFIRINOX) or nanoliposome irinotecan plus fluorouracil and leucovorin has been proposed in recent years, and patients treated with combination therapy have longer overall survival than those treated with gemcitabine alone [9, 10]. However, even though adjuvant therapy has been improved, the mortality rate of PC patients has remained stubbornly high, which is associated to chemotherapy resistance. Admittedly, PC cells can develop resistance to gemcitabine in a variety of ways [11], and most of the research on the chemical resistance of advanced PC has focused on gemcitabine, while the research on other drugs is still in its infancy [12]. Therefore, there is an urgent demand to elucidate the mechanism of gemcitabine resistance in PC cells for the treatment.
Long non coding RNAs (lncRNAs) are transcripts larger than 200 nucleotides with no or limited protein-coding potential. More than 68% of the genes expressed in the human transcriptome are transcribed into lncRNAs [13]. LncRNAs can be involved in regulating various physiological and pathological cell activities. In the nucleus, lncRNAs act as enhancers, decoys, scaffolds or guides to directly interact with DNA or chromatin regulatory factors, such as transcription factors and RNA-binding proteins, to control gene expression. Whereas in the cytoplasm, lncRNAs can competitively bind with miRNA to regulate mRNA stability and translation or recruit cofactors and influence the activity of related enzymes to regulate the transcription and translation process [14-16]. Nowadays, lncRNAs have attracted more and more attention, and there is sufficient evidence that lncRNA is related to multiple diseases, especially cancer. In cancer cells, abnormally expressed lncRNA has been viewed as a classical oncogene or tumor suppressor gene, with variation in lncRNA expression species, amount of expression and even efficacy in different cancers and at different times of progression [17, 18]. In our previous study, we found that in PC, there are lncRNAs simultaneously regulating Wnt/β-catenin and autophagy, which can lead to gemcitabine resistance, and lncRNAs that can be used as biomarkers for prognosis analysis were also screened through data analysis [19, 20]. Therefore, we believe that lncRNAs have the potential to regulate PC progression and lead to drug resistance from both Wnt/β-catenin and autophagy pathways.
In this review, we collect lncRNAs related to Wnt/β-catenin and autophagy pathways in PC, discuss their roles in processes regulating cell metabolism and effects on chemoresistance, from which we identify the shared pathways with core factors to provide new targets and research directions for PC prevention and treatment.
LncRNAs regulate Wnt/β-catenin signaling pathway in PC
Wnt/β-catenin signaling pathway is of vital importance in regulating various physiological activities of cells, including tissue homeostasis, cell proliferation, cell differentiation and cell death [21-23]. The canonical Wnt/β-catenin pathway is activated by the binding of endocrine or paracrine Wnt ligands to Frizzled (FZD) and low density lipoprotein receptor-related protein (LRP) family membrane receptors on the cell surface. In the absence of Wnt ligand, β-catenin is captured by a complex consisting of adenomatous polyposis coli (APC), axis inhibitor (AXIN), casein kinase 1 (CK1) and glycogen synthase kinase-3 beta (GSK‑3β), resulting in degradation of β-catenin. Upon Wnt activation, the complex is recruited to the plasma membrane through interaction with the FZD, thus losing its ability to degrade β-catenin. After that, β-catenin would translocate from cytoplasm to nucleus and interact with the transcriptional response element TCF/LEF (T-cell factor/lymphoid enhancer-binding factor) to activate the transcription of target genes [24]. Wnt/β-catenin signaling regulates PC from multiple perspectives including initiation, progression, propagation and treatment resistance [25, 26]. In addition, the abnormal activation of Wnt promotes the immunosuppression of PC [27], promotes the development and differentiation of pancreatic cancer stem cells (PaCSCs) [28], and is associated with poor prognosis [29].
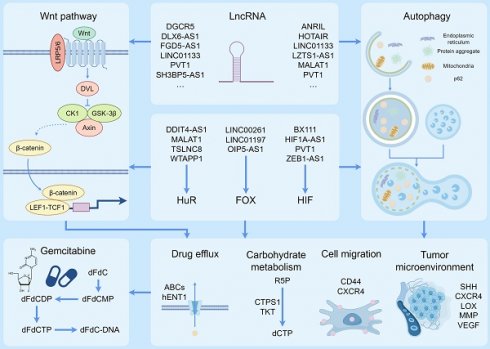
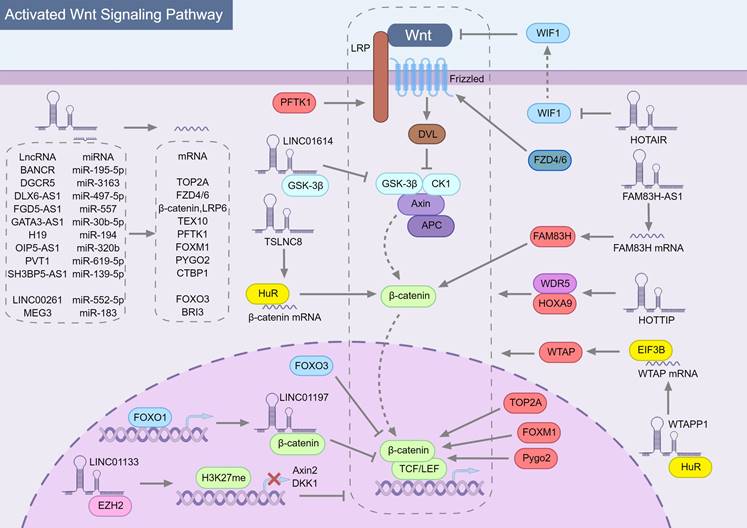
In general, lncRNAs can regulate the Wnt/β-catenin pathway in multiple ways (Fig. 1), but mainly by spongating miRNA to stabilize mRNA and then up-regulate the expression of the corresponding protein. Among these studies shown in Table 1 (23 articles), nearly half (11 articles) described that lncRNAs could act as competitive endogenous RNAs (ceRNAs) to competitively adsorb miRNAs, resulting in loss or attenuation of miRNA function and promoting the expression of target genes acting on different links in the Wnt/β-catenin pathway. Some target genes are integral members of the Wnt/β-catenin pathway, such as FZD4/6, LPR6 and β-catenin, which are up-regulated by lncRNA FYVE, RhoGEF, and PH domain containing 5 antisense RNA 1 (FGD5-AS1) [30] and lncRNA distal-less homeobox 6 antisense RNA 1 (DLX6-AS1) [31] to enhance Wnt signaling. Other target genes indirectly affect the Wnt/β-catenin pathway. Cyclin dependent kinase 14 (CDK14, PFTK1), a serine/threonine protein kinase that can induce phosphorylation of LRP5/6 to activate the Wnt/β-catenin pathway [32, 33], can be up-regulated by lncRNA H19 imprinted maternally expressed transcript (H19) [34]. In the nucleus, forkhead box M1 (FOXM1), forkhead box O1 (FOXO3), pygopus family PHD finger 2 (PYGO2) and DNA topoisomerase II alpha (TOP2A) can regulate the transcription of Wnt/β-catenin pathway downstream genes through interacting with β-catenin. Inhibiting the original activity of lncRNAs by binding specific proteins is another way to regulate the Wnt/β-catenin pathway. Both lncRNA long intergenic non-protein coding RNA 01614 (LINC01614) [35] binding GSK‑3β and lncRNA LINC01197[36] binding β-catenin can affect the original function of the protein to regulate Wnt signal transduction.
What is special in these studies is the regulation of Wnt/β-catenin pathway by lncRNAs through enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2) and human antigen R (HuR). EZH2 is a catalytic subunit of polycomb repressive complex 2 (PRC2) that can restrain transcription of target genes by triggering trimethylation of methylation of histone H3 at lysine 27 (H3K27me) [37, 38]. LncRNA LINC01133 can recruite methylated EZH2 to mediate histone methylation and up-regulate dickkopf Wnt signaling pathway inhibitor 1 (DKK1) [39] and AXIN2[40] promoter methylation, which inhibited DKK1 and AXIN2 to activate Wnt/β-catenin signaling pathway. The homeobox transcript antisense intergenic RNA (HOTAIR) is one of the most extensively studied lncRNAs found dysregulated in human cancer. Although the mechanism of lncRNA HOTAIR in PC is unknown, HOTAIR can increase the radioresistance of PC cells by down-regulating Wnt inhibitory factor 1 (WIF1) [41], which inhibits the activation of Wnt/β-catenin signaling pathway by binding Wnt protein and inhibiting its signal transduction activity in the intercellular space [42]. In esophageal squamous cell carcinoma cells [43] and human chondrosarcoma cells [44], HOTAIR inhibits WIF1 expression by promoting trimethylation of H3K27 in the WIF1 promoter, thereby activating Wnt/ β-catenin pathway. In PC, HOTAIR may also regulate WIF1 in a similar way.
The role of lncRNAs in modulating the Wnt/β-catenin signaling pathway in pancreatic cancer. Most lncRNAs indirectly regulate Wnt/β-catenin signaling pathway through sponging miRNA, among which the top half of lncRNAs have an activation effect and the bottom two have an inhibitory effect. The targets of miRNAs are directly the components of the Wnt/β-catenin pathway, while other target proteins regulate the components of the Wnt/β-catenin pathway. Among them, factors that can activate Wnt/β-catenin are marked in red, and those that inhibit Wnt/β-catenin are marked in blue. In addition, there are also lncRNAs (such as LINC01614 and LINC01197) that directly bind to Wnt/β-catenin pathway proteins and inhibit their physiological activity. Specifically, HuR can stabilize RNA, including mRNA and lncRNA. The combination of TSLNC8, HuR and β-catenin mRNA can promote β-catenin translation and HuR can stabilize WTAPP1, which enhances WTAP translation by collecting EIF3B into WTAP mRNA and finally regulates Wnt/β-catenin pathway.
LncRNA regulates PC via Wnt/β-catenin signaling pathway
| LcnRNA | Gene and pathway | Interaction with wnt signaling | Cancer phenotype | Reference |
|---|---|---|---|---|
| BANCR | miR-195-5p | Activate | Promote PC cell proliferation, invasion and migration | [129] |
| DGCR5 | miR-3163/TOP2A | Activate | Promote PC cell migration, invasion EMT and gemcitabine resistance | [369] |
| DLX6-AS1 | miR-497-5p/FZD4/FZD6 | Activate | Promote PC cell proliferation, invasion, migration and inhibit apoptosis Promote PC growth and metastasis in vivo | [31] |
| FAM83H-AS1 | FAM83H/ β-catenin | Activate | Promote PDAC cell proliferation, migration and invasion in vitro and in vivo | [51] |
| FGD5‑AS1 | miR-577/β-catenin, LRP6 | Activate | Promote PC cell proliferation, invasion and migration | [30] |
| GATA3-AS1 | miR-30b-5p/TEX10 | Activate | Promote PC cell viability, proliferation, invasion, stemness and inhibit apoptosis | [370] |
| H19 | miR-194/PFTK1/LRP5/6 | Activate | Promote PDAC cell proliferation and migration | [34] |
| HOTAIR | WIF1 | Activate | Enhance the radiosensitivity of PDAC cells, reduce the proliferation, and increase the apoptosis of cells after radiation | [41] |
| HOTAIR | Wnt | Activate | Promote PC cell proliferation, migration, invasion and EMT | [371] |
| HOTTIP | WDR5/HOXA9/Wnt | Activate | Enhance CSC properties and promote PDAC tumorigenesis | [372] |
| HULC | Wnt | Activate | Promote PC cell proliferation and invasion and inhibit apoptosis | [373] |
| LINC01133 | EZH2/ H3K27me/AXIN2 | Activate | Promote PC cell proliferation, migration, invasion, EMT and inhibit apoptosis | [40] |
| LINC01133 | DKK1 | Activate | Promote PC cell growth, proliferation, migration, metastasis and invasion | [39] |
| LINC01614 | GSK‑3β/AXIN1 | Activate | Promote PC cell proliferation, migration, invasion in vitro and tumor proliferation in vitro and in vivo | [35] |
| OIP5-AS1 | miR-320b/FOXM1 | Activate | Promote PC cell proliferation, migration and invasion | [374] |
| PVT1 | miR-619-5p/PYGO2 | Activate | Promote PC cell viability and gemcitabine resistance in vitro and in vivo | [20] |
| SH3BP5-AS1 | miR-139-5p/CTBP1 | Activate | Promote PC cell migration, invasion and gemcitabine resistance | [375] |
| TSLNC8 | HuR/β-catenin | Activate | Promoted PC cell proliferation and invasion in vitro and enhance PC growth and metastasis in vivo | [46] |
| WTAPP1 | WTAP | Activate | Promotes PC cell proliferation and invasiveness | [49] |
| LINC00261 | miR-552-5p/FOXO3 | Inhibit | Inhibit PC cell migration, invasion and EMT | [157] |
| LINC01197 | FOXO1/LINC01197/β-catenin | Inhibit | Inhibit PC cell proliferation and growth | [36] |
| MEG3 | miR-183/BRI3 | Inhibit | Inhibit pNET cell viability, invasion and migration and induce apoptosis. | [376] |
| NEN885 | Wnt | Inhibit | Inhibit GEP - NEN cell migration, invasion and EMT | [377] |
HuR is an RNA binding protein that binds to adenylate/uridylate-rich regions primarily in the 3′ UTR and regulates mRNA stability and translation [45]. In PC, the combination of tumor suppressive lncRNA on chromosome 8p12 (TSLNC8), HuR and β-catenin mRNA can promote β-catenin translation and thus activate Wnt signaling [46]. Cellular nucleic acid binding protein (CNBP) is a conserved single-stranded nucleic acid-binding protein that acts as both a transcription regulator and a translational regulator [47]. Protein tyrosine kinase 7 (PTK7), T-cell-specific transcription factor 4 (TCF4) and CDK14 are direct transcriptional targets of CNBP, which can directly or indirectly participate in Wnt/β-catenin pathway regulation [48]. In PC, CNBP could recognize N6-methyladenosine (m6A) lncRNA WT1 associated protein pseudogene 1 (WTAPP1) and recruit HuR to promote WTAPP1 stability. Furthermore, WTAPP1 can enhance WTAP translation by recruiting eukaryotic translation initiation factor 3 subunit B (EIF3B) to WTAP mRNA, induce carcinogenic Wnt signaling and promote PC progression [49]. Interestingly, the m6A modification of RNA is dependent on dedicated methyltransferases (METTL), the core of which is the METTL3-METTL14-WTAP complex [50]. Then m6A-modified WTAPP1 can promote WTAP expression levels, and WTAP may also form methyltransferase to promote m6A modification of WTAPP1. There may be a positive feedback regulation between WTAPP1 and WTAP. Furthermore, lncRNA family with sequence similarity 83 member H antisense RNA 1 (FAM83H-AS1) is similar to TSLNC8 and FGD5-AS1 in that it can up-regulate the level of β-catenin to activate Wnt/β-catenin signaling, but the difference is that FAM83H-AS1 induces FAM83H expression by stabilizing FAM83H mRNA, thus enhances the ability of FAM83H binding to β-catenin and inhibiting its degradation, and ultimately promotes the proliferation, invasion and metastasis of PC cells [51]. The precise mechanism by which FAM83H-AS1 stabilizes FAM83H mRNA is currently unknown, but in ovarian cancer, FAM83H-AS1 could interact with HuR and increase the stability of HuR protein, which has certain guiding significance [52].
LncRNAs regulate autophagy pathway in PC
There are two sides in the process of transformation and malignant development of cancer cells by autophagy. On the one hand, autophagy removes damaged organelles, peroxides, endogenous bacteria and viruses, prevents normal cells from producing excessive oxidative stress damage to DNA, maintains normal metabolic level, and thus maintains homeostasis of intracellular environment and prevents malignant transformation; on the other hand, while maintaining cell homeostasis, autophagy can also remove various tumor suppressor factors and even therapeutic drugs, increasing drug resistance, which is conducive to the maintenance of tumor and its malignant process[53-55]. Current studies have shown that the level of autophagy in PC cells is universally elevated which is highly activated autonomously in the late stage from the formation of intraductal tumors to PC, and a high level of autophagy is required for sustained malignant growth in vivo and in vitro [56].
The high level of autophagy is associated with the abnormal expression of MIT/TFE family in PC, among which melanocyte inducing transcription factor (MITF), transcription factor binding to IGHM enhancer 3 (TFE3) and transcription factor EB (TFEB) are strongly correlated with autophagic lysosomal characteristics [57, 58]. Under nutrient replete conditions, mechanistic target of rapamycin complex 1 (mTORC1) at the lysosomal membrane phosphorylates MIT/TFE proteins, leading to their association with 14-3-3 proteins and retention in the cytoplasm, whereas mTORC1 inactivation upon starvation allows its nuclear translocation [59-62]. Autophagy related (ATG) genes regulated by different MIT/TFE family members varied, but the genes ATG9B, ATG16L1, GABA type A receptor associated protein like 1 (GABARAPL1), WD repeat domain, phosphoinositide interacting 1 (WIPI1), and UV radiation resistance associated (UVRAG), which can enhance the biosynthesis of autophagosomes and lysosomes and activate their function, were more than 2-fold expressed when MITF, TFE3, or TFEB were overexpressed, indicating the vital role of the MIT/TFE family in autophagy induction[62]. However, in PC, the nucleoplasmic transport protein, importin 8 (IPO8) can bind to stabilize MIT/TFE factors and translocate to the nucleus, leading to activation of transcription of target genes [57]. Current studies on lncRNAs regulating autophagy pathway in PC are shown in the following table (Table 2).
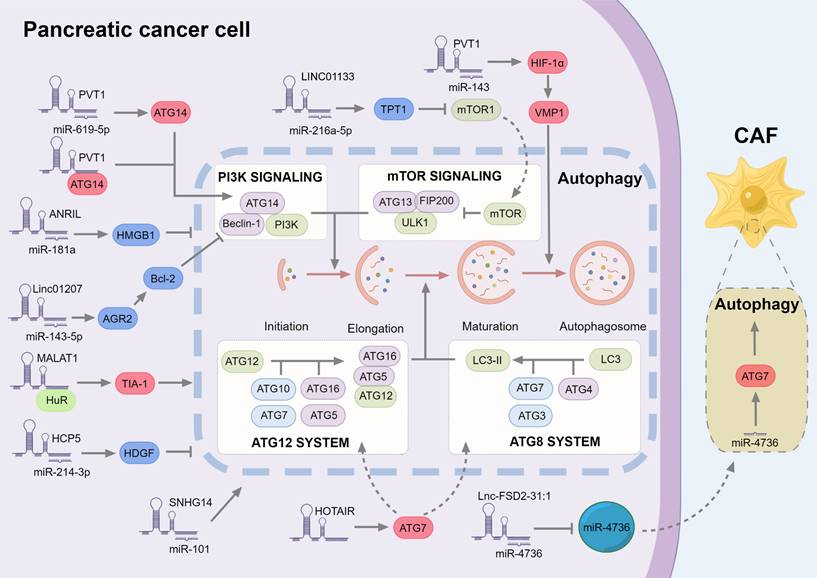
In the research of autophagy regulation in PC, most lncRNAs also play a role by sponging miRNA to stabilize mRNA. For lncRNAs that directly regulate autophagy pathway factors, similar to the regulation of Wnt pathway, lncRNA plasmacytoma variant translocation 1 (PVT1) also regulates autophagy through the target miR-619-5p [20]. ATG14 could be down-regulated by miR-619-5p but also activate autophagy by binding to PVT1. Vacuole membrane protein 1 (VMP1) is the downstream target of hypoxia inducible factor 1 subunit alpha (HIF-1α), which can promote the separation of the isolation membrane from the endoplasmic reticulum, and then form free autophagosomes [63]. On the other hand, PVT1 can promote autophagy and reduce gemcitabine sensitivity in PC by regulating the miR-143/HIF-1α/VMP1 axis [64]. In addition to ATG14, there are lncRNAs that affect autophagy through ATG7. HOTAIR can promote autophagy and down-regulate the radiosensitivity of PC cells by targeting ATG7. In liver ischemia-reperfusion injury [94] and acute lung injury [95], HOTAIR can up-regulate ATG7 and promote autophagy by sponging miR-17-5p and miR-20b-5p. As a key component of the tumor microenvironment (TME), cancer-associated fibroblasts (CAFs) have complex functions to protect cancer cells, which are generally believed to promote cancer and may also inhibit tumor progression in some circumstances [65, 66]. In PC cells, lnc-FSD2-31:1 can down-regulate miR‐4736 in extracellular vesicles and up-regulate ATG7, the target of miR‐4736 in CAF, leading to promotion of CAF autophagy and inhibition of CAF fibrosis [67].
LncRNA regulates PC via autophagy
| LcnRNA | Gene and pathway | Interaction with autophagy | Cancer phenotype | Reference |
|---|---|---|---|---|
| HOTAIR | ATG7 | Activate | Reduce the radiosensitivity of PC cells | [378] |
| Lnc-FSD2-31 | miR-4736/ATG7 | Activate | Inhibit PC cell growth | [67] |
| MALAT1 | HuR/TIA1 | Activate | Promote PC proliferation and metastasis | [92] |
| PVT1 | miR-619-5p/ATG14 | Activate | Promote PC cell viability and gemcitabine resistance | [20] |
| PVT1 | miR-143/HIF-1α/VMP1 | Activate | Promote PC cell viability and gemcitabine resistance | [64] |
| SNHG14 | miR-101 | Activate | Promote PC cell viability, proliferation and gemcitabine resistance | [379] |
| ANRIL | miR-181a/HMGB1 | Repress | Promote PC cell proliferation, migration, invasion and gemcitabine resistance | [68] |
| LINC01207 | miR-143-5p/AGR2 | Repress | Promote PC cell growth and inhibit apoptosis | [380] |
| LINC01133 | miR-216a-5p/TPT1/mTORC1 | Repress | Promote PC cell proliferation and metastasis | [381] |
| LZTS1-AS1 | LZTS1-AS1/miR-532/TWIST1 | Repress | Promote PC cell proliferation, migration, invasion and oncogenicity, inhibit apoptosis and autophagy | [86] |
| UCA1 | UCA1/MAPK/ERK | Repress | Promote PC cell mitochondrial fusion and migration, inhibit mitophagy | [89] |
| HCP5 | miR-214-3p/HDGF | —— | Promote PC-GR cell proliferation, invasion and migration, inhibit cell apoptosis and increase gemcitabine resistance | [237] |
Among the factors that indirectly regulate the autophagy pathway, the regulation of HMGB1(high mobility group box 1) and TWIST1(twist family bHLH transcription factor 1) on autophagy is controversial. In the study of Chen et al. [68], the knockdown of lncRNA cyclin dependent kinase inhibitor 2B antisense RNA 1 (ANRIL) up-regulated miR-181a and down-regulated HMGB1 at mRNA and protein levels, which made cancer cells sensitive to gemcitabine and showed inhibition of tumor activity and promotion of autophagy. Interference of miR-181a and overexpression of HMGB1 showed opposite biological effects. Therefore, HGMB1 appears to promote PC progression and inhibit autophagy. As a cancer-promoting factor, HMGB1 is also regulated by lncRNA small nucleolar RNA host gene 16 (SNHG16)/miR-218-5p [69] and lncRNA zinc finger E-box binding homeobox 2 antisense RNA 1 (ZEB2-AS1)/miR-204 [70] axis in PC. Extracellular HMGB1 can regulate inflammation and PC progression through the receptor of advanced glycosylation end-product specific receptor (AGER) [71, 72] and Toll-like receptor 4 (TLR4) [73]. However, in the nucleus, HMGB1 can repair damaged DNA [74]. At the same time, low expression of HMGB1 can promote the progression of PC, and the decreased expression of HMGB1 in the pancreas is related to poor survival [75]. The reason may be associated with the subcellular localization of HMGB1. Under normal conditions, most HMGB1 is localized in the nucleus, and there is little HMGB1 in the cytoplasm. While under various stresses, HMGB1 is transferred from the nucleus to the cytoplasm, and its extracellular transport mainly depends on the active secretion of living inflammatory cells (such as macrophages) or the passive release of necrotic cells [76]. In the aspect of autophagy, HMGB1 is generally regarded as an autophagy inducing factor [77], and cytosolic HMGB1 promotes autophagy by directly binding to beclin-1[78], contrary to the phenomenon studied by Chen et al. [68]. This may be related to the potential role of lncRNAs.
TWIST1 is a transcriptional regulator that has been identified to play vital roles in angiogenesis, chemotherapy resistance, metastasis, senescence and stemness in various cancers, including PC [79, 80]. The relationship between TWIST1 and autophagy is peculiar. Autophagy deficiency can up-regulate TWIST1[81], P62 (Sequestosome 1, SQSTM1) can bind and stabilize TWIST1 [82], and the formation of B-cell lymphoma 2 (BCL2)/TWIST1 complex can promote the nuclear transport of TWIST1 [83]. In contrast, TWIST1 silencing can activate AMP-activated protein kinase (AMPK) and inhibit mTOR signaling [84], and its target gene eva-1 homolog A (EVA1A) can promote endothelial cell apoptosis and inflammatory activation through autophagy regulation [85]. LncRNA leucine zipper tumor suppressor 1 antisense RNA 1 (LZTS1-AS1) promotes the proliferation, metastasis and oncogenicity of PC cells and inhibits autophagy through LZTS1-AS1/miR-532/TWIST1 axis [86]. The mechanism of TWIST regulating autophagy needs further study.
In addition to macroautophagy, mitophagy in PC cells can also be regulated by lncRNAs. The MAPK/ERK pathway is closely related to mitochondrial dynamics [87], which in turn is related to tumor progression [88]. In PC, lncRNA urothelial cancer associated 1 (UCA1), which is up-regulated and can enhance the migration ability of cancer cells, regulates mitochondrial dynamics through the activation of MAPK/ERK pathway, including up-regulating mitochondrial membrane potential, enhancing mitochondrial fusion and reducing mitochondrial fission to inhibit mitophagy [89].
LncRNAs can also regulate autophagy through HuR in ways that do not regulate autophagy through miRNAs. TIA1 cytotoxic granule associated RNA binding protein (TIA1) is the same RNA binding protein (RBP) as HuR and has similar regulatory effects on RNA [90, 91]. Li et al. [92] illustrated that knockdown of lncRNA metastasis associated lung adenocarcinoma transcript 1 (MALAT1) reduced HuR expression, and a direct interaction between MALAT1 and HuR was found. On the other hand, knockdown of MALAT1 had no significant effect on TIA1 accumulation, but enhanced its activity after transcription, while suppressing the expression of autophagy. Therefore HuR can regulate autophagy as an endogenous messenger between MALAT1 and TIA1 to influence tumor proliferation and metastasis. Unfortunately, this article did not validate the interaction between HuR and TIA1, but other studies have shown that HuR can contribute to the maintenance of elevated TIA1 mRNA levels and therefore maintain TIA1 expression by binding to TIA1 3' UTR [93].
In general, although most lncRNAs regulate autophagy in different directions, they generally promote the process of PC, which is related to the duality of autophagy, and the generation of drug resistance is often related to high levels of autophagy. LncRNA can regulate autophagy in a similar way to Wnt/β-catenin pathway, mainly as ceRNA binding miRNA to regulate downstream targets (Fig. 2).
The role of lncRNAs in regulating autophagy in pancreatic cancer. The autophagic process is divided into multiple systems that regulate autophagosome formation at different stages. Most lncRNAs also regulate autophagy by sponge miRNA, in which the factors promoting autophagy are marked in red, and the inhibitors are marked in red. Among them, PVT1 can not only promote the expression of ATG14, but also directly bind to ATG14 to promote the formation of autophagosomes. In addition, PVT1 further increased the expression of VMP1 by up-regulating HIF-1α, and VMP1 promoted the separation of the isolation membrane from the endoplasmic reticulum, thereby forming free autophagosomes. Finally, although Lnc-FSD2-31:1 inhibits miR-4736, miR-4736 regulates autophagy of CAFs in the form of exosomes.
LncRNA acts as a cross node in autophagy and Wnt/β-catenin pathways
Molecular basis between Wnt/β-catenin pathway and autophagy
Wnt/β-catenin pathway and autophagy are important pathways to regulate cell physiological processes and maintain cell homeostasis. In addition to the PI3K-AKT-mTOR pathway, AMPK pathway and EGFR pathway, which can simultaneously regulate Wnt/β-catenin pathway and autophagy [94-97], there is signal crosstalk between the two pathways, and their components also interact with each other at the molecular level to affect their activity and protein level.
β-catenin is a transcriptional regulator downstream of the Wnt/β-catenin pathway, which negatively regulates autophagy and down-regulates the expression of P62 [98-100]. Under starvation, microtubule associated protein 1 light chain 3 (LC3) directly can target β-catenin for autophagic degradation, resulting in reduced binding of β-catenin to the P62 promoter and increased transcription of P62 to promote autophagy [98, 101]. In addition, BCL2, an important target gene of β-catenin [97-99], inhibits beclin-1-dependent autophagy [102]. Inhibition of Wnt/β-catenin signaling pathway in PC can down-regulate BCL2, destroy mitochondrial homeostasis, and inhibit tumor growth and development [103].
In the Wnt/β-catenin pathway, GSK‑3β is recruited by AXIN to form a complex with APC and CK1α to phosphorylate β-catenin, leading to its ubiquitination and degradation by the proteasome [104, 105]. In the autophagy pathway, GSK‑3β phosphorylates unc-51 like autophagy activating kinase 1 (ULK1) or acetylates ULK1 through the GSK‑3β-TIP60 (lysine acetyltransferase 5, KAT5)-ULK1 axis to activate autophagy [106-108]. Meanwhile, AMPK and GSK3 coordinate to phosphorylate tuberous sclerosis complex (TSC) to inhibit mTOR signaling and promote lysosomal acidification [109-112]. In addition, GSK3 regulates FOXK1 phosphorylation and inhibits nuclear translocation of FOXK1[113, 114], while FOXK1 inhibits the expression of autophagy genes through recruitment of Sin3A complex [115, 116].
Dishevelled segment polarity protein (DVL) is involved in both canonical and noncanonical Wnt/β-catenin signaling pathways and DVL is recruited to the plasma membrane to bind FZD receptors to initiate Wnt/β-catenin signaling [117, 118]. In the autophagy degradation pathway, DVL can directly bind to LC3 and be degraded by P62, thereby inhibiting Wnt signaling [119]. The function of DVL is also related to ULK1 kinase activity. DVL can be phosphorylated by ULK1, and Wnt5a-mediated autophagy promotes bacterial clearance in macrophages dependent on DVL and ULK1 [120, 121]. DVL can also enter the nucleus to form protein complexes with β-catenin and TCF, and induce the transcriptional expression of target genes [122], while DVL phosphorylation by ULK1 inhibits the formation of DVL complexes [121].
In general, the Wnt/ β-catenin pathway and autophagy are mutually inhibitory in normal cells. But the Wnt/β-catenin pathway and autophagy are highly activated in PC compared with normal tissues, and the proliferation, migration, invasion, EMT (epithelial-mesenchymal transition) and drug resistance of cancer cells all depend on high levels of Wnt and autophagy [25, 123]. There are other factors affecting both Wnt/β-catenin pathway and autophagy in PC. A tandem mechanism between Wnt/ β-catenin signaling and autophagy has been reported to regulate the progression of PC [20, 95, 124, 125]. According to our previous experimental results and research analysis, lncRNA has the potential to simultaneously regulate Wnt/β-catenin pathway and autophagy.
Competitive endogenous RNA hypothesis
The ceRNA hypothesis considers miRNAs as miRNA recognition elements (MREs) that bind to RNA transcripts via complementary sequences [126]. All types of RNA transcripts have different MRE binding sites, and they can communicate with each other according to the shared MRE, and interact with each other to affect the properties and functions of RNA transcripts [126].
In addition to miR-619-5p and miR-143 mentioned above, PVT1 can also regulate the progression of PC through PVT1/miR-20b/CCND1 (cyclin D1) [127] and PVT1/miR-519d-3p/HIF-1α [128] axes. This means that the same lncRNA can regulate the same mRNA through different MREs, and also can regulate different mRNAs through the same MRE. MiR-195-5p as a communication factor, BRAF-activated non-protein coding RNA (BANCR) can activate the Wnt/β-catenin sighting pathway through miR-195-5p [129]. Similarly, miR-195-5p is the target of lncRNA LINC00473, which can drive the progression of PC [130], and WIPI2, a key autophagy regulator [131], can be up-regulated by lncRNA ceramide synthase 6 antisense RNA 1 (CERS6-AS1) to regulate the migration and apoptosis of PC cells via miR-195-5p [132]. In summary, a huge communication network is formed between lncRNA-miRNA-mRNA.
Although the ceRNA hypothesis has several limitations: miRNAs target a variety of RNAs, they exert varying degrees of repression on all of them. The differences in RNA types, expression levels, and subcellular localization among different types of cells make ceRNA networks complex and diverse [126, 133]. These do not negate the reference value of ceRNA hypothesis. Key lncRNAs and miRNAs can still be found in the ceRNA network of PC cells.
Protein-lncRNA interaction
Protein-RNA interaction is universal [134-136]. In addition to acting as sponges for miRNAs, lncRNAs can also bind individual proteins or protein complexes and regulate their function [135]. Among them, lncRNAs can directly bind to proteins to induce them to target specific sites, and can also serve as scaffolds for the assembly of protein complexes [136]. We summarized three types of proteins, which can be regulated by lncRNA, have the potential to simultaneously regulate autophagy and Wnt/β-catenin signaling pathways, and are related to the formation of drug resistance in PC.
HuR
RBP can control the metabolism of massive transcripts and is a key factor that regulates gene transcription to translation [137]. HuR is one of the most prominent and well-studied factors in RBPs [138, 139]. HuR is mainly localized in the nucleus, while the abundance in the cytoplasm varies with the cell cycle, starting to increase at S phase until the cell enters the G2/M phase and decreasing again when the cell re-enters the G1 phase [140]. The nucleo-cytoplasmic shuttling ability of HuR is closely related to its physiological function [138].
In PC, HuR would target specific mRNAs in response to external stresses such as gemcitabine [141], hypoxia [142], apoptosis [143], hypoglycemia [144], DNA damage [145, 146], and poly ADP-ribose polymerase (PARP) inhibitors [146]. PC cells interfered with HuR siRNA had decreased migratory ability and the tumors formed in vitro became smaller [147]. The knockdown of HuR induced more PC cell death, while the xenograft tumor experiment using HuR knockout PC cell lines showed no growth at all of the subcutaneous tumors [148]. To verify that the absence of HuR resulted in the failure of tumorigenesis, PC cells supplemented with HuR cDNA restored the tumorigenic ability in a nude mouse model [148].
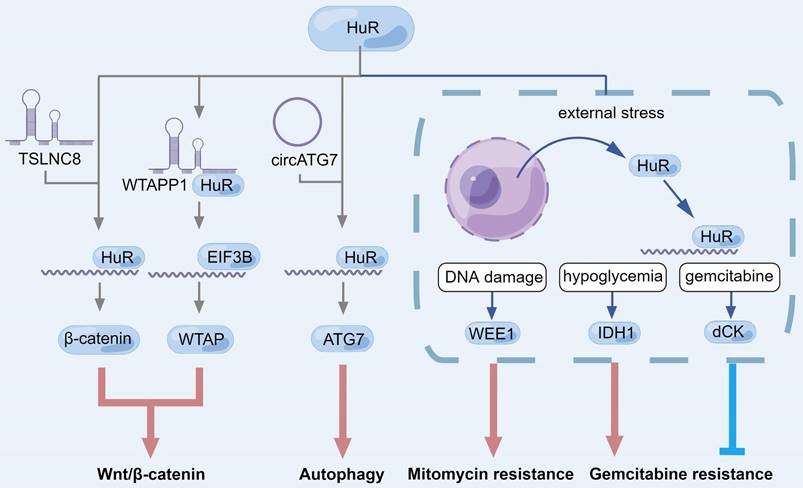
In research on lncRNA, in addition to TSLNC8, MALAT1, and WTAPP1 mentioned earlier, HuR can stabilize lncRNA DNA damage inducible transcript 4 antisense RNA 1 (DDIT4-AS1) by binding to the m6A site and up-regulated DDIT4-AS1 can mediate downregulation of DDIT4 mRNA through upstream frameshift 1 (UPF1), an RNA helicase, thereby activating the mTOR signaling pathway, enhancing PC dryness and inhibiting chemical sensitivity to gemcitabine [149]. For mRNA, lncRNA nicotinamide nucleotide transhydrogenase antisense RNA 1 (NNT-AS1) can recognize and stabilize m6A-modified integrin subunit beta 1 (ITGB1) mRNA through METTL3-HuR to activate the MAPK/ERK/PDL1 (programmed cell death 1 ligand 1) signaling pathway and finally promote the immune escape of PC cells [150]. In PC, HuR has the potential to recognize and stabilize m6A-modified RNA. In addition, variant subcellular localization of circATG7 regulated ATG7 mRNA level through different pathways [151]. In the cytoplasm, circATG7 stabilizes ATG7 mRNA by sponging miR-766-5p, and up-regulates ATG7 mRNA level by recruiting HuR in the nucleus, which eventually leads to the promotion of PC cell proliferation, metastasis and autophagy [151]. These all demonstrated that HuR can regulate Wnt/β-catenin signaling and autophagy through multiple pathways and is related to the progression of PC (Fig. 3). However, it should be noted that there are abundant modification sites on HuR including phosphorylation, methylation, and ubiquitination, which directly affect HuR subcellular localization and RNA binding activity [138]. Further studies are needed to investigate post-translational modification of HuR in PC.
FOX transcription factor family
FOX (forkhead box) family proteins are evolutionarily conserved DNA-binding proteins [152, 153]. Structurally, FOX proteins possess the conserved winged helix forkhead domain, also known as the DNA-binding domain, for attaching DNA [154]. The extra-FOX protein-protein interaction domain interacts with other factors to regulate DNA transcription and repair [155].
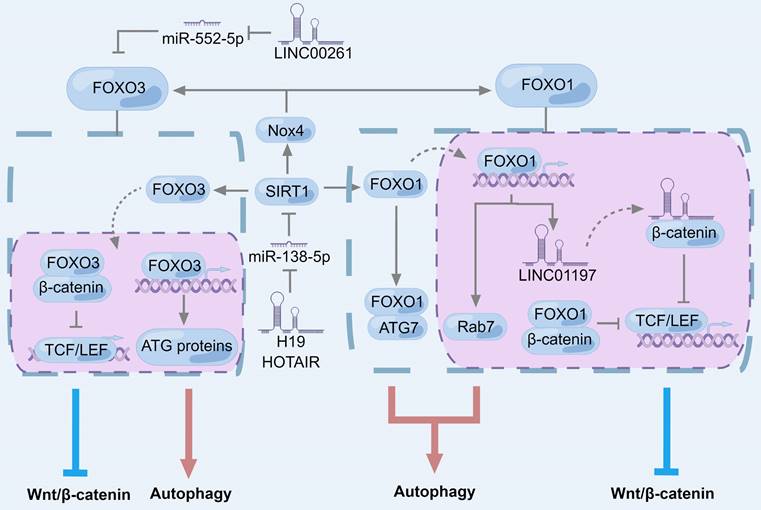
Here we focused on the FOXO subfamily and FOXM1(Fig. 4 and 5). In the nucleus, β-catenin can interact with TCF/LEF to induce transcription of target genes [24], but FOXO can competitively bind with β-catenin to inhibit the activity of β-catenin/TCF [156]. Both FOXO1 and FOXO3 are down-regulated in PC and inhibit the progression of PC [157-159]. Overexpression of LINC00261 can up-regulate FOXO3, the target gene of miR-552-5p, and inhibit the Wnt/β-catenin signaling pathway in PC [157]. LINC01197 is a target gene of FOXO1, which inhibits Wnt/β-catenin signaling activity by transcribing LINC01197, allowing it to bind to catenin and disrupt the interaction of catenin with TCF4 in PC cells [36].
For autophagy, sirtuin 1 (SIRT1) is a deacetylase, which can not only directly regulate the deacetylation of autophagy-related factors, such as beclin-1, ATG5, ATG7 and LC3 to induce autophagy [160-162], but also regulate autophagy through the FOXO family members FOXO1 and FOXO3[163, 164]. Sirt1-mediated FOXO1 deacetylation could activate its function and nuclear translocation to ultimately promote autophagy, while activation of FOXO1 can enhance the expression of RAS-related GTP-binding protein 7 (Rab7) [163], a small GTPase that mediates late autophagosome-lysosome fusion [165]. In addition, acetylated FOXO1 can directly bind ATG7 to induce autophagy [166]. In multiple cell lines, including PC cells, it has been confirmed that miR-138-5p specifically targets SIRT1 3' untranslated region and inhibits autophagy by reducing SIRT1[164, 167, 168]. Furthermore, knockdown of Rab7 or FOXO1 in PC inhibited the SIRT1-mediated increase of autophagic flux, suggesting that SIRT1 regulates autophagy in PC via FOXO1/Rab7 axis [164]. However, miR-138-5p can be spongy by lncRNA H19[169] and HOTAIR [170], which are up-regulated in PC [41, 171]. lncRNA may affect autophagy in PC by regulating SIRT1.
FOXO3 could up-regulate the transcription of multiple autophagy genes, such as ULK2, beclin-1, phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3), BCL2 interacting protein 3 (BNIP3), ATG4B, ATG4C, ATG5, ATG7, ATG12, ATG13, ATG14, ATG16L1, LC3, and GABARAPL1 [172-174]. FOXO3 deacetylated by SIRT1 can drive the transcription of BNIP3 and induce mitochondrial autophagy [175, 176]. In the study of skeletal dystrophic cachexia of PC, SIRT1 can indirectly regulate the expression of FOXO1 and FOXO3 by nuclear transcription factor-kappa B (NF-kB) signaling [177]. SIRT1 knockout induced NF-kB signaling and enhanced NADPH oxidase 4 (NOX4) transcription in cachexia muscles caused by PC, leading to increased reactive oxygen species (ROS) levels and FOXO expression [177]. Metformin differentially can regulate cellular ROS levels through AMPK-FOXO3-MnSOD (manganese superoxide dismutase, SOD2) pathway, especially in PC cells [178]. After combined administration with apigenin, ROS levels were further increased and exerted anticancer activity through DNA damaging-induced apoptosis, autophagy, and necrosis [178].
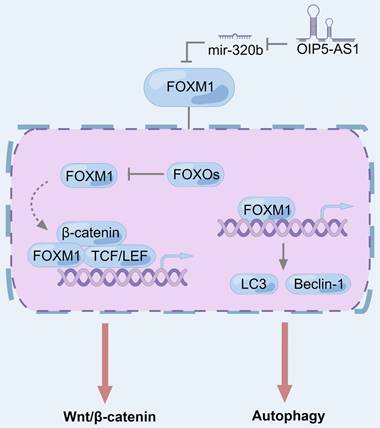
FOXM1 expression is elevated in a wide range of cancer cell lines and cancer types and can be used as a biomarker for cancer diagnosis, treatment, and prognosis [179-181]. In PC, LINC00857 acts as a protein scaffold to bind FOXM1 to ovarian tumor family deubiquitinase ubiquitin aldehyde binding 1 (OTUB1), thereby inhibiting FOXM1 degradation through the ubiquitin-proteasome pathway [182]. In the Wnt/β-catenin pathway, FOXM1 competes with FOXO and combines with β-catenin to regulate the transcription of downstream factors, thus modulating Wnt signal [183]. In terms of autophagy, FOXM1 is shown to up-regulate LC3-II/LC3-I and beclin-1 and promote autophagy in bladder cancer [184], liver cancer [185], prostate cancer [186], and gastric cancer [187], and it is determined in triple-negative breast cancer cells that FOXM1 directly binds to the promoter of LC3 and beclin-1 genes to promote transcription [188].
HIF family
In both primary and metastatic tumors, PC has characteristics of high levels of fibrosis [189]. The resulting hyperplasia of connective tissue forms a mechanical barrier around tumor cells, restricting the generation of blood vessels. Moreover, PC cells have a high level of metabolism, so the cancer microenvironmen is severely hypoxic, which is another characteristic of PC [190, 191]. The mechanism of biological adaptation to hypoxia is mediated by hypoxia-inducible factor (HIF) [192], which is regulated by a dimer composed of α subunits (Hif-1α, HIF-2α and HIF-3α) and β subunits (HIF-1β) [193]. Under normal oxygen conditions, HIF-α protein subunit is unstable and rapidly degraded by proteasome. Under hypoxic conditions, HIF-α is stable and translocates to the nucleus to bind to HIF-1β, where it is induced to the regulatory regions of target genes and modulates their transcription [194]. HIF-1α and HIF-2α are two major isoforms in mammalian cells. HIF-1α is widely distributed in almost all types of cells and is also regarded as a widely used hypoxia marker [195], while HIF-2α is expressed in certain cell types such as hepatocytes and endothelial cells [196].
The role of HuR during Wnt/β-catenin signaling pathway, autophagy and drug resistance in pancreatic cancer. LncRNA can induce HuR to bind to mRNA to promote the translation of downstream factors, or directly bind to HuR to recruit EIF3B to the mRNA translation initiation site, which ultimately manifests as autophagy and Wnt/β-catenin pathway activation. In response to different external stresses, HuR is translocated from the nucleus to the cytoplasm, where HuR can up-regulate specific factors to regulate drug resistance of pancreatic cancer cells under the conditions of DNA damage, hypoglycemia and gemcitabine treatment.
The role of FOXO1 and FOXO3 in regulating Wnt/β-catenin signaling pathway and autophagy in pancreatic cancer. Sirt1-mediated deacetylation of FOXO1 and FOXO3 can activate themselves, leading to their nuclear translocation and promoting autophagy. SIRT1 deficiency can induce NF-κB signaling in pancreatic cancer-induced cachexia muscles, enhance Nox4 transcription, and induce FOXO expression. In the nucleus, FOXO1 and FOXO3 bind to β-catenin to inhibit its transcriptional activity. LINC01197, as a target gene of FOXO1, can also bind to β-catenin to inhibit Wnt / β-catenin signaling. In addition to transcription of autophagy-related proteins, acetylated FOXO1 can directly bind to ATG7 to induce autophagy.
The role of FOXM1 in regulating Wnt/β-catenin signaling pathway and autophagy in pancreatic cancer. FOXM1 is inhibited by FOXO subfamily and its interaction with β-catenin can promote the binding of β-catenin to TCF/LEF and activate Wnt/β-catenin signaling. For autophagy, overexpression of FOXM1 and nuclear displacement can regulate the expression of autophagy-related genes, which is manifested as promoting autophagy.
PC cells can adapt to the extreme conditions of hypoxia by activating HIF, which in turn transcribe genes related to angiogenesis and glycolysis [197]. According to recent studies, HIF-1α has feedback regulation with various lncRNAs in PC, and promotes PC progression and drug resistance (Fig. 6). LncRNA CF129145. 1 (CF129), a downstream target gene of HIF-1α, was inhibited by HIF-1α under hypoxic conditions, and CF129 reduction inhibited the degradation of tumor protein P53 (P53, TP53) by makorin ring finger protein 1 (MKRN1) ubiquitination. After P53 translocation into the nucleus and transcription of FOXC2, FOXC2 can bind to the HIF-1α promoter to activate its transcription. Thus, during hypoxia, HIF-1α/CF129/P53/FOXC2 forms a feedback loop and promotes PC progression [198].
LncRNA HIF1A-AS1 is an antisense RNA of HIF-1α, and HIF-1α can also activate HIF1A-AS1 transcription [199]. In the cytoplasm, HIF1A-AS1 can induce Y box binding protein 1 (YB1) to interact with the serine/threonine kinase AKT, leading to phosphorylation of YB1 (pYB1). Meanwhile, pYB1 is recruited by HIF1A-AS1 to bind to HIF-1α mRNA, thereby promoting the translation of HIF-1α. Thus, the positive feedback between HIF1A-AS1 and HIF-1α makes them highly expressed in PC and promotes the resistance to gemcitabine [199].
Similarly, lncRNA NR2F1-AS1 is an antisense RNA of nuclear receptor subfamily 2, group F, member 1 (NR2F1), which is the target of NR2F1-AS1 and is positively regulated by NR2F1-AS1. NR2F1 can activate AKT/mTOR pathway and up-regulate HIF-1α in PC. NR2F1-AS1 is also a hypoxia-responsive lncRNA in PC cells, which can be transcription by HIF-1α under hypoxic conditions. Therefore, NR2F1-AS1 forms a positive feedback with HIF-1α via the NR2F1/AKT/mTOR axis [200].
ZEB1(zinc finger E-box binding homeobox 1) is an EMT activator and a key regulator of PC cell plasticity, metastasis and drug resistance [201, 202]. By binding with histone deacetylase 1 (HDAC1) and HIF-1α, ZEB1 can inhibit the acetylation of HIF-1α and further maintain the stability of HIF-1α [203]. In PC, ZEB1-AS1 is an antisense RNA of ZEB1, which can up-regulate the mRNA and protein levels of ZEB1[203], and lncRNA ZEB1 transcriptional regulator RNA (BX111, ZEBTR) induces ZEB1 transcription by recruiting YB1[204]. Under hypoxia, both ZEB1-AS1 and BX111 are transcriptized and up-regulated by HIF-1α, which stabilizes HIF-1α through ZEB1, thus forming a positive feedback loop with HIF-1α [203, 204]. Moreover, similar to ZEB1, metastasis associated 1 family member 2 (MTA2) can form a complex with HDAC1 to deacetylate and stabilize HIF-1α [205]. LncRNA MTA2 transcriptional regulator RNA (MTA2TR) is regulated by HIF-1α transcription in PC and recruits activating transcription factor 3 (ATF3) to the MTA2 promoter to promote MTA2 transcription, forming a HIF-1α/MTA2TR/MTA2 positive feedback loop [206].
The positive feedback loop between HIF-1α and lncRNAs in pancreatic cancer. LncRNAs can be divided into three categories according to the steps of HIF-1α expression. In the HIF-1α transcription stage, HIF-1α can inhibit the transcription of CF129 and then inhibit the ubiquitination of P53. P53 enters the nucleus and transcribe FOXC2, which in turn transcribe the target gene HIF-1α. PVT1 can bind to the HIF-1α promoter and promote the expression of HIF-1α. In the HIF-1α translation stage, HIF1A-AS1 can induce YB1 phosphorylation by AKT and recruit pYB1 to HIF-1αmRNA, thereby promoting HIF-1α translation. PVT1 spongifies miR-519d-3p and miR-143 to stabilize HIF-1αmRNA. After HIF-1α translation, MTA2TR, BX111 and ZEB1-AS1 induce the expression of MTA2 and ZEB1 which in turn cooperate with HDACs to deacetylate HIF-1α. Nr2f1-as1 induces NR2F1 to up-regulate HIF-1α by activating AKT/mTOR signaling. PVT1 interacts with HIF-1α and maintains its protein level.
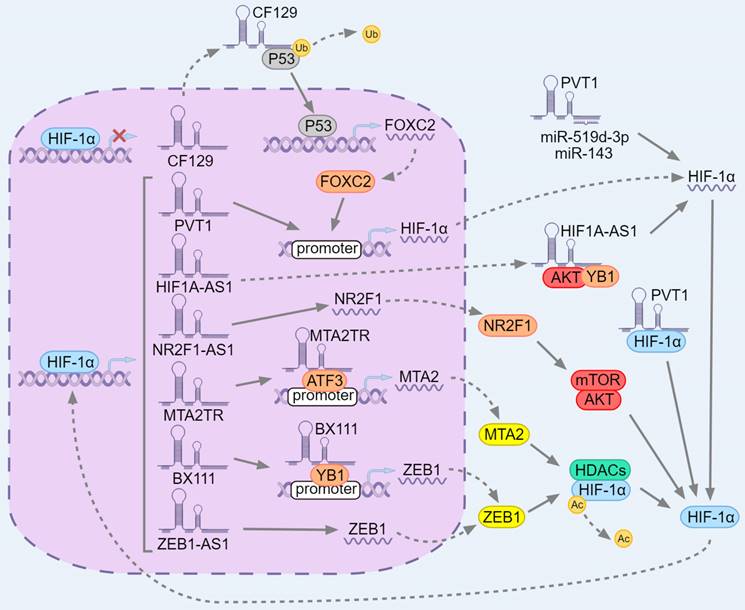
In addition to targeting miR-143 [64] and miR-519d-3p [128] to regulate HIF-1α expression, there is a positive feedback loop between PVT1 and HIF-1α. [207] On the one hand, PVT1 can bind to the HIF-1α promoter and activate its transcription, and bind to HIF-1α protein to up-regulate the level of HIF-1α after translation. On the other hand, PVT1 is also a downstream target of HIF-1α, and HIF-1α can bind to the PVT1 promoter to activate its transcription. Moreover, the expression level of PVT1 and HIF-1α can stabilize each other after transcription, HIF-1α can down-regulate the attenuation rate of PVT1, and PVT1 can inhibit the proteasome-dependent degradation of HIF-1α. In this way, the positive feedback loop between PVT1 and HIF-1α promotes the progression of PC [207].
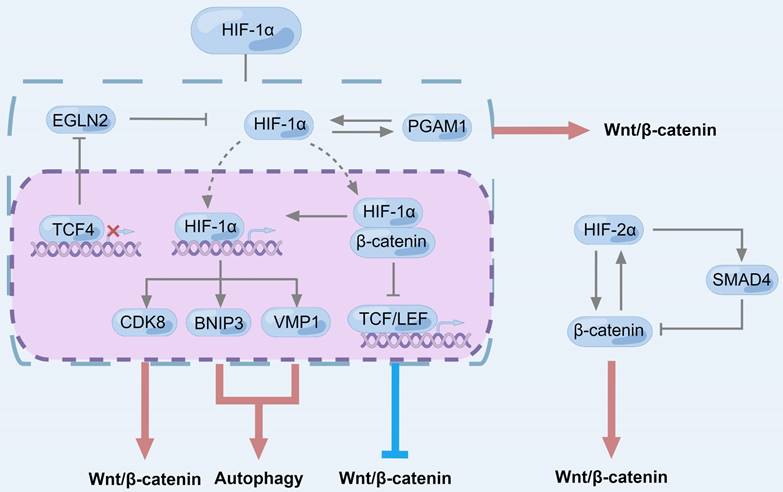
HIF-1α has been widely confirmed to promote autophagy [208, 209], and its downstream factor BNIP3 has been identified in a variety of cells [210-213]. Furthermore, HIF-1α has been shown to promote the transformation of non-stem cell PC cells into prominin 1 positive (CD133+, PROM1 positive) PC stem cell-like cells under intermittent hypoxia by inducing autophagy [214], and to promote the EMT and metastatic capacity of stem cells [215]. On the other hand, HIF-1α is also generally believed to be activated by the Wnt/β-catenin pathway [216, 217] (Fig. 7). In PC, as a transcriptional chaperone of β-catenin, TCF4 positively can regulate aerobic glycolysis by inhibiting egl-9 family hypoxia-inducible factor 2 (EGLN2), leading to up-regulation of HIF-1α [218]. In esophageal squamous cell carcinoma, HIF-1α can directly bind to the promoter region of TCF4 and promote the expression of TCF4[219]. HIF-1α can also directly bind to β-catenin [219-221], enhance the transcriptional activity of HIF-1α [220, 221], but inhibit the interaction between TCF4 and β-catenin, making β-catenin lose the ability to transduce signals [220, 221]. In addition to affecting the transcriptional activity of β-catenin, mutant Kirsten rat sarcoma viral oncogene homolog (K-RAS) can up-regulate HIF-1α in PC cells, and HIF-1α overexpression increases the protein level of CDK8. Furthermore, CDK8 stabilizes β-catenin and activates Wnt/β-catenin pathway by regulating AXIN2 and GSK‑3β [222]. Similarly, phosphoglycerate mutase 1 (PGAM1), a key glycolytic protein, is significantly overexpressed in PC metastases and associated with poor prognosis [223], while the use of an allosteric PGAM1 inhibitor restrains PC progression [224]. Among them, PGAM1 mainly exists in the cytoplasm and cell membrane and interacts with HIF-1α to positively regulate each other. The up-regulated PGAM1 promotes EMT by activating Wnt/β-catenin signaling pathway [223]. Thus, there may be complex feedback regulation between HIF and Wnt/β-catenin signaling pathways.
Regarding HIF-2α, CAF cells with specific deletion of HIF-2α inhibited PC tumor progression and growth and increased the survival of experimental mice by 50% [225]. Down-regulation of HIF-2α in CAF induced tumor fibrosis and significantly reduced the intratumoral recruitment of immunosuppressive M2 macrophages and regulatory T cells, and improved the immunosuppressive effect of TME [225]. The interaction between HIF-2α and β-catenin in PC leads to increased activity of classical Wnt/β-catenin, and also promotes HIF-2α transcriptional activity by stabilizing HIF-2α [226]. HIF-2α is associated with the early development of PC. In normal human pancreas, HIF-2α is easily degraded to a very low level, but hypoxic conditions induce the stabilization of HIF-2α, leading to the development of chronic pancreatitis [227]. In the context of oncogenic K-RAS, pancreatic cells further develop into cysts similar to mucinous cystic neoplasms [227], the formation of which is associated with the activation of Wnt/β-catenin signaling [228]. Knockdown of HIF-2α in low-grade pancreatic intraepithelial neoplasia (PanIN) increased the number of cell lesions, but these lesions failed to progress to high-grade PanIN and showed decreased protein levels of β-catenin and drosophila mothers against decapentaplegic protein 4 (SMAD4) [229]. Interestingly, the expression of β-catenin was negatively regulated by SMAD4 [230], and HIF-2α could regulate the expression of β-catenin and SMAD4 independently in different ways [229]. The two pathways are competitive, and HIF-2α is more likely to up-regulate β-catenin to activate the canonical Wnt/β-catenin signaling pathway in the early progression of PC [229] (Fig. 7).
Resistance to gemcitabine
Mechanisms of action of gemcitabine
Gemcitabine is the first-line drug for the treatment of advanced PC, and the current research on drug resistance focuses on gemcitabine [12]. Gemcitabine is a cytosine nucleoside derivative, also known as 2',2'-difluoro-2'deoxycytidine (dFdC), and its mechanism of action is related to the multiple effects of its intracellular metabolites on DNA synthesis [231]. After entering the cell via nucleoside transporters, gemcitabine is progressively phosphorylated to gemcitabine monophosphate (dFdCMP), gemcitabine diphosphate (dFdCDP) and gemcitabine triphosphate (dFdCTP) [231]. Among them, dFdCTP can be involved in DNA synthesis. After dFdCTP is incorporated into the DNA chain and ligated with another deoxynucleotide, the DNA strand stops extending, which is called "masked chain termination". [231, 232] Similarly, this effect contributes to the inability of DNA repair enzymes to recognize dFdCTP, which interferes with the normal DNA repair function of cells, so that gemcitabine continues to exert the function of inhibiting DNA synthesis [232]. In addition, there are other mechanisms by which gemcitabine interferes with cellular regulation. Different metabolites increase each other's physiological activities and enhance the ability to inhibit cell growth as a whole. This interaction is called "self-enhancement"[232]. dFdCTP competes with deoxycytidine triphosphate (dCTP) for binding to DNA polymerase to inhibit its activity [231]. As a ribonucleoside reductase (RR) inhibitor, dFdCDP can regulate RR activity by limiting the formation of nucleoside triphosphate (NTP), which reduces cytidine diphosphate (CDP) to deoxycytidine diphosphate (dCDP)[233], leading to depletion of the deoxyribonucleotide pool required for DNA synthesis and enhancing the effect of dFdCTP[231]. dFdCTP can also inhibit the effect of deoxycytidine monophosphate deaminase (dCMP) on dFdCMP, preventing its conversion to 2',2'-difluorodeoxyuridine monophosphate (dFdUMP), which is then discharged from cells [231, 234, 235].
The role of HIF in regulating Wnt/β-catenin signaling pathway and autophagy in pancreatic cancer. CDK8, BNIP3 and VMP1 are the downstream factors of HIF-1α, which activate autophagy and Wnt/β-catenin signaling. HIF-1α binds to β-catenin and inhibits the transcriptional activity of β-catenin but up-regulates the activity of HIF-1α. As a β-catenin transcriptional chaperone, TCF4 can inhibit EGLN2 to positively regulate aerobic glycolysis, leading to up-regulation of HIF-1α. PGAM1 mainly exists in cytoplasm and cell membrane and interacts with HIF-1α to positively regulate each other, while up-regulated PGAM1 promotes EMT of pancreatic cancer cells by activating Wnt/β-catenin signaling pathway. Different from HIF-1α, the transcriptional activity of HIF-2α is upregulated after binding to β-catenin. HIF-2α could regulate β-catenin and SMAD4 independently in different ways. There is a competitive relationship between the two pathways, and HIF-2α is more inclined to up-regulate β-catenin to activate the classical wnt signaling pathway in the early progression of pancreatic cancer
Regulatory pathways for gemcitabine
In PC, the intracellular metabolism of gemcitabine requires the co-regulation of multiple enzymes, which are regulated by a variety of miRNAs [236]. According to the ceRNA hypothesis [126], these miRNAs can be used as the mediators of lncRNA regulation of downstream factors, and lncRNA also has the potential to regulate the metabolism of gemcitabine cells.
Besides these pathways, the susceptibility of PC cells to gemcitabine is inversely proportional to the levels of Wnt/β-catenin and autophagy pathways. LncRNA can affect the resistance of PC cells to gemcitabine through autophagy and Wnt/β-catenin pathways. In addition to PVT1 mentioned above, lncRNA histocompatibility leukocyte antigen complex P5 (HCP5) can act as ceRNA to inhibit the expression of miR-214-3p and target heparin binding growth factor (HDGF) to regulate the proliferation, invasion, migration, apoptosis and autophagy of PC cells, thus promoting gemcitabine resistance [237]. In pancreatic stellate cell (PSC), HIF-1α can induce the expression of HDGF and the increased level of HDGF shows anti-apoptotic and pro-fibrotic effects, which can maintain tumor lesions [238]. Interestingly, HDGF was more commonly found to regulate Wnt/β-catenin pathway than autophagy in cancer cells [239-242]. The expression of Wnt/β-catenin pathway genes was increased in the xenograft model of primary non-small cell lung cancer (NSCLC), while Wnt/β-catenin pathway genes were significantly down-regulated after anti-HDGF treatment, especially Wnt1 and FZD were severely inhibited. HDGF may be a target for inhibiting the proliferation of cancer stem cells and preventing the recurrence of lung cancer after chemotherapy [243].
HuR
In addition to being targeted by lncRNAs to regulate autophagy and Wnt/β-catenin in PC, HuR also shows a strong correlation with gemcitabine resistance, which can be used as a marker for the treatment and prognosis of PC [244-246] (Fig. 3).
In PC cells, HuR associates with deoxycytidine kinase (dCK) mRNA, which encodes a metabolic enzyme that can activate gemcitabine [141, 247]. Inducible factors including gemcitabine can increase the translocation of HuR from the nucleus to the cytoplasm, leading to the strengthening of the association between HuR and dCK mRNA, resulting in the attenuation of gemcitabine resistance [141, 145, 247, 248]. On the other hand, HuR can up-regulate tumor resistance to other chemotherapeutic agents. WEE1 G2 checkpoint kinase (WEE1) mRNA, a mitotic inhibitor kinase, can be stabilized by HuR [145] to regulate DNA damage repair pathways [249, 250]. Similarly, when PC cells were under external environmental stress, such as DNA damage, HuR translocalized from the nucleus to the cytoplasm and then up-regulating WEE1. Positive regulation of WEE1 by HuR can increase H2A. X variant histone phosphorylation at serine-139 (γH2AX) levels, induce CDK1 phosphorylation, and promote cell cycle arrest at G2-M transition [145]. Different from the effect on gemcitabine, the increase of HuR contributes to DNA damage repair and resistance to cytotoxic therapy [145].
HuR can also affect the drug resistance in PC by regulating glucose metabolism. Although low nutrient levels slow down the growth of PC cells, they promote chemotherapy resistance [144]. Acute glucose deprivation can act as a potent stimulus for HuR translocation from the nucleus to the cytoplasm, where HuR stabilizes its target mRNA. These transcripts encode enzymes essential for glucose metabolism, and these targets are essential for the survival of cancer cells in the metabolically impaired TME [251]. Silencing HuR attenuated drug resistance in PC cells, and drug resistance would be further reduced under conditions of nutrient deficiency. This is mainly due to the fact that HuR can export from the nucleus to the cytoplasm to stabilize isocitrate dehydrogenase (NADP(+)) 1 (IDH1) expression and enhance ROS scavenging, which enhances the reducing capacity of PC and protects PC under nutrient deficiency [251]. Similarly, IDH1 overexpression also enhanced gemcitabine resistance in PC cells [144].
FOXO3-FOXM1 axis
FOXO3 and FOXM1 are a pair of transcription factors with opposite functions, which not only compete for binding to promoters of the same DNA motif, but also have opposite transcriptional effects on target genes [252]. In addition, FOXM1 is also a downstream target of FOXO3, and FOXM1 is negatively regulated by FOXO3[253, 254]. FOXO3-FOXM1 axis is a key regulatory target of cancer drug resistance [255]. The dysregulation of FOXO3-FOXM1 axis outside the autophagy and Wnt/β-catenin pathways can lead to drug resistance by regulating drug efflux and DNA repair [255, 256] (Fig. 8). Membrane ATP binding cassette (ABC) transporters are tightly linked to drug transport, acting as protein pumps for drug efflux that drive the development of multidrug resistance (MDR) in cancer cells [257]. FOX proteins are transcription factors that can directly regulate the expression of different ABC proteins, and the target genes of FOXM1 include ABCC4 [258], ABCC5 [259, 260], ABCC10 [261], ABCG2 [262], FOXO3 include ABCA6 [263], ABCB1 [264], FOXO1 include ABCA1 [265], ABCA6 [263], ABCA9 [266], ABCC2 [265]. Meanwhile, the up-regulated FOX proteins in cancer cells can generate drug resistance in an ABC protein-dependent manner [256, 258-262, 264, 265]. On the other hand, chemotherapeutic agents can mediate their cytotoxic and cytostatic functions through FOXO3 and FOXM1. For example, paclitaxel induced FOXO3 nuclear translocation to mediate its cytotoxicity and in turn promoted breast cancer cell death due to the fact that paclitaxel can promote the nuclear translocation of FOXO3 by activating c-Jun NH2 terminal kinase 1/2 (JNK1/2) in combination with inhibiting the AKT pathway and activating the pro apoptotic molecule BCL2-interacting mediator of cell death (BIM, BCL2L11) to trigger apoptosis [267, 268]. Similarly, paclitaxel can down-regulate the mRNA and protein levels of FOXM1 and induce mitotic arrest and senescence in cancer cells partly by down-regulating FOXM1 [269].
The role of FOXO3-FOXM1 axis in regulating drug resistance in pancreatic cancer. A variety of ABC transporters are the target genes of FOXO1, FOXO3 and FOXM1, which can make pancreatic cancer cells resistant to multiple drugs. During FOXM1 deubiquitination, FAT10 competitively binds with ubiquitin to inhibit FOXM1's ubiquitination, while UCHL3 can deubiquitinate the ubiquitinated FOXM1, both of which reduce the sensitivity of pancreatic cancer cells to gemcitabine. PHB1 is a downstream factor of FOXM1, which can activate the RAS-RAF-MEK-ERK pathway to generate a positive feedback loop and lead to the development of paclitaxel resistance. MiR-223 targets FOXO3 to acquire CDDP resistance in cancer cells. cGMP inhibited FOXO3 and affected the CSC phenotype of pancreatic cancer cells through the FOXO3/LKB1/AMPK/PGC-1β/PDHA1/CD44 axis. FOXO3 can maintain CD44 expression and make CSC acquire drug resistance.
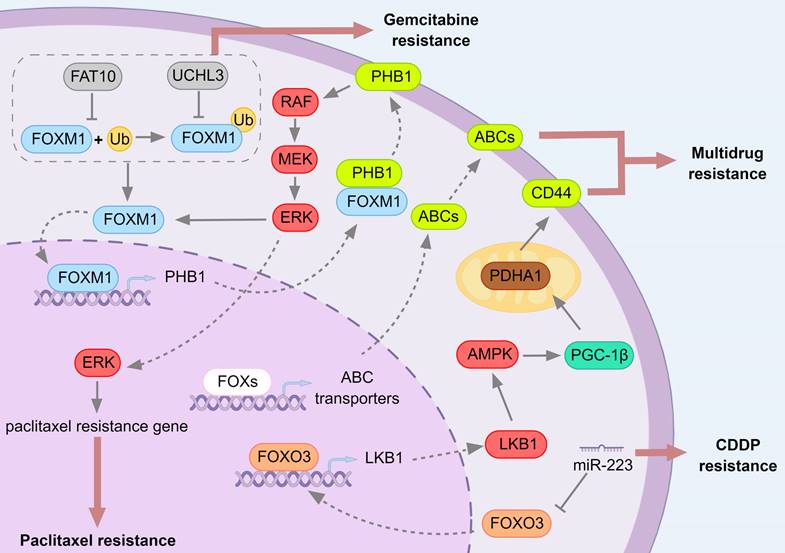
In PC, gemcitabine resistance is associated with FOXM1 stability [270, 271]. Ubiquitin C-terminal hydrolase L3 (UCHL3) as a deubiquitinating enzyme is able to inhibit FOXM1 ubiquitination and degradation [270], while Human leukocyte antigen F-associated transcript 10 (FAT10) as a ubiquitin like protein is able to inhibit FOXM1 ubiquitination and stabilize FOXM1 expression by competing with ubiquitin for binding to FOXM1 [271]. The down-regulation of UCHL3 and FAT10 increased FOXM1 ubiquitination, and also promoted the sensitivity of PC cells to gemcitabine [270, 271]. FOXM1 expression was also up-regulated in the gemcitabine-resistant cell line model [272].
For other chemotherapeutic drugs, such as paclitaxel, FOXM1 can induce paclitaxel resistance in PC through different pathways. Prohibitin 1 (PHB1) is a downstream factor of FOXM1, which can recruit Raf-1 proto-oncogene (RAF1) to Caveolin-1-enriched lipid rafts and activates the RAS-RAF-MEK-ERK pathway leading to drug resistance [273]. In the cytoplasm, FOXM1 can bind to PHB1 and stabilize PHB1 to shift it to the cell membrane. PHB1 depletion can also down-regulate the level of FOXM1, and there is a positive feedback regulation of FOXM1/PHB1/RAF-MEK-ERK [273, 274]. In addition, ABCA2 is also regulated by FOXM1/PHB1/RAF-MEK-ERK pathway. Overexpression of FOXM1 or depletion of PHB1 affected ABCA2 protein and mRNA levels [273]. The expression levels of FOXM1, PHB1, ABCA2 and p-ERK1/2 were up-regulated in PC cells and paclitaxel-resistant cell lines, and were also proportional to the drug dose [273]. Genistein is a natural isoflavone found in legumes with antitumor effects [275]. It has been reported to affect the incidence of PC [276, 277] and inhibit the growth of cancer cells in vitro and in vivo [278], which is a potential chemopreventive and therapeutic agent for PC. After treatment with genistein, the expression of FOXM1 and its downstream genes was down-regulated, and the growth and invasion of PC cells were inhibited [279]. Whereas overexpression of FOXM1 reduced genistein-induced cell growth inhibition and apoptosis [279]. NOSH-aspirin, a novel anti-inflammatory agent, also has anticancer effects and can significantly reduce the growth and progression of PC [280, 281]. The high level FOXM1 of tumors in a xenograft mouse model of PC was also observably inhibited by NOSH-aspirin [282]. Dacomitinib can inhibit the growth and proliferation of PC cells by down-regulating FOXM1 and its downstream targets, such as polo-like kinase 1 (PLK1), survivin, cyclin B1 (CCNB1), c-Myc and aurora kinase B (AURKB) [283].
In contrast to FOXM1, FOXO3 is regarded as a tumor suppressor and the overexpression of FOXO3 can inhibit the proliferation, tumorigenic potential and invasiveness of various cancer cells [284]. Similarly, downregulation of FOXO3 can promote PC development [158]. But the difference is that the role of FOXO3 on drug sensitivity may be multifaceted.
In cholangiocarcinoma (CCA), LINC01714 was shown to be recurrently down-regulated in clinical samples and significantly correlated with overall survival in patients, and overexpression of LINC01714 inhibited the proliferation, migration and invasion of cancer cells both in vitro and in vivo [285]. Moreover, LINC01714 interacted with FOXO3 with increased FOXO3 protein level but decreased FOXO3 phosphorylation at Ser318 site. Interestingly, LINC01714 enhanced gemcitabine sensitivity of CCA tumor cells by regulating FOXO3 phosphorylation [285]. This may be related to the output of FOXO3 phosphorylation from the nucleus and the loss of its transcriptional activity [286]. In NSCLC, both tripartite motif containing 22 (TRIM22) and troponin C1 (TNNC1), downstream factors negatively regulated by FOXO3, are up-regulated in cancer cells and gemcitabine resistant cell lines and confer gemcitabine resistance by protective autophagy [287, 288].
In different cancers, the variant expression of FOXO3 has variant effects on prognosis. High expression of FOXO3 has a favorable prognosis in acute myeloid leukemia [289], breast cancer [290], bladder cancer [291], gastric cancer [292], nasopharyngeal carcinoma [293] and human ovarian cancer [294]. But in hepatocellular carcinoma [295, 296] and PC [297], high levels of activated FOXO3 lead to poor patient prognosis. This discrepancy may be due to P53 mutations. In the case of P53 mutations, FOXO3 acts as a tumor suppressor, and wild-type P53 alters the site at which FOXO3 recognizes target promoters, thereby inhibiting FOXO3 induced apoptosis and instead potentiating chemoresistance and survival of cells [298].
For PC, FOXO3 is a target of miR-223, and downregulation of FOXO3 by miR-233 leads to the proliferation, apoptosis, and cisplatin resistance in cancer cells [299]. Furthermore, FOXO3 is essential for cluster of differentiation-44 (CD44) expression and cancer stem cell (CSC) properties, and inhibition of FOXO3 by cyclic guanosine monophosphate (cGMP) [297] as well as through FOXO3/LKB1/AMPK/PGC-1β (peroxisome proliferator-activated receptor gamma, coactivator 1 beta)/PDHA1 (pyruvate dehydrogenase E1 subunit alpha 1)/CD44 axis influences the CSC phenotype of PC cells [300]. FOXO3 to maintain CD44 expression enables CSCs to acquire drug resistance [297, 300]. Cardamonin (CAR), a flavonoid present in the genus arangal, inhibits PC cell growth and promotes apoptosis [301, 302]. After controlled experiments, CAR can promote the chemosensitivity of PC cells to gemcitabine, and cell viability is further decreased after combined use of gemcitabine. These functions are achieved by CAR through FOXO3 promotion and FOXM1 inhibition [302].
Hypoxia signaling pathway
The main role of HIFs in mediating tumor biology is caused by hypoxia [192] and the hypoxic environment of PC is in turn associated with its extreme TME [190, 191]. The function of HIFs to induce drug resistance is tightly linked to hypoxia and the TME (Fig. 9). Under hypoxic conditions, HIF-α remains stable and translocates to the nucleus to bind to HIF-1β, which in turn induces downstream target gene transcription [194]. Typical among the factors downstream of HIF that have been associated with drug resistance are the ABC transporters, which act as protein pumps to efflux drugs enabling cells to acquire drug resistance [257]. Hypoxia-induced HIF-1α and HIF-2α have a promoting effect on the expression of ABC transporters, including ABCB1, ABCB5, ABCC1, and ABCG2, and induce drug resistance in cancer cells [303].
HIF-1α can directly bind to the promoters of ABCC1 [304], ABCB1 [305], ABCB6 [306], ABCA1 [307] and ABCG2 [308], and HIF-2α can directly bind to the promoter of ABCG2 [309] to promote the transcriptional expression of ABC proteins. In addition, HIF-1α can also transcribe specific protein 1 (SP1), which can activate the ABCC8 promoter [310]. For PC, HIF-1 has been confirmed to make cancer cells resistant to gemcitabine and 5-Fluorouracil (5-FU) by regulating ABCB1 and ABCG2 [308, 311]. While quercetin, a flavonoid that can inhibit the efflux activity of ABCB1, in combination with gemcitabine can down-regulate HIF-1α and up-regulate the apoptosis regulator P53, enhancing the cytotoxic activity of gemcitabine [312].
The role of HIF-1α in regulating drug resistance in pancreatic cancer. HIF-1α can make pancreatic cancer resistant in three ways. First, multiple ABC transporters are downstream target genes of HIF-1α, and HIF-1α also upregulates the transcription factor Sp1, which transcribes ABCC8. Up-regulation of ABC transporters promote drug efflux and make cells acquire drug resistance. Secondly, HIF-1α can regulate glucose metabolism in pancreatic cancer cells hENT1 promotes the nuclear translocation of HIF-1α, and MUC1 promotes the nuclear displacement of NF-κB and HIF-1α, thereby altering the glucose metabolism level of pancreatic cancer cells. After inducing the transcription of glycolytic genes (TKT and CTPS1), the synthesis of dCTP is increased, which can compete with gemcitabine for DNA synthesis and inhibit the cytotoxicity of gemcitabine. Finally, a variety of proteins in the target genes of HIF-1α are transferred to the cell membrane or secreted to the extracellular to participate in the regulation of tumor microenvironment. Tumor microenvironment affects HIF-1α expression and drug resistance by maintaining hypoxic environment, regulating intercellular signaling and forming physical barriers.
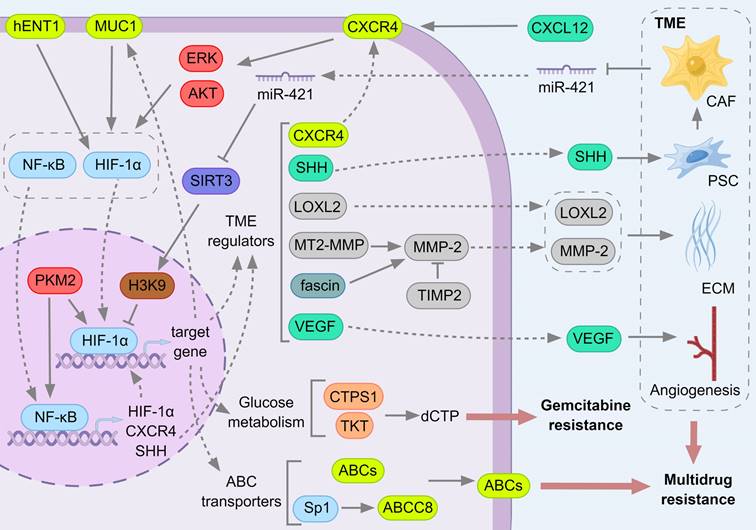
Besides, hypoxia-induced HIF stabilization can reprogram the metabolic way of cancer cells and produce drug resistance. HIF-1 can activate the transcription of glucose transporters and glycolytic enzymes and increase glucose metabolism through the glycolytic pathway, but reduce glucose entry into the tricarboxylic acid cycle (TCA cycle) [313], and this metabolic pattern is beneficial to cancer cell proliferation. Under hypoxic and glucose deprivation conditions, HIF can activate anaerobic metabolism of PC cells and inhibit their apoptosis [314]. Meanwhile, mucin 1 (MUC1), a polymorphic mucin-like protein that is overexpressed in PC, can stabilize HIF-1α and promote HIF-1α recruitment to glycolytic gene promoters (transketolase (TKT) and CTP synthase 1 (CTPS1)) in a hypoxia-dependent manner [315]. MUC1 is also a target gene of HIF-1α [316], and MUC1 and HIF-1α can regulate each other in a positive feedback manner. MUC1 and HIF-1α can synergistically regulate glucose metabolism and pyrimidine biosynthesis in gemcitabine-resistant PC cells, resulting in increased nucleotide synthesis and accumulation of dCTP, which can cause competitive inhibition of active gemcitabine, thus producing gemcitabine resistance [317].
The level of human equilibrative nucleoside transporter 1 (hENT1) is tightly associated with the sensitivity of PC cells to gemcitabine and can be used as a biological indicator to predict the efficacy and prognosis of gemcitabine [318-321]. HENT1 was down-regulated in gemcitabine resistant PC cells, and overexpression of hENT1 significantly reversed chemoresistance. Specifically, hENT1 can induce the development of drug resistance by regulating glucose transport and glycolysis through HIF-1α and c-Myc, low hENT1 and c-Myc expression in drug-resistant cells, highly active HIF-1α potentiates glycolysis and generates chemoresistance, whereas overexpression of hENT1 elevated c-Myc expression, suppressed HIF-1α, restored glucose transport and glycolysis in cells and reversed gemcitabine induced drug resistance [322].
The effects of TME and PC cells are reciprocal. In addition to maintaining the hypoxic environment of cancer cells to stabilize HIF-1α and activate its downstream target genes, CAF also regulate cancer cell metabolism through paracrine pathways. Exosome miR-421 secreted by CAF can down-regulate SIRT3 in PC cells [323]. SIRT3 is a target of miR-421 and also located upstream of HIF-1α. It can inhibit the expression and activity of HIF-1α in different cancer cells through AMPK/mTOR axis [324], FOXO3 [325] and reducing ROS [326]. In PC, SIRT3 inhibited the transcription of HIF-1α by deacetylating acetylation of Histone H3 at lysine9 (H3K9ac), and miR-421 with high expression of CAF promoted the proliferation of PC cells [323].
In contrast, HIF-1α can generate a desmoplastic response that solidifies the TME of PC [327]. The sonic hedgehog ligand (SHH) is a member of the hedgehog proteins and a frequently used signaling transmitter in mediating intercellular communication [328]. In PC, SHH can regulate the physiological activities of PC cells, including desmoplasia, cancer cell metastasis, and lymphatic vessel formation, through paracrine secretion [329, 330]. It has been reported that SHH is expressed in PC cells in a HIF dependent manner and induces the desmoplastic response of CAFs through a paracrine manner [327]. While SHH activates PSCs to secrete high levels of perineural invasion (PNI) -related molecules to promote PNI in PC [331]. The desmoplastic connective tissue and PNI further maintain the hypoxic environment of the tumor and stabilizes HIF, allowing PC cells to acquire drug resistance.
In the same way, CXCL12/CXCR4 (C-X-C motif chemokine ligand 12/C-X-C motif chemokine receptor 4) signaling axis can confer drug resistance to PC cells and participate in the invasion and metastasis of PC [332, 333]. CXCL12 secreted by stromal cells binded to the receptor CXCR4 on PC cells, thereby activating AKT and ERK, leading to nuclear accumulation of NF-kB. As a result, nuclear NF-kB directly binded to the SHH promoter and induces SHH expression [334]. When PC cells were treated with gemcitabine, ROS was up-regulated and mediated the nuclear translocation of NF-kB and HIF-1α by activating ERK1/ 2 and AKT. Then NF-kB and HIF-1α bind to the CXCR4 promoter, up-regulate CXCR4 and lead to enhanced motility and invasion of PC cells [335]. The adverse reactions of gemcitabine treatment were reflected. First, gemcitabine may enhance the anti-apoptotic pathway downstream of CXCR4, thereby making cancer cells resistant. In addition, gemcitabine may enhance the metastasis of PC cells to other CXCL12 overexpression environments [335].
Lysyl oxidase (LOX) and lysyl oxidase-like protein (LOXL) are copper-dependent amine oxidases that catalyze the covalent cross-linking of collagen and elastin in extracellular matrix (ECM), which are related to the progression of cancer [336-338]. There is a hypoxia response element (HRE) on the promoter of human LOX gene to respond to HIF and transcribe LOX protein [339]. LOX and LOXL levels in PC cells and PSCs increased with HIF-1α activity [340, 341]. LOXL2 can reduce the drug concentration in the tumor during chemotherapy. This may be due to LOXL2 forming a physical barrier to inhibit the diffusion of gemcitabine by increasing fibrous collagen and then increasing ECM stiffness, or it may be due to the collapse of blood vessels in the tumor caused by LOXL2, which can limit the transport of gemcitabine into the tumor interior [342].
Correspondingly, matrix metalloproteinase (MMP) is a kind of proteolytic enzyme, which can decompose ECM protein, promote angiogenesis, and is related to tumor proliferation and metastasis [343, 344]. MMP has the opposite effect to LOX, and inhibition of LOX can promote the expression of MMP [345, 346]. However, LOX has little effect on the activity of MMP in rat aortic smooth muscle cells [347], and even in gastric cancer [348], cervical cancer [349], colorectal cancer [350], non-small cell lung cancer [351], breast cancer [352], LOX is positively correlated with MMPs or increases MMPs activity, while in liver cancer, lysyl oxidase propeptide (LOX-PP) inhibits liver cancer cell migration by down-regulating MMPs expression [353]. The relationship between MMP and LOX remains unclear.
In PC, HIF-1α can promote the transcription of membrane type 2 matrix metalloproteinase (MT2-MMP) [354, 355], and further, MT2-MMP participates in the progression of PC by activating MMP-2[356]. MMP-2 in PC cells and PSC also depends on HIF-1α regulation [340]. In addition, HIF-1α binds to the HRE on the fascin promoter and activates its mRNA transcription, and overexpression of fascin can increase MMP-2 expression and promote PC cell migration and invasion [357]. Moreover, ROS/MMP-3 signaling pathway is activated by high glucose and up-regulates ribonucleotide reductase catalytic subunit M1 (RRM1) expression, a member of ribonucleoside reductase, inducing gemcitabine resistance in PC [358]. MT1-MMP up-regulates the expression of high mobility group A2 (HMGA2), a non-histone DNA-binding nuclear protein involved in chromatin remodeling and gene transcription, to attenuate the therapeutic effect of gemcitabine in PC [359]. In addition, the use of MMP-2 inhibitor tissue inhibitor metalloproteinase 2 (TIMP2) and MMP-9 inhibitor TIMP1 increased the inhibitory effect of hyperthermia combined with gemcitabine on the invasion of gemcitabine-resistant PC cells [360].
Vascular endothelial-derived growth factor (VEGF) is another key factor in pancreatic tumor angiogenesis. VEGF plays an important role in the whole process of angiogenesis, and its expression is mainly regulated by HIF [361]. In PC, hypoxia can induce the translocation of pyruvate kinase M2 (PKM2) and nuclear factor kappa B subunit 3 (p65, NFKB3) to the nucleus of PC cells, and PKM2 can activate NF-kB to mediate the transcription of HIF-1α and its target gene VEGF [197]. In addition, PKM2 is a co-activator of HIF-1α and its downstream target gene [362]. VEGF can stimulate PC cells to up-regulate HIF-1α and enhance glycolysis [363]. In the PC transplantation model, gemcitabine can reduce the protein levels of VEGF, VEGFR2, platelet and endothelial cell adhesion molecule 1 (CD31, PECAM1) and HIF-1α to reduce blood vessel formation, thereby reducing tumor growth [364]. While using other drugs, the small molecule compound LB-100 enhanced the cytotoxicity of doxorubicin by increasing angiogenesis through the HIF-1α/VEGF axis [365]. However, inositol tripyrophosphate (ITPP) can down-regulate HIF-1α, VEGF and LOX and improve the sensitivity of gemcitabine treatment, which may be caused by the improvement of vascular structure and the reduction of connective tissue proliferation [366]. Metformin can inhibit the mTOR/HIF-1α/VEGF signaling cascade to inhibit the progression of gemcitabine-resistant PC [367]. At the same time, metformin can also down-regulate SHH to show the effect of of inhibiting PSCs. Among the downstream effects, the inhibition of VEGF leads to the reduction of neovascularization, the weakening of fibroproliferative reaction, and the enhancement of the anti-tumor effect of gemcitabine [368].
In general, the effect of TME on tumor has two sides, and therapeutic drugs also start from both sides. By reducing angiogenesis and promoting connective tissue proliferation, the cancer cells are kept in a state of hypoxia and malnutrition for a long time, and at the same time, the metastasis of cancer cells is blocked at the physical level to achieve the effect of tumor inhibition. On the contrary, improving the vascular structure and inhibiting the proliferation of connective tissue can make the drug directly reach the inside of the tumor, which is beneficial to the anti-tumor effect of the drug.
Conclusion
LncRNA can regulate autophagy and Wnt/β-catenin pathway from multiple perspectives. Through the common MRE elements, the information exchange between lncRNA-miRNA-mRNA can be realized. In addition to sponging miRNA, lncRNA can also directly bind to proteins to regulate cell physiological activities. Extensive studies have determined that lncRNAs can regulate intermediate mediators, such as HuR, FOX and HIF, in the process of PC, while regulating autophagy and Wnt/β-catenin pathway, and further regulate chemotherapy resistance of PC cells. Although studies on PC resistance have mainly focused on gemcitabine, these several intermediate mediators also have an impact on the efficacy of other drugs. In PC, not only the differentially expressed lncRNAs may serve as PC markers and targets for therapy, intermediate mediators hold the same potential. Furthermore, lncRNAs show strong specific expression in different tissues and cancers. Whether the specific expression of lncRNA is consistent with the drug resistance pathway of PC remains to be further investigated.
Abbreviations
5-FU: 5-Fluorouracil; ABC: ATP binding cassette; AGER: advanced glycosylation end-product specific receptor; AGR2: anterior gradient 2, protein disulphide isomerase family member; AMPK: AMP-activated protein kinase; ANRIL: cyclin dependent kinase inhibitor 2B antisense RNA 1; APC: adenomatous polyposis coli; ATF3: activating transcription factor 3; ATG: autophagy related gene; AURKB: aurora kinase B; AXIN: axis inhibitor; BANCR: BRAF-activated non-protein coding RNA; BCL2: B-cell lymphoma 2; BIM: BCL2-interacting mediator of cell death, BCL2L11; BNIP3: BCL2 interacting protein 3; BRI3: brain protein I3; BX111: ZEB1 transcriptional regulator RNA, ZEBTR; CAF: cancer-associated fibroblast; CAR: cardamonin; CCA: cholangiocarcinoma; CCNB1: cyclin B1; CCND1: cyclin D1; CD133+: prominin 1, PROM1 positive; CD31: platelet and endothelial cell adhesion molecule 1, PECAM1; CD44: cluster of differentiation-44; CDK: cyclin dependent kinase; ceRNA: competitive endogenous RNA; CERS6-AS1: ceramide synthase 6 antisense RNA 1; cGMP: cyclic guanosine monophosphate; CK1: casein kinase 1; CNBP: cellular nucleic acid binding protein; CSC: cancer stem cell; CTBP1: C-terminal binding protein 1; CTPS1: CTP synthase 1; CXCL12: C-X-C motif chemokine ligand 12; CXCR4: C-X-C motif chemokine receptor 4; DDIT4-AS1: DNA damage inducible transcript 4 antisense RNA 1; DKK1: dickkopf Wnt signaling pathway inhibitor 1; DLX6-AS1: distal-less homeobox 6 antisense RNA 1; DVL: dishevelled segment polarity protein; ECM: extracellular matrix; EGLN2: egl-9 family hypoxia-inducible factor 2; EIF3B: eukaryotic translation initiation factor 3 subunit B; EMT: epithelial-mesenchymal transition; EVA1A: eva-1 homolog A; EZH2: enhancer of zeste 2 polycomb repressive complex 2 subunit; FAM83H: family with sequence similarity 83 member H; FAT10: human leukocyte antigen F-associated transcript 10; FGD5-AS1: FYVE, RhoGEF, and PH domain containing 5 antisense RNA 1; FOLFIRINOX: combination chemotherapy with fluorouracil, leucovorin, irinotecan, and oxaliplatin; FOX: forkhead box; FZD: Frizzled; GABARAPL1: GABA type A receptor associated protein like 1; GEP-NEN: gastroenteropancreatic neuroendocrine neoplasm; GSK3: glycogen synthase kinase-3; H19: H19 imprinted maternally expressed transcript; H2AX: H2A. X variant histone; H3K27me: methylation of Histone H3 at lysine 27; H3K9ac: acetylation of Histone H3 at lysine 9; HCP5: histocompatibility leukocyte antigen complex P5; HDAC1: histone deacetylase 1; HDGF: heparin binding growth factor; hENT1: human equilibrative nucleoside transporter 1; HIF: hypoxia inducible factor; HMGA2: high mobility group A2; HMGB1: high mobility group box 1; HOTAIR: homeobox transcript antisense intergenic RNA; HOXA9: homeobox A9; HRE: hypoxia response element; HuR: human antigen R; IDH1: isocitrate dehydrogenase (NADP(+)) 1; IPO8: importin 8; ITGB1: integrin subunit beta 1; ITPP: inositol tripyrophosphate; JNK1/2: C-Jun NH2 terminal kinase 1/2; K-RAS: Kirsten rat sarcoma viral oncogene homolog; LC3: microtubule associated protein 1 light chain 3; LEF: lymphoid enhancer-binding factor; LINC: long intergenic non-protein coding RNA; lncRNA: long non coding RNA; LOX: lysyl oxidase; LRP: low density lipoprotein receptor related protein; LZTS1-AS1: leucine zipper tumor suppressor 1 antisense RNA 1; m6A: N6-methyladenosine; MALAT1: metastasis associated lung adenocarcinoma transcript 1; MDR: multidrug resistance; METTL: methyltransferase; MITF: melanocyte inducing transcription factor; MKRN1: makorin ring finger protein 1; MMP: matrix metalloproteinase; MnSOD: manganese superoxide dismutase, SOD2; MRE: miRNA recognition element; MTA2: metastasis associated 1 family member 2; MTA2TR: MTA2 transcriptional regulator RNA; MT2-MMP: Membrane type 2 matrix metalloproteinase; mTOR: mammalian target of rapamycin; MUC1: mucin 1; NF-kB: nuclear transcription factor-kappa B; NNT-AS1: nicotinamide nucleotide transhydrogenase antisense RNA 1; NOX4: NADPH oxidase 4; NR2F1: nuclear receptor subfamily 2, group F, member 1; NSCLC: non-small cell lung cancer; OTUB1: ovarian tumor family deubiquitinase ubiquitin aldehyde binding 1; P53: tumor protein P53, TP53; P62: sequestosome 1, SQSTM1; p65: nuclear factor kappa B subunit 3, NFKB3; PaCSC: pancreatic cancer stem cell; PanIN: pancreatic intraepithelial neoplasia; PARP: poly ADP-ribose polymerase; PC: pancreatic cancer; PDAC: pancreatic ductal adenocarcinoma; PDHA1: pyruvate dehydrogenase E1 subunit alpha 1; PDL1: programmed cell death 1 ligand 1; PGAM1: phosphoglycerate mutase 1; PGC-1β: peroxisome proliferator-activated receptor gamma, coactivator 1 beta; PHB1: prohibitin 1; PIK3C3: phosphatidylinositol 3-kinase catalytic subunit type 3; PKM2: pyruvate kinase M2; PLK1: polo-like kinase 1; pNET: pancreatic neuroendocrine tumor; PNI: perineural invasion; PRC2: polycomb repressive complex 2; PSC: pancreatic stellate cell; PTK7: protein tyrosine kinase 7; PVT1: plasmacytoma variant translocation 1; PYGO2: pygopus family PHD finger 2; Rab7: RAS-related GTP-binding protein 7; RAF1: Raf-1 proto-oncogene; RBP: RNA binding proteins; ROS: reactive oxygen species; RR: ribonucleoside reductase; RRM1: ribonucleotide reductase catalytic subunit M1; SHH: sonic hedgehog ligand; SIRT1: sirtuin 1; SMAD4: drosophila mothers against decapentaplegic protein 4; SNHG16: small nucleolar RNA host gene 16; SP1: specific protein 1; TCA cycle: tricarboxylic acid cycle; TCF: transcription factor; TCF4: T-cell-specific transcription factor 4; TEX10: testis expressed 10; TFE3: transcription factor binding to IGHM enhancer 3; TFEB: transcription factor EB; TIA1: TIA1 cytotoxic granule associated RNA binding protein; TIMP1: tissue inhibitor metalloproteinase 1; TIP60: lysine acetyltransferase 5, KAT5; TKT: transketolase; TLR4: Toll-like receptor 4; TME: tumor microenvironment; TNNC1: troponin C1; TOP2A: DNA topoisomerase II alpha; TPT1: tumor protein, translationally-controlled 1; TRIM22: tripartite motif containing 22; TSC: tuberous sclerosis complex; TSLNC8: tumor suppressive lncRNA on chromosome 8p12; TWIST1: twist family bHLH transcription factor 1; UCA1: urothelial cancer associated 1; UCHL3: ubiquitin C-terminal hydrolase L3; ULK1: Unc-51 like autophagy activating kinase 1; UPF1: upstream frameshift 1; UVRAG: UV radiation resistance associated; VEGF: vascular endothelial-derived growth factor; VMP1: vacuole membrane protein 1; WDR5: WD repeat domain 5; WEE1: WEE1 G2 checkpoint kinase; WIF1: Wnt inhibitory factor 1; WIPI1: WD repeat domain, phosphoinositide interacting 1; WTAP: WT1 associated protein; WTAPP1: WT1 associated protein pseudogene 1; YB1: Y box binding protein 1; ZEB1: Zinc finger E-box binding homeobox 1; ZEB2-AS1: zinc finger E-box binding homeobox 2 antisense RNA 1
Acknowledgements
We thank the Hubei University of Technology for the research equipment and technical support for this research. We thank Figdraw (www.figdraw.com) for its graphical assistance during the preparation of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (82273970 and 32070726 to JT, 32270768 to CZ, 82370715 to XC), International Science and Technology Cooperation Project of Hubei Province (2022EHB038 to CZ), Wuhan Science and Technology Project (2022020801020272 to CZ), Innovation Group Project of Hubei Province (2023AFA026) and the National Key R&D Program of China(2023YFC2507900).
Author contributions
JT, XC and CZ provided financial support for the publication of this review. YH drafted the manuscript, prepared the figure and edited the manuscript. JT, CZ, RZ, HL, SX, DG and XC helped revise the review and modify the contents of the initial draft. JT, XC and CZ conceptualized and reviewed the manuscript. All authors approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Klimstra DS. Nonductal neoplasms of the pancreas. Mod Pathol. 2007;20(Suppl 1):S94-112
2. Adsay NV, Basturk O, Cheng JD, Andea AA. Ductal neoplasia of the pancreas: nosologic, clinicopathologic, and biologic aspects. Semin Radiat Oncol. 2005;15:254-64
3. Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin AV. et al. Pancreatic cancer. Nat Rev Dis Primers. 2016;2:16022
4. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17-48
5. Torphy RJ, Fujiwara Y, Schulick RD. Pancreatic cancer treatment: better, but a long way to go. Surg Today. 2020;50:1117-25
6. Burris HA 3rd, Moore MJ, Andersen J, Green MR, Rothenberg ML, Modiano MR. et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403-13
7. Neoptolemos JP, Palmer DH, Ghaneh P, Psarelli EE, Valle JW, Halloran CM. et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial. Lancet. 2017;389:1011-24
8. Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M. et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691-703
9. Conroy T, Hammel P, Hebbar M, Ben Abdelghani M, Wei AC, Raoul JL. et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N Engl J Med. 2018;379:2395-406
10. Wang-Gillam A, Li CP, Bodoky G, Dean A, Shan YS, Jameson G. et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): a global, randomised, open-label, phase 3 trial. Lancet. 2016;387:545-57
11. Hsu SK, Jadhao M, Liao WT, Chang WT, Hung CT, Chiu CC. Culprits of PDAC resistance to gemcitabine and immune checkpoint inhibitor: Tumour microenvironment components. Front Mol Biosci. 2022;9:1020888
12. Zeng S, Pöttler M, Lan B, Grützmann R, Pilarsky C, Yang H. Chemoresistance in Pancreatic Cancer. Int J Mol Sci. 2019 20
13. Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208
14. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96-118
15. Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021 220
16. Fang Y, Fullwood MJ. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genomics Proteomics Bioinformatics. 2016;14:42-54
17. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661-7
18. Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017;77:3965-81
19. Lyu H, Zhang J, Wei Q, Huang Y, Zhang R, Xiao S. et al. Identification of Wnt/β-Catenin- and Autophagy-Related lncRNA Signature for Predicting Immune Efficacy in Pancreatic Adenocarcinoma. Biology (Basel). 2023 12
20. Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X. et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/beta-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 2020;19:118
21. Steinhart Z, Angers S. Wnt signaling in development and tissue homeostasis. Development. 2018 145
22. Albrecht LV, Tejeda-Muñoz N, De Robertis EM. Cell Biology of Canonical Wnt Signaling. Annu Rev Cell Dev Biol. 2021;37:369-89
23. Park HB, Kim JW, Baek KH. Regulation of Wnt Signaling through Ubiquitination and Deubiquitination in Cancers. Int J Mol Sci. 2020 21
24. Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X. et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7:3
25. Ram Makena M, Gatla H, Verlekar D, Sukhavasi S, M KP, K CP. Wnt/β-Catenin Signaling: The Culprit in Pancreatic Carcinogenesis and Therapeutic Resistance. Int J Mol Sci. 2019 20
26. Aguilera KY, Dawson DW. WNT Ligand Dependencies in Pancreatic Cancer. Front Cell Dev Biol. 2021;9:671022
27. Du W, Menjivar RE, Donahue KL, Kadiyala P, Velez-Delgado A, Brown KL. et al. WNT signaling in the tumor microenvironment promotes immunosuppression in murine pancreatic cancer. J Exp Med. 2023 220
28. Barman S, Fatima I, Singh AB, Dhawan P. Pancreatic Cancer and Therapy: Role and Regulation of Cancer Stem Cells. Int J Mol Sci. 2021 22
29. Nakamoto M, Hisaoka M. Clinicopathological Implications of Wingless/int1 (WNT) Signaling Pathway in Pancreatic Ductal Adenocarcinoma. J uoeh. 2016;38:1-8
30. Zhang WT, Zhang JJ, Shao Q, Wang YK, Jia JP, Qian B. et al. FGD5-AS1 is an oncogenic lncRNA in pancreatic cancer and regulates the Wnt/β-catenin signaling pathway via miR-577. Oncol Rep. 2022 47
31. Yang J, Ye Z, Mei D, Gu H, Zhang J. Long noncoding RNA DLX6-AS1 promotes tumorigenesis by modulating miR-497-5p/FZD4/FZD6/Wnt/β-catenin pathway in pancreatic cancer. Cancer Manag Res. 2019;11:4209-21
32. Wang X, Jia Y, Fei C, Song X, Li L. Activation/Proliferation-associated Protein 2 (Caprin-2) Positively Regulates CDK14/Cyclin Y-mediated Lipoprotein Receptor-related Protein 5 and 6 (LRP5/6) Constitutive Phosphorylation. J Biol Chem. 2016;291:26427-34
33. Niehrs C, Shen J. Regulation of Lrp6 phosphorylation. Cell Mol Life Sci. 2010;67:2551-62
34. Sun Y, Zhu Q, Yang W, Shan Y, Yu Z, Zhang Q. et al. LncRNA H19/miR-194/PFTK1 axis modulates the cell proliferation and migration of pancreatic cancer. J Cell Biochem. 2019;120:3874-86
35. Chen LJ, Wu L, Wang W, Zhai LL, Xiang F, Li WB. et al. Long non-coding RNA 01614 hyperactivates WNT/β-catenin signaling to promote pancreatic cancer progression by suppressing GSK-3β. Int J Oncol. 2022 61
36. Ling J, Wang F, Liu C, Dong X, Xue Y, Jia X. et al. FOXO1-regulated lncRNA LINC01197 inhibits pancreatic adenocarcinoma cell proliferation by restraining Wnt/β-catenin signaling. J Exp Clin Cancer Res. 2019;38:179
37. Mirzaei S, Gholami MH, Hushmandi K, Hashemi F, Zabolian A, Canadas I. et al. The long and short non-coding RNAs modulating EZH2 signaling in cancer. J Hematol Oncol. 2022;15:18
38. Gaballa JM, Braga Neto MB, Ramos GP, Bamidele AO, Gonzalez MM, Sagstetter MR. et al. The Role of Histone Methyltransferases and Long Non-coding RNAs in the Regulation of T Cell Fate Decisions. Front Immunol. 2018;9:2955
39. Weng YC, Ma J, Zhang J, Wang JC. Long non-coding RNA LINC01133 silencing exerts antioncogenic effect in pancreatic cancer through the methylation of DKK1 promoter and the activation of Wnt signaling pathway. Cancer Biol Ther. 2019;20:368-80
40. Liu Y, Tang T, Yang X, Qin P, Wang P, Zhang H. et al. Tumor-derived exosomal long noncoding RNA LINC01133, regulated by Periostin, contributes to pancreatic ductal adenocarcinoma epithelial-mesenchymal transition through the Wnt/β-catenin pathway by silencing AXIN2. Oncogene. 2021;40:3164-79
41. Jiang Y, Li Z, Zheng S, Chen H, Zhao X, Gao W. et al. The long non-coding RNA HOTAIR affects the radiosensitivity of pancreatic ductal adenocarcinoma by regulating the expression of Wnt inhibitory factor 1. Tumour Biol. 2016;37:3957-67
42. Malinauskas T, Jones EY. Extracellular modulators of Wnt signalling. Curr Opin Struct Biol. 2014;29:77-84
43. Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ. et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675-82
44. Yang Y, Xing D, Wang Y, Jia H, Li B, Li JJ. A long non-coding RNA, HOTAIR, promotes cartilage degradation in osteoarthritis by inhibiting WIF-1 expression and activating Wnt pathway. BMC Mol Cell Biol. 2020;21:53
45. Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B. Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis. Int J Mol Sci. 2013;14:10015-41
46. Chai W, Liu R, Li F, Zhang Z, Lei B. Long noncoding RNA TSLNC8 enhances pancreatic cancer aggressiveness by regulating CTNNB1 expression via association with HuR. Hum Cell. 2021;34:165-76
47. Armas P, Coux G, Weiner AMJ, Calcaterra NB. What's new about CNBP?. Divergent functions and activities for a conserved nucleic acid binding protein. Biochim Biophys Acta Gen Subj. 2021;1865:129996
48. Margarit E, Armas P, García Siburu N, Calcaterra NB. CNBP modulates the transcription of Wnt signaling pathway components. Biochim Biophys Acta. 2014;1839:1151-60
49. Deng J, Zhang J, Ye Y, Liu K, Zeng L, Huang J. et al. N(6) -methyladenosine-Mediated Upregulation of WTAPP1 Promotes WTAP Translation and Wnt Signaling to Facilitate Pancreatic Cancer Progression. Cancer Res. 2021;81:5268-83
50. Huang H, Weng H, Chen J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell. 2020;37:270-88
51. Zhou M, Pan S, Qin T, Zhao C, Yin T, Gao Y. et al. LncRNA FAM83H-AS1 promotes the malignant progression of pancreatic ductal adenocarcinoma by stabilizing FAM83H mRNA to protect β-catenin from degradation. J Exp Clin Cancer Res. 2022;41:288
52. Dou Q, Xu Y, Zhu Y, Hu Y, Yan Y, Yan H. LncRNA FAM83H-AS1 contributes to the radioresistance, proliferation, and metastasis in ovarian cancer through stabilizing HuR protein. Eur J Pharmacol. 2019;852:134-41
53. Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F. et al. Autophagy in malignant transformation and cancer progression. Embo j. 2015;34:856-80
54. Rybstein MD, Bravo-San Pedro JM, Kroemer G, Galluzzi L. The autophagic network and cancer. Nat Cell Biol. 2018;20:243-51
55. Sun L, Wang X, Chen L, Gao Z, Xu S, Hu C. et al. CPT1A mediates chemoresistance in human hypopharyngeal squamous cell carcinoma via ATG16L1-dependent cellular autophagy. Cell Insight. 2023;2:100127
56. Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H. et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717-29
57. Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M. et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361-5
58. Perera RM, Di Malta C, Ballabio A. MiT/TFE Family of Transcription Factors, Lysosomes, and Cancer. Annu Rev Cancer Biol. 2019;3:203-22
59. Yang M, Liu E, Tang L, Lei Y, Sun X, Hu J. et al. Emerging roles and regulation of MiT/TFE transcriptional factors. Cell Commun Signal. 2018;16:31
60. Bronisz A, Sharma SM, Hu R, Godlewski J, Tzivion G, Mansky KC. et al. Microphthalmia-associated transcription factor interactions with 14-3-3 modulate differentiation of committed myeloid precursors. Mol Biol Cell. 2006;17:3897-906
61. Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903-14
62. Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N. et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014;7:ra9
63. Wang P, Kou D, Le W. Roles of VMP1 in Autophagy and ER-Membrane Contact: Potential Implications in Neurodegenerative Disorders. Front Mol Neurosci. 2020;13:42
64. Liu YF, Luo D, Li X, Li ZQ, Yu X, Zhu HW. PVT1 Knockdown Inhibits Autophagy and Improves Gemcitabine Sensitivity by Regulating the MiR-143/HIF-1α/VMP1 Axis in Pancreatic Cancer. Pancreas. 2021;50:227-34
65. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM. et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174-86
66. Biffi G, Tuveson DA. Diversity and Biology of Cancer-Associated Fibroblasts. Physiol Rev. 2021;101:147-76
67. Geng X, Li L, Luo Y, Yang W, Hu J, Zhao Z. et al. Tumor Cell Derived Lnc-FSD2-31:1 Contributes to Cancer-Associated Fibroblasts Activation in Pancreatic Ductal Adenocarcinoma Progression through Extracellular Vesicles Cargo MiR-4736. Adv Sci (Weinh). 2023: e2203324.
68. Wang L, Bi R, Li L, Zhou K, Yin H. lncRNA ANRIL aggravates the chemoresistance of pancreatic cancer cells to gemcitabine by targeting inhibition of miR-181a and targeting HMGB1-induced autophagy. Aging (Albany NY). 2021;13:19272-81
69. Liu S, Zhang W, Liu K, Liu Y. LncRNA SNHG16 promotes tumor growth of pancreatic cancer by targeting miR-218-5p. Biomed Pharmacother. 2019;114:108862
70. Gao H, Gong N, Ma Z, Miao X, Chen J, Cao Y. et al. LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth and invasion through regulating the miR-204/HMGB1 axis. Int J Biol Macromol. 2018;116:545-51
71. Kang R, Tang D, Schapiro NE, Loux T, Livesey KM, Billiar TR. et al. The HMGB1/RAGE inflammatory pathway promotes pancreatic tumor growth by regulating mitochondrial bioenergetics. Oncogene. 2014;33:567-77
72. He C, Sun S, Zhang Y, Xie F, Li S. The role of irreversible electroporation in promoting M1 macrophage polarization via regulating the HMGB1-RAGE-MAPK axis in pancreatic cancer. Oncoimmunology. 2021;10:1897295
73. Liu Y, Pu X, Qin X, Gong J, Huang Z, Luo Y. et al. Neutrophil Extracellular Traps Regulate HMGB1 Translocation and Kupffer Cell M1 Polarization During Acute Liver Transplantation Rejection. Front Immunol. 2022;13:823511
74. Lange SS, Vasquez KM. HMGB1: the jack-of-all-trades protein is a master DNA repair mechanic. Mol Carcinog. 2009;48:571-80
75. Kang R, Xie Y, Zhang Q, Hou W, Jiang Q, Zhu S. et al. Intracellular HMGB1 as a novel tumor suppressor of pancreatic cancer. Cell Res. 2017;27:916-32
76. Tang D, Billiar TR, Lotze MT. A Janus tale of two active high mobility group box 1 (HMGB1) redox states. Mol Med. 2012;18:1360-2
77. Frisardi V, Matrone C, Street ME. Metabolic Syndrome and Autophagy: Focus on HMGB1 Protein. Front Cell Dev Biol. 2021;9:654913
78. Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P. et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881-92
79. Ghafouri-Fard S, Abak A, Bahroudi Z, Shoorei H, Abbas Raza SH, Taheri M. The interplay between non-coding RNAs and Twist1 signaling contribute to human disorders. Biomed Pharmacother. 2021;135:111220
80. Zhao Z, Rahman MA, Chen ZG, Shin DM. Multiple biological functions of Twist1 in various cancers. Oncotarget. 2017;8:20380-93
81. Shi YX, Sun ZW, Jia DL, Wang HB. Autophagy deficiency promotes lung metastasis of prostate cancer via stabilization of TWIST1. Clin Transl Oncol. 2022;24:1403-12
82. Lai R, Ji L, Zhang X, Xu Y, Zhong Y, Chen L. et al. Stanniocalcin2 inhibits the epithelial-mesenchymal transition and invasion of trophoblasts via activation of autophagy under high-glucose conditions. Mol Cell Endocrinol. 2022;547:111598
83. Devanand P, Sundaramoorthy S, Ryu MS, Jayabalan AK, Ohn T, Lim IK. Translational downregulation of Twist1 expression by antiproliferative gene, B-cell translocation gene 2, in the triple negative breast cancer cells. Cell Death Dis. 2019;10:410
84. Jin HO, Hong SE, Woo SH, Lee JH, Choe TB, Kim EK. et al. Silencing of Twist1 sensitizes NSCLC cells to cisplatin via AMPK-activated mTOR inhibition. Cell Death Dis. 2012;3:e319
85. Canham L, Sendac S, Diagbouga MR, Wolodimeroff E, Pirri D, Tardajos Ayllon B. et al. EVA1A (Eva-1 Homolog A) Promotes Endothelial Apoptosis and Inflammatory Activation Under Disturbed Flow Via Regulation of Autophagy. Arterioscler Thromb Vasc Biol. 2023;43:547-61
86. Wu H, Li A, Zheng Q, Gu J, Zhou W. LncRNA LZTS1-AS1 induces proliferation, metastasis and inhibits autophagy of pancreatic cancer cells through the miR-532 /TWIST1 signaling pathway. Cancer Cell Int. 2023;23:130
87. Liu H, Ho PW, Leung CT, Pang SY, Chang EES, Choi ZY. et al. Aberrant mitochondrial morphology and function associated with impaired mitophagy and DNM1L-MAPK/ERK signaling are found in aged mutant Parkinsonian LRRK2(R1441G) mice. Autophagy. 2021;17:3196-220
88. Zampieri LX, Silva-Almeida C, Rondeau JD, Sonveaux P. Mitochondrial Transfer in Cancer: A Comprehensive Review. Int J Mol Sci. 2021 22
89. Wang H, Ding Y, He Y, Yu Z, Zhou Y, Gong A. et al. LncRNA UCA1 promotes pancreatic cancer cell migration by regulating mitochondrial dynamics via the MAPK pathway. Arch Biochem Biophys. 2023;748:109783
90. Qin H, Ni H, Liu Y, Yuan Y, Xi T, Li X. et al. RNA-binding proteins in tumor progression. J Hematol Oncol. 2020;13:90
91. Sidali A, Teotia V, Solaiman NS, Bashir N, Kanagaraj R, Murphy JJ. et al. AU-Rich Element RNA Binding Proteins: At the Crossroads of Post-Transcriptional Regulation and Genome Integrity. Int J Mol Sci. 2021 23
92. Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH. et al. Long Noncoding RNA MALAT1 Promotes Aggressive Pancreatic Cancer Proliferation and Metastasis via the Stimulation of Autophagy. Mol Cancer Ther. 2016;15:2232-43
93. Pullmann R Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X. et al. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol Cell Biol. 2007;27:6265-78
94. Shorning BY, Dass MS, Smalley MJ, Pearson HB. The PI3K-AKT-mTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int J Mol Sci. 2020 21
95. Zhou C, Liang Y, Zhou L, Yan Y, Liu N, Zhang R. et al. TSPAN1 promotes autophagy flux and mediates cooperation between WNT-CTNNB1 signaling and autophagy via the MIR454-FAM83A-TSPAN1 axis in pancreatic cancer. Autophagy. 2021;17:3175-95
96. Hu T, Li C. Convergence between Wnt-β-catenin and EGFR signaling in cancer. Mol Cancer. 2010;9:236
97. Zhou C, Dong X, Wang M, Qian X, Hu M, Liang K. et al. Phosphorylated STYK1 restrains the inhibitory role of EGFR in autophagy initiation and EGFR-TKIs sensitivity. Cell Insight. 2022;1:100045
98. Petherick KJ, Williams AC, Lane JD, Ordóñez-Morán P, Huelsken J, Collard TJ. et al. Autolysosomal β-catenin degradation regulates Wnt-autophagy-p62 crosstalk. Embo j. 2013;32:1903-16
99. Nàger M, Sallán MC, Visa A, Pushparaj C, Santacana M, Macià A. et al. Inhibition of WNT-CTNNB1 signaling upregulates SQSTM1 and sensitizes glioblastoma cells to autophagy blockers. Autophagy. 2018;14:619-36
100. Chu CW, Ko HJ, Chou CH, Cheng TS, Cheng HW, Liang YH. et al. Thioridazine Enhances P62-Mediated Autophagy and Apoptosis Through Wnt/β-Catenin Signaling Pathway in Glioma Cells. Int J Mol Sci. 2019 20
101. Jia Z, Wang J, Wang W, Tian Y, XiangWei W, Chen P. et al. Autophagy eliminates cytoplasmic β-catenin and NICD to promote the cardiac differentiation of P19CL6 cells. Cell Signal. 2014;26:2299-305
102. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N. et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927-39
103. Roy SK, Ma Y, Lam BQ, Shrivastava A, Srivastav S, Shankar S. et al. Riluzole regulates pancreatic cancer cell metabolism by suppressing the Wnt-β-catenin pathway. Sci Rep. 2022;12:11062
104. Nong J, Kang K, Shi Q, Zhu X, Tao Q, Chen YG. Phase separation of Axin organizes the β-catenin destruction complex. J Cell Biol. 2021 220
105. Zhang X, Wu N, Huang H, Li S, Liu S, Zhang R. et al. Phosphorylated PTTG1 switches its subcellular distribution and promotes β-catenin stabilization and subsequent transcription activity. Oncogene. 2023;42:2439-55
106. Ryu HY, Kim LE, Jeong H, Yeo BK, Lee JW, Nam H. et al. GSK3B induces autophagy by phosphorylating ULK1. Exp Mol Med. 2021;53:369-83
107. Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y. et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477-81
108. Nie T, Yang S, Ma H, Zhang L, Lu F, Tao K. et al. Regulation of ER stress-induced autophagy by GSK3β-TIP60-ULK1 pathway. Cell Death Dis. 2016;7:e2563
109. Azoulay-Alfaguter I, Elya R, Avrahami L, Katz A, Eldar-Finkelman H. Combined regulation of mTORC1 and lysosomal acidification by GSK-3 suppresses autophagy and contributes to cancer cell growth. Oncogene. 2015;34:4613-23
110. Avrahami L, Paz R, Dominko K, Hecimovic S, Bucci C, Eldar-Finkelman H. GSK-3-TSC axis governs lysosomal acidification through autophagy and endocytic pathways. Cell Signal. 2020;71:109597
111. Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X. et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955-68
112. Yuan W, Fang W, Zhang R, Lyu H, Xiao S, Guo D. et al. Therapeutic strategies targeting AMPK-dependent autophagy in cancer cells. Biochim Biophys Acta Mol Cell Res. 2023;1870:119537
113. He L, Gomes AP, Wang X, Yoon SO, Lee G, Nagiec MJ. et al. mTORC1 Promotes Metabolic Reprogramming by the Suppression of GSK3-Dependent Foxk1 Phosphorylation. Mol Cell. 2018;70:949-60.e4
114. Sakaguchi M, Cai W, Wang CH, Cederquist CT, Damasio M, Homan EP. et al. FoxK1 and FoxK2 in insulin regulation of cellular and mitochondrial metabolism. Nat Commun. 2019;10:1582
115. Bowman CJ, Ayer DE, Dynlacht BD. Foxk proteins repress the initiation of starvation-induced atrophy and autophagy programs. Nat Cell Biol. 2014;16:1202-14
116. Chen Y, Wu J, Liang G, Geng G, Zhao F, Yin P. et al. CHK2-FOXK axis promotes transcriptional control of autophagy programs. Sci Adv. 2020;6:eaax5819
117. Gao C, Chen YG. Dishevelled: The hub of Wnt signaling. Cell Signal. 2010;22:717-27
118. Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68-75
119. Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T. et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat Cell Biol. 2010;12:781-90
120. Jati S, Kundu S, Chakraborty A, Mahata SK, Nizet V, Sen M. Wnt5A Signaling Promotes Defense Against Bacterial Pathogens by Activating a Host Autophagy Circuit. Front Immunol. 2018;9:679
121. Hwang SH, Bang S, Kang KS, Kang D, Chung J. ULK1 negatively regulates Wnt signaling by phosphorylating Dishevelled. Biochem Biophys Res Commun. 2019;508:308-13
122. Gan XQ, Wang JY, Xi Y, Wu ZL, Li YP, Li L. Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J Cell Biol. 2008;180:1087-100
123. Wang Y, Qin C, Yang G, Zhao B, Wang W. The role of autophagy in pancreatic cancer progression. Biochim Biophys Acta Rev Cancer. 2021;1876:188592
124. Gong J, Belinsky G, Sagheer U, Zhang X, Grippo PJ, Chung C. Pigment Epithelium-derived Factor (PEDF) Blocks Wnt3a Protein-induced Autophagy in Pancreatic Intraepithelial Neoplasms. J Biol Chem. 2016;291:22074-85
125. Zhou C, Zhu X, Liu N, Dong X, Zhang X, Huang H. et al. B-lymphoid tyrosine kinase-mediated FAM83A phosphorylation elevates pancreatic tumorigenesis through interacting with β-catenin. Signal Transduct Target Ther. 2023;8:66
126. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-8
127. Zu F, Liu P, Wang H, Zhu T, Sun J, Sheng W. et al. Integrated analysis identifies a pathway-related competing endogenous RNA network in the progression of pancreatic cancer. BMC Cancer. 2020;20:958
128. Sun J, Zhang P, Yin T, Zhang F, Wang W. Upregulation of LncRNA PVT1 Facilitates Pancreatic Ductal Adenocarcinoma Cell Progression and Glycolysis by Regulating MiR-519d-3p and HIF-1A. J Cancer. 2020;11:2572-9
129. Wu X, Xia T, Cao M, Zhang P, Shi G, Chen L. et al. LncRNA BANCR Promotes Pancreatic Cancer Tumorigenesis via Modulating MiR-195-5p/Wnt/β-Catenin Signaling Pathway. Technol Cancer Res Treat. 2019;18:1533033819887962
130. Zhou WY, Zhang MM, Liu C, Kang Y, Wang JO, Yang XH. Long noncoding RNA LINC00473 drives the progression of pancreatic cancer via upregulating programmed death-ligand 1 by sponging microRNA-195-5p. J Cell Physiol. 2019;234:23176-89
131. Almannai M, Marafi D, El-Hattab AW. WIPI proteins: Biological functions and related syndromes. Front Mol Neurosci. 2022;15:1011918
132. Gao KF, Zhao YF, Liao WJ, Xu GL, Zhang JD. CERS6-AS1 promotes cell proliferation and represses cell apoptosis in pancreatic cancer via miR-195-5p/WIPI2 axis. Kaohsiung J Med Sci. 2022;38:542-53
133. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344-52
134. Zhu J, Fu H, Wu Y, Zheng X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci. 2013;56:876-85
135. Ferrè F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17:106-16
136. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18:5-18
137. Pereira B, Billaud M, Almeida R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer. 2017;3:506-28
138. Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017 8
139. Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR). Wiley Interdiscip Rev RNA. 2020;11:e1581
140. Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. Embo j. 2000;19:2340-50
141. Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A. et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009;69:4567-72
142. Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L. et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene. 2016;35:2529-41
143. Chae MJ, Sung HY, Kim EH, Lee M, Kwak H, Chae CH. et al. Chemical inhibitors destabilize HuR binding to the AU-rich element of TNF-alpha mRNA. Exp Mol Med. 2009;41:824-31
144. Zarei M, Lal S, Parker SJ, Nevler A, Vaziri-Gohar A, Dukleska K. et al. Posttranscriptional Upregulation of IDH1 by HuR Establishes a Powerful Survival Phenotype in Pancreatic Cancer Cells. Cancer Res. 2017;77:4460-71
145. Lal S, Burkhart RA, Beeharry N, Bhattacharjee V, Londin ER, Cozzitorto JA. et al. HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer Res. 2014;74:1128-40
146. Chand SN, Zarei M, Schiewer MJ, Kamath AR, Romeo C, Lal S. et al. Posttranscriptional Regulation of PARG mRNA by HuR Facilitates DNA Repair and Resistance to PARP Inhibitors. Cancer Res. 2017;77:5011-25
147. Jimbo M, Blanco FF, Huang YH, Telonis AG, Screnci BA, Cosma GL. et al. Targeting the mRNA-binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells. Oncotarget. 2015;6:27312-31
148. Lal S, Cheung EC, Zarei M, Preet R, Chand SN, Mambelli-Lisboa NC. et al. CRISPR Knockout of the HuR Gene Causes a Xenograft Lethal Phenotype. Mol Cancer Res. 2017;15:696-707
149. Zhang Y, Liu X, Wang Y, Lai S, Wang Z, Yang Y. et al. The m(6)A demethylase ALKBH5-mediated upregulation of DDIT4-AS1 maintains pancreatic cancer stemness and suppresses chemosensitivity by activating the mTOR pathway. Mol Cancer. 2022;21:174
150. Lu Y, Chen Q, Zhu S, Gong X. Hypoxia promotes immune escape of pancreatic cancer cells by lncRNA NNT-AS1/METTL3-HuR-mediated ITGB1 m(6)A modification. Exp Cell Res. 2023;432:113764
151. He Z, Cai K, Zeng Z, Lei S, Cao W, Li X. Autophagy-associated circRNA circATG7 facilitates autophagy and promotes pancreatic cancer progression. Cell Death Dis. 2022;13:233
152. Larroux C, Luke GN, Koopman P, Rokhsar DS, Shimeld SM, Degnan BM. Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol. 2008;25:980-96
153. Herman L, Todeschini AL, Veitia RA. Forkhead Transcription Factors in Health and Disease. Trends Genet. 2021;37:460-75
154. Pierrou S, Hellqvist M, Samuelsson L, Enerbäck S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. Embo j. 1994;13:5002-12
155. Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198-206
156. Hoogeboom D, Essers MA, Polderman PE, Voets E, Smits LM, Burgering BM. Interaction of FOXO with beta-catenin inhibits beta-catenin/T cell factor activity. J Biol Chem. 2008;283:9224-30
157. Chen T, Lei S, Zeng Z, Zhang J, Xue Y, Sun Y. et al. Linc00261 inhibits metastasis and the WNT signaling pathway of pancreatic cancer by regulating a miR-552-5p/FOXO3 axis. Oncol Rep. 2020;43:930-42
158. Yan H, Li Q, Wu J, Hu W, Jiang J, Shi L. et al. MiR-629 promotes human pancreatic cancer progression by targeting FOXO3. Cell Death Dis. 2017;8:e3154
159. Ling J, Dong X, Wang L, Xue Y, Jia X, Song W. et al. MiR-27a-regulated FOXO1 promotes pancreatic ductal adenocarcinoma cell progression by enhancing Wnt/β-catenin signaling activity. Am J Transl Res. 2019;11:3069-80
160. Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE. et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374-9
161. Sun T, Li X, Zhang P, Chen WD, Zhang HL, Li DD. et al. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat Commun. 2015;6:7215
162. Deng Z, Sun M, Wu J, Fang H, Cai S, An S. et al. SIRT1 attenuates sepsis-induced acute kidney injury via Beclin1 deacetylation-mediated autophagy activation. Cell Death Dis. 2021;12:217
163. Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ Res. 2010;107:1470-82
164. Tian S, Guo X, Yu C, Sun C, Jiang J. miR-138-5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget. 2017;8:11071-82
165. Jäger S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P. et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837-48
166. Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S. et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665-75
167. Luan B, Sun C. MiR-138-5p affects insulin resistance to regulate type 2 diabetes progression through inducing autophagy in HepG2 cells by regulating SIRT1. Nutr Res. 2018;59:90-8
168. Ma J, Zhang Y, Ji H, Chen L, Chen T, Guo C. et al. Overexpression of miR-138-5p suppresses MnCl(2) -induced autophagy by targeting SIRT1 in SH-SY5Y cells. Environ Toxicol. 2019;34:539-47
169. Ou L, Wang D, Zhang H, Yu Q, Hua F. Decreased Expression of miR-138-5p by lncRNA H19 in Cervical Cancer Promotes Tumor Proliferation. Oncol Res. 2018;26:401-10
170. Zhang Y, Ai H, Fan X, Chen S, Wang Y, Liu L. Knockdown of long non-coding RNA HOTAIR reverses cisplatin resistance of ovarian cancer cells through inhibiting miR-138-5p-regulated EZH2 and SIRT1. Biol Res. 2020;53:18
171. Wang J, Zhao L, Shang K, Liu F, Che J, Li H. et al. Long non-coding RNA H19, a novel therapeutic target for pancreatic cancer. Mol Med. 2020;26:30
172. Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280-93
173. Lee JW, Nam H, Kim LE, Jeon Y, Min H, Ha S. et al. TLR4 (toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy. 2019;15:753-70
174. Zhao M, Gao J, Cui C, Zhang Y, Jiang X, Cui J. Inhibition of PTEN Ameliorates Secondary Hippocampal Injury and Cognitive Deficits after Intracerebral Hemorrhage: Involvement of AKT/FoxO3a/ATG-Mediated Autophagy. Oxid Med Cell Longev. 2021;2021:5472605
175. Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S. et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest. 2010;120:1043-55
176. Yao J, Wang J, Xu Y, Guo Q, Sun Y, Liu J. et al. CDK9 inhibition blocks the initiation of PINK1-PRKN-mediated mitophagy by regulating the SIRT1-FOXO3-BNIP3 axis and enhances the therapeutic effects involving mitochondrial dysfunction in hepatocellular carcinoma. Autophagy. 2022;18:1879-97
177. Dasgupta A, Shukla SK, Vernucci E, King RJ, Abrego J, Mulder SE. et al. SIRT1-NOX4 signaling axis regulates cancer cachexia. J Exp Med. 2020 217
178. Warkad MS, Kim CH, Kang BG, Park SH, Jung JS, Feng JH. et al. Metformin-induced ROS upregulation as amplified by apigenin causes profound anticancer activity while sparing normal cells. Sci Rep. 2021;11:14002
179. Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92-102
180. Halasi M, Gartel AL. FOX(M1) news-it is cancer. Mol Cancer Ther. 2013;12:245-54
181. Nandi D, Cheema PS, Jaiswal N, Nag A. FoxM1: Repurposing an oncogene as a biomarker. Semin Cancer Biol. 2018;52:74-84
182. Zhang W, Qian W, Gu J, Gong M, Zhang W, Zhang S. et al. Mutant p53 driven-LINC00857, a protein scaffold between FOXM1 and deubiquitinase OTUB1, promotes the metastasis of pancreatic cancer. Cancer Lett. 2023;552:215976
183. Koch S. Regulation of Wnt Signaling by FOX Transcription Factors in Cancer. Cancers (Basel). 2021 13
184. Huang X, Huang W, Wu K, Lin Q, Chen G. PRRX1/FOXM1 reduces gemcitabin-induced cytotoxicity by regulating autophagy in bladder cancer. Transl Androl Urol. 2022;11:1116-29
185. Xiang X, Fu Y, Zhao K, Miao R, Zhang X, Ma X. et al. Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated PHB2. Theranostics. 2021;11:4929-44
186. Lin JZ, Wang WW, Hu TT, Zhu GY, Li LN, Zhang CY. et al. FOXM1 contributes to docetaxel resistance in castration-resistant prostate cancer by inducing AMPK/mTOR-mediated autophagy. Cancer Lett. 2020;469:481-9
187. Xin L, Zhou Q, Yuan YW, Zhou LQ, Liu L, Li SH. et al. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol. 2019;145:2507-17
188. Hamurcu Z, Delibaşı N, Nalbantoglu U, Sener EF, Nurdinov N, Tascı B. et al. FOXM1 plays a role in autophagy by transcriptionally regulating Beclin-1 and LC3 genes in human triple-negative breast cancer cells. J Mol Med (Berl). 2019;97:491-508
189. Whatcott CJ, Diep CH, Jiang P, Watanabe A, LoBello J, Sima C. et al. Desmoplasia in Primary Tumors and Metastatic Lesions of Pancreatic Cancer. Clin Cancer Res. 2015;21:3561-8
190. Koong AC, Mehta VK, Le QT, Fisher GA, Terris DJ, Brown JM. et al. Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys. 2000;48:919-22
191. Erkan M, Hausmann S, Michalski CW, Fingerle AA, Dobritz M, Kleeff J. et al. The role of stroma in pancreatic cancer: diagnostic and therapeutic implications. Nat Rev Gastroenterol Hepatol. 2012;9:454-67
192. Tirpe AA, Gulei D, Ciortea SM, Crivii C, Berindan-Neagoe I. Hypoxia: Overview on Hypoxia-Mediated Mechanisms with a Focus on the Role of HIF Genes. Int J Mol Sci. 2019 20
193. Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev. 2012;92:967-1003
194. Ziello JE, Jovin IS, Huang Y. Hypoxia-Inducible Factor (HIF)-1 regulatory pathway and its potential for therapeutic intervention in malignancy and ischemia. Yale J Biol Med. 2007;80:51-60
195. Jun JC, Rathore A, Younas H, Gilkes D, Polotsky VY. Hypoxia-Inducible Factors and Cancer. Curr Sleep Med Rep. 2017;3:1-10
196. Wiesener MS, Jürgensen JS, Rosenberger C, Scholze CK, Hörstrup JH, Warnecke C. et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. Faseb j. 2003;17:271-3
197. Azoitei N, Becher A, Steinestel K, Rouhi A, Diepold K, Genze F. et al. PKM2 promotes tumor angiogenesis by regulating HIF-1α through NF-κB activation. Mol Cancer. 2016;15:3
198. Liu M, Zhong J, Zeng Z, Huang K, Ye Z, Deng S. et al. Hypoxia-induced feedback of HIF-1α and lncRNA-CF129 contributes to pancreatic cancer progression through stabilization of p53 protein. Theranostics. 2019;9:4795-810
199. Xu F, Huang M, Chen Q, Niu Y, Hu Y, Hu P. et al. LncRNA HIF1A-AS1 Promotes Gemcitabine Resistance of Pancreatic Cancer by Enhancing Glycolysis through Modulating the AKT/YB1/HIF1α Pathway. Cancer Res. 2021;81:5678-91
200. Liu Y, Chen S, Cai K, Zheng D, Zhu C, Li L. et al. Hypoxia-induced long noncoding RNA NR2F1-AS1 maintains pancreatic cancer proliferation, migration, and invasion by activating the NR2F1/AKT/mTOR axis. Cell Death Dis. 2022;13:232
201. Krebs AM, Mitschke J, Lasierra Losada M, Schmalhofer O, Boerries M, Busch H. et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518-29
202. Meidhof S, Brabletz S, Lehmann W, Preca BT, Mock K, Ruh M. et al. ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med. 2015;7:831-47
203. Jin Y, Zhang Z, Yu Q, Zeng Z, Song H, Huang X. et al. Positive Reciprocal Feedback of lncRNA ZEB1-AS1 and HIF-1α Contributes to Hypoxia-Promoted Tumorigenesis and Metastasis of Pancreatic Cancer. Front Oncol. 2021;11:761979
204. Deng SJ, Chen HY, Ye Z, Deng SC, Zhu S, Zeng Z. et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811-28
205. Zhu S, Deng S, He C, Liu M, Chen H, Zeng Z. et al. Reciprocal loop of hypoxia-inducible factor-1α (HIF-1α) and metastasis-associated protein 2 (MTA2) contributes to the progression of pancreatic carcinoma by suppressing E-cadherin transcription. J Pathol. 2018;245:349-60
206. Zeng Z, Xu FY, Zheng H, Cheng P, Chen QY, Ye Z. et al. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1α. Theranostics. 2019;9:5298-314
207. Zhu Y, Wu F, Gui W, Zhang N, Matro E, Zhu L. et al. A positive feedback regulatory loop involving the lncRNA PVT1 and HIF-1α in pancreatic cancer. J Mol Cell Biol. 2021;13:676-89
208. Xia Y, Jiang L, Zhong T. The role of HIF-1α in chemo-/radioresistant tumors. Onco Targets Ther. 2018;11:3003-11
209. Jing X, Yang F, Shao C, Wei K, Xie M, Shen H. et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer. 2019;18:157
210. Zhang Y, Liu D, Hu H, Zhang P, Xie R, Cui W. HIF-1α/BNIP3 signaling pathway-induced-autophagy plays protective role during myocardial ischemia-reperfusion injury. Biomed Pharmacother. 2019;120:109464
211. Fu ZJ, Wang ZY, Xu L, Chen XH, Li XX, Liao WT. et al. HIF-1α-BNIP3-mediated mitophagy in tubular cells protects against renal ischemia/reperfusion injury. Redox Biol. 2020;36:101671
212. Lu J, Peng Y, Zou J, Wang J, Lu S, Fu T. et al. Hypoxia Inducible Factor-1α Is a Regulator of Autophagy in Osteoarthritic Chondrocytes. Cartilage. 2021;13:1030s-40s
213. Tang Z, Zhang Z, Lin Q, Xu R, Chen J, Wang Y. et al. HIF-1α/BNIP3-Mediated Autophagy Contributes to the Luteinization of Granulosa Cells During the Formation of Corpus Luteum. Front Cell Dev Biol. 2020;8:619924
214. Zhu H, Wang D, Liu Y, Su Z, Zhang L, Chen F. et al. Role of the Hypoxia-inducible factor-1 alpha induced autophagy in the conversion of non-stem pancreatic cancer cells into CD133+ pancreatic cancer stem-like cells. Cancer Cell Int. 2013;13:119
215. Zhu H, Wang D, Zhang L, Xie X, Wu Y, Liu Y. et al. Upregulation of autophagy by hypoxia-inducible factor-1α promotes EMT and metastatic ability of CD133+ pancreatic cancer stem-like cells during intermittent hypoxia. Oncol Rep. 2014;32:935-42
216. Vallée A, Guillevin R, Vallée JN. Vasculogenesis and angiogenesis initiation under normoxic conditions through Wnt/β-catenin pathway in gliomas. Rev Neurosci. 2018;29:71-91
217. Vallée A, Lecarpentier Y, Vallée JN. The Key Role of the WNT/β-Catenin Pathway in Metabolic Reprogramming in Cancers under Normoxic Conditions. Cancers (Basel). 2021 13
218. Xiang J, Hu Q, Qin Y, Ji S, Xu W, Liu W. et al. TCF7L2 positively regulates aerobic glycolysis via the EGLN2/HIF-1α axis and indicates prognosis in pancreatic cancer. Cell Death Dis. 2018;9:321
219. Tang K, Toyozumi T, Murakami K, Sakata H, Kano M, Endo S. et al. HIF-1α stimulates the progression of oesophageal squamous cell carcinoma by activating the Wnt/β-catenin signalling pathway. Br J Cancer. 2022;127:474-87
220. Kaidi A, Williams AC, Paraskeva C. Interaction between beta-catenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol. 2007;9:210-7
221. Zhang Q, Bai X, Chen W, Ma T, Hu Q, Liang C. et al. Wnt/β-catenin signaling enhances hypoxia-induced epithelial-mesenchymal transition in hepatocellular carcinoma via crosstalk with hif-1α signaling. Carcinogenesis. 2013;34:962-73
222. Xu W, Wang Z, Zhang W, Qian K, Li H, Kong D. et al. Mutated K-ras activates CDK8 to stimulate the epithelial-to-mesenchymal transition in pancreatic cancer in part via the Wnt/β-catenin signaling pathway. Cancer Lett. 2015;356:613-27
223. Liu X, Tan X, Liu P, Wu Y, Qian S, Zhang X. Phosphoglycerate Mutase 1 (PGAM1) Promotes Pancreatic Ductal Adenocarcinoma (PDAC) Metastasis by Acting as a Novel Downstream Target of the PI3K/Akt/mTOR Pathway. Oncol Res. 2018;26:1123-31
224. Wen CL, Huang K, Jiang LL, Lu XX, Dai YT, Shi MM. et al. An allosteric PGAM1 inhibitor effectively suppresses pancreatic ductal adenocarcinoma. Proc Natl Acad Sci U S A. 2019;116:23264-73
225. Garcia Garcia CJ, Huang Y, Fuentes NR, Turner MC, Monberg ME, Lin D. et al. Stromal HIF2 Regulates Immune Suppression in the Pancreatic Cancer Microenvironment. Gastroenterology. 2022;162:2018-31
226. Zhang Q, Lou Y, Zhang J, Fu Q, Wei T, Sun X. et al. Hypoxia-inducible factor-2α promotes tumor progression and has crosstalk with Wnt/β-catenin signaling in pancreatic cancer. Mol Cancer. 2017;16:119
227. Schofield HK, Tandon M, Park MJ, Halbrook CJ, Ramakrishnan SK, Kim EC. et al. Pancreatic HIF2α Stabilization Leads to Chronic Pancreatitis and Predisposes to Mucinous Cystic Neoplasm. Cell Mol Gastroenterol Hepatol. 2018;5:169-85.e2
228. Sano M, Driscoll DR, De Jesus-Monge WE, Klimstra DS, Lewis BC. Activated wnt signaling in stroma contributes to development of pancreatic mucinous cystic neoplasms. Gastroenterology. 2014;146:257-67
229. Criscimanna A, Duan LJ, Rhodes JA, Fendrich V, Wickline E, Hartman DJ. et al. PanIN-specific regulation of Wnt signaling by HIF2α during early pancreatic tumorigenesis. Cancer Res. 2013;73:4781-90
230. Freeman TJ, Smith JJ, Chen X, Washington MK, Roland JT, Means AL. et al. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of β-catenin. Gastroenterology. 2012;142:562-71.e2
231. Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17(Suppl 5):v7-12
232. Plunkett W, Huang P, Xu YZ, Heinemann V, Grunewald R, Gandhi V. Gemcitabine: metabolism, mechanisms of action, and self-potentiation. Semin Oncol. 1995;22:3-10
233. Elledge SJ, Zhou Z, Allen JB. Ribonucleotide reductase: regulation, regulation, regulation. Trends Biochem Sci. 1992;17:119-23
234. Heinemann V, Xu YZ, Chubb S, Sen A, Hertel LW, Grindey GB. et al. Cellular elimination of 2',2'-difluorodeoxycytidine 5'-triphosphate: a mechanism of self-potentiation. Cancer Res. 1992;52:533-9
235. Gilbert JA, Salavaggione OE, Ji Y, Pelleymounter LL, Eckloff BW, Wieben ED. et al. Gemcitabine pharmacogenomics: cytidine deaminase and deoxycytidylate deaminase gene resequencing and functional genomics. Clin Cancer Res. 2006;12:1794-803
236. Funamizu N, Honjo M, Tamura K, Sakamoto K, Ogawa K, Takada Y. microRNAs Associated with Gemcitabine Resistance via EMT, TME, and Drug Metabolism in Pancreatic Cancer. Cancers (Basel). 2023 15
237. Liu Y, Wang J, Dong L, Xia L, Zhu H, Li Z. et al. Long Noncoding RNA HCP5 Regulates Pancreatic Cancer Gemcitabine (GEM) Resistance By Sponging Hsa-miR-214-3p To Target HDGF. Onco Targets Ther. 2019;12:8207-16
238. Chen YT, Chen FW, Chang TH, Wang TW, Hsu TP, Chi JY. et al. Hepatoma-derived growth factor supports the antiapoptosis and profibrosis of pancreatic stellate cells. Cancer Lett. 2019;457:180-90
239. Huge N, Reinkens T, Buurman R, Sandbothe M, Bergmann A, Wallaschek H. et al. MiR-129-5p exerts Wnt signaling-dependent tumor-suppressive functions in hepatocellular carcinoma by directly targeting hepatoma-derived growth factor HDGF. Cancer Cell Int. 2022;22:192
240. Zheng Y, Lu S, Xu Y, Zheng J. Long non-coding RNA AGAP2-AS1 promotes the proliferation of glioma cells by sponging miR-15a/b-5p to upregulate the expression of HDGF and activating Wnt/β-catenin signaling pathway. Int J Biol Macromol. 2019;128:521-30
241. Xia C, Li Q, Cheng X, Wu T, Gao P. miR-4323 targets hepatoma-derived growth factor (HDGF) to suppress colorectal cancer cell proliferation. Pathol Res Pract. 2021;225:153544
242. Cheng C, Li W, Peng X, Liu X, Zhang Z, Liu Z. et al. miR-1254 induced by NESG1 inactivates HDGF/DDX5-stimulated nuclear translocation of β-catenin and suppresses NPC metastasis. Mol Ther Methods Clin Dev. 2021;20:615-24
243. Zhao J, Ma MZ, Ren H, Liu Z, Edelman MJ, Pan H. et al. Anti-HDGF targets cancer and cancer stromal stem cells resistant to chemotherapy. Clin Cancer Res. 2013;19:3567-76
244. Richards NG, Rittenhouse DW, Freydin B, Cozzitorto JA, Grenda D, Rui H. et al. HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Ann Surg. 2010;252:499-505 discussion -6
245. Toyota K, Murakami Y, Kondo N, Uemura K, Nakagawa N, Takahashi S. et al. Cytoplasmic Hu-Antigen R (HuR) Expression is Associated with Poor Survival in Patients with Surgically Resected Cholangiocarcinoma Treated with Adjuvant Gemcitabine-Based Chemotherapy. Ann Surg Oncol. 2018;25:1202-10
246. Jakstaite A, Maziukiene A, Silkuniene G, Kmieliute K, Gulbinas A, Dambrauskas Z. HuR mediated post-transcriptional regulation as a new potential adjuvant therapeutic target in chemotherapy for pancreatic cancer. World J Gastroenterol. 2015;21:13004-19
247. Pineda DM, Rittenhouse DW, Valley CC, Cozzitorto JA, Burkhart RA, Leiby B. et al. HuR's post-transcriptional regulation of Death Receptor 5 in pancreatic cancer cells. Cancer Biol Ther. 2012;13:946-55
248. McAllister F, Pineda DM, Jimbo M, Lal S, Burkhart RA, Moughan J. et al. dCK expression correlates with 5-fluorouracil efficacy and HuR cytoplasmic expression in pancreatic cancer: a dual-institutional follow-up with the RTOG 9704 trial. Cancer Biol Ther. 2014;15:688-98
249. Ghelli Luserna di Rorà A, Cerchione C, Martinelli G, Simonetti G. A WEE1 family business: regulation of mitosis, cancer progression, and therapeutic target. J Hematol Oncol. 2020;13:126
250. Pilié PG, Tang C, Mills GB, Yap TA. State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 2019;16:81-104
251. Burkhart RA, Pineda DM, Chand SN, Romeo C, Londin ER, Karoly ED. et al. HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer. RNA Biol. 2013;10:1312-23
252. Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13:482-95
253. Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B. et al. Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol Cell Biol. 2007;27:4917-30
254. Karadedou CT, Gomes AR, Chen J, Petkovic M, Ho KK, Zwolinska AK. et al. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene. 2012;31:1845-58
255. Yao S, Fan LY, Lam EW. The FOXO3-FOXM1 axis: A key cancer drug target and a modulator of cancer drug resistance. Semin Cancer Biol. 2018;50:77-89
256. Ghosh S, Singh R, Vanwinkle ZM, Guo H, Vemula PK, Goel A. et al. Microbial metabolite restricts 5-fluorouracil-resistant colonic tumor progression by sensitizing drug transporters via regulation of FOXO3-FOXM1 axis. Theranostics. 2022;12:5574-95
257. Robey RW, Pluchino KM, Hall MD, Fojo AT, Bates SE, Gottesman MM. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat Rev Cancer. 2018;18:452-64
258. Zhu X, Xue L, Yao Y, Wang K, Tan C, Zhuang M. et al. The FoxM1-ABCC4 axis mediates carboplatin resistance in human retinoblastoma Y-79 cells. Acta Biochim Biophys Sin (Shanghai). 2018;50:914-20
259. Hou Y, Dong Z, Zhong W, Yin L, Li X, Kuerban G. et al. FOXM1 Promotes Drug Resistance in Cervical Cancer Cells by Regulating ABCC5 Gene Transcription. Biomed Res Int. 2022;2022:3032590
260. Hou Y, Zhu Q, Li Z, Peng Y, Yu X, Yuan B. et al. The FOXM1-ABCC5 axis contributes to paclitaxel resistance in nasopharyngeal carcinoma cells. Cell Death Dis. 2017;8:e2659
261. Xie T, Geng J, Wang Y, Wang L, Huang M, Chen J. et al. FOXM1 evokes 5-fluorouracil resistance in colorectal cancer depending on ABCC10. Oncotarget. 2017;8:8574-89
262. Roh YG, Mun MH, Jeong MS, Kim WT, Lee SR, Chung JW. et al. Drug resistance of bladder cancer cells through activation of ABCG2 by FOXM1. BMB Rep. 2018;51:98-103
263. Gai J, Ji M, Shi C, Li W, Chen S, Wang Y. et al. FoxO regulates expression of ABCA6, an intracellular ATP-binding-cassette transporter responsive to cholesterol. Int J Biochem Cell Biol. 2013;45:2651-9
264. Hui RC, Francis RE, Guest SK, Costa JR, Gomes AR, Myatt SS. et al. Doxorubicin activates FOXO3a to induce the expression of multidrug resistance gene ABCB1 (MDR1) in K562 leukemic cells. Mol Cancer Ther. 2008;7:670-8
265. Choi HK, Cho KB, Phuong NT, Han CY, Han HK, Hien TT. et al. SIRT1-mediated FoxO1 deacetylation is essential for multidrug resistance-associated protein 2 expression in tamoxifen-resistant breast cancer cells. Mol Pharm. 2013;10:2517-27
266. Hwang HJ, Lee KH, Cho JY. ABCA9, an ER cholesterol transporter, inhibits breast cancer cell proliferation via SREBP-2 signaling. Cancer Sci. 2023;114:1451-63
267. Sunters A, Madureira PA, Pomeranz KM, Aubert M, Brosens JJ, Cook SJ. et al. Paclitaxel-induced nuclear translocation of FOXO3a in breast cancer cells is mediated by c-Jun NH2-terminal kinase and Akt. Cancer Res. 2006;66:212-20
268. Sunters A, Fernández de Mattos S, Stahl M, Brosens JJ, Zoumpoulidou G, Saunders CA. et al. FoxO3a transcriptional regulation of Bim controls apoptosis in paclitaxel-treated breast cancer cell lines. J Biol Chem. 2003;278:49795-805
269. Khongkow P, Gomes AR, Gong C, Man EP, Tsang JW, Zhao F. et al. Paclitaxel targets FOXM1 to regulate KIF20A in mitotic catastrophe and breast cancer paclitaxel resistance. Oncogene. 2016;35:990-1002
270. Song Z, Li J, Zhang L, Deng J, Fang Z, Xiang X. et al. UCHL3 promotes pancreatic cancer progression and chemo-resistance through FOXM1 stabilization. Am J Cancer Res. 2019;9:1970-81
271. Zhu J, Zhao J, Luo C, Zhu Z, Peng X, Zhu X. et al. FAT10 promotes chemotherapeutic resistance in pancreatic cancer by inducing epithelial-mesenchymal transition via stabilization of FOXM1 expression. Cell Death Dis. 2022;13:497
272. Liu C, Shi J, Li Q, Li Z, Lou C, Zhao Q. et al. STAT1-mediated inhibition of FOXM1 enhances gemcitabine sensitivity in pancreatic cancer. Clin Sci (Lond). 2019;133:645-63
273. Huang C, Zhang X, Jiang L, Zhang L, Xiang M, Ren H. FoxM1 Induced Paclitaxel Resistance via Activation of the FoxM1/PHB1/RAF-MEK-ERK Pathway and Enhancement of the ABCA2 Transporter. Mol Ther Oncolytics. 2019;14:196-212
274. Lee K, Yu H, Shouse S, Kong B, Lee J, Lee SH. et al. RNA-Seq Reveals Different Gene Expression in Liver-Specific Prohibitin 1 Knock-Out Mice. Front Physiol. 2021;12:717911
275. Spagnuolo C, Russo GL, Orhan IE, Habtemariam S, Daglia M, Sureda A. et al. Genistein and cancer: current status, challenges, and future directions. Adv Nutr. 2015;6:408-19
276. Sarkar FH, Li Y. The role of isoflavones in cancer chemoprevention. Front Biosci. 2004;9:2714-24
277. Sarkar FH, Li Y. Mechanisms of cancer chemoprevention by soy isoflavone genistein. Cancer Metastasis Rev. 2002;21:265-80
278. Bi YL, Min M, Shen W, Liu Y. Genistein induced anticancer effects on pancreatic cancer cell lines involves mitochondrial apoptosis, G(0)/G(1)cell cycle arrest and regulation of STAT3 signalling pathway. Phytomedicine. 2018;39:10-6
279. Wang Z, Ahmad A, Banerjee S, Azmi A, Kong D, Li Y. et al. FoxM1 is a novel target of a natural agent in pancreatic cancer. Pharm Res. 2010;27:1159-68
280. Kodela R, Chattopadhyay M, Kashfi K. NOSH-Aspirin: A Novel Nitric Oxide-Hydrogen Sulfide-Releasing Hybrid: A New Class of Anti-inflammatory Pharmaceuticals. ACS Med Chem Lett. 2012;3:257-62
281. Kashfi K. Utility Of Nitric Oxide And Hydrogen Sulfide-Releasing Chimeras As Anticancer Agents. Redox Biol. 2015;5:420
282. Chattopadhyay M, Kodela R, Santiago G, Le TTC, Nath N, Kashfi K. NOSH-aspirin (NBS-1120) inhibits pancreatic cancer cell growth in a xenograft mouse model: Modulation of FoxM1, p53, NF-κB, iNOS, caspase-3 and ROS. Biochem Pharmacol. 2020;176:113857
283. Momeny M, Esmaeili F, Hamzehlou S, Yousefi H, Javadikooshesh S, Vahdatirad V. et al. The ERBB receptor inhibitor dacomitinib suppresses proliferation and invasion of pancreatic ductal adenocarcinoma cells. Cell Oncol (Dordr). 2019;42:491-504
284. Liu Y, Ao X, Ding W, Ponnusamy M, Wu W, Hao X. et al. Critical role of FOXO3a in carcinogenesis. Mol Cancer. 2018;17:104
285. Shen S, Wang J, Zheng B, Tao Y, Li M, Wang Y. et al. LINC01714 Enhances Gemcitabine Sensitivity by Modulating FOXO3 Phosphorylation in Cholangiocarcinoma. Mol Ther Nucleic Acids. 2020;19:446-57
286. Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS. et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857-68
287. Wang Y, Liang HX, Zhang CM, Zou M, Zou BB, Wei W. et al. FOXO3/TRIM22 axis abated the antitumor effect of gemcitabine in non-small cell lung cancer via autophagy induction. Transl Cancer Res. 2020;9:937-48
288. Ye X, Xie G, Liu Z, Tang J, Cui M, Wang C. et al. TNNC1 Reduced Gemcitabine Sensitivity of Nonsmall-Cell Lung Cancer by Increasing Autophagy. Med Sci Monit. 2020;26:e922703
289. Zhou J, Zhou LY, Tang X, Zhang J, Zhai LL, Yi YY. et al. Circ-Foxo3 is positively associated with the Foxo3 gene and leads to better prognosis of acute myeloid leukemia patients. BMC Cancer. 2019;19:930
290. Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y. et al. Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis. Cell Death Differ. 2020;27:966-83
291. Wang Y, Kang XL, Zeng FC, Xu CJ, Zhou JQ, Luo DN. Correlations of Foxo3 and Foxo4 expressions with clinicopathological features and prognosis of bladder cancer. Pathol Res Pract. 2017;213:766-72
292. An Y, Wang B, Wang X, Dong G, Jia J, Yang Q. SIRT1 inhibits chemoresistance and cancer stemness of gastric cancer by initiating an AMPK/FOXO3 positive feedback loop. Cell Death Dis. 2020;11:115
293. Shou Z, Lin L, Liang J, Li JL, Chen HY. Expression and prognosis of FOXO3a and HIF-1α in nasopharyngeal carcinoma. J Cancer Res Clin Oncol. 2012;138:585-93
294. Lu M, Zhao Y, Xu F, Wang Y, Xiang J, Chen D. The expression and prognosis of FOXO3a and Skp2 in human ovarian cancer. Med Oncol. 2012;29:3409-15
295. Fondevila F, Fernández-Palanca P, Méndez-Blanco C, Payo-Serafín T, Lozano E, Marin JJG. et al. Association of FOXO3 Expression with Tumor Pathogenesis, Prognosis and Clinicopathological Features in Hepatocellular Carcinoma: A Systematic Review with Meta-Analysis. Cancers (Basel). 2021 13
296. Song SS, Ying JF, Zhang YN, Pan HY, He XL, Hu ZM. et al. High expression of FOXO3 is associated with poor prognosis in patients with hepatocellular carcinoma. Oncol Lett. 2020;19:3181-8
297. Kumazoe M, Takai M, Bae J, Hiroi S, Huang Y, Takamatsu K. et al. FOXO3 is essential for CD44 expression in pancreatic cancer cells. Oncogene. 2017;36:2643-54
298. Rupp M, Hagenbuchner J, Rass B, Fiegl H, Kiechl-Kohlendorfer U, Obexer P. et al. FOXO3-mediated chemo-protection in high-stage neuroblastoma depends on wild-type TP53 and SESN3. Oncogene. 2017;36:6190-203
299. Huang R, Song X, Wang CM. MiR-223 regulates CDDP resistance in pancreatic cancer via targeting FoxO3a. Eur Rev Med Pharmacol Sci. 2019;23:7892-8
300. Kumazoe M, Takai M, Hiroi S, Takeuchi C, Kadomatsu M, Nojiri T. et al. The FOXO3/PGC-1β signaling axis is essential for cancer stem cell properties of pancreatic ductal adenocarcinoma. J Biol Chem. 2017;292:10813-23
301. Hossan MS, Break MKB, Bradshaw TD, Collins HM, Wiart C, Khoo TJ. et al. Novel Semi-Synthetic Cu (II)-Cardamonin Complex Exerts Potent Anticancer Activity against Triple-Negative Breast and Pancreatic Cancer Cells via Inhibition of the Akt Signaling Pathway. Molecules. 2021 26
302. Sun H, Zhang N, Jin Y, Xu H. Cardamonin Promotes the Apoptosis and Chemotherapy Sensitivity to Gemcitabine of Pancreatic Cancer Through Modulating the FOXO3a-FOXM1 Axis. Dose Response. 2021;19:15593258211042163
303. Erin N, Grahovac J, Brozovic A, Efferth T. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resist Updat. 2020;53:100715
304. Lv Y, Zhao S, Han J, Zheng L, Yang Z, Zhao L. Hypoxia-inducible factor-1α induces multidrug resistance protein in colon cancer. Onco Targets Ther. 2015;8:1941-8
305. Comerford KM, Wallace TJ, Karhausen J, Louis NA, Montalto MC, Colgan SP. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res. 2002;62:3387-94
306. Xiang L, Wang Y, Lan J, Na F, Wu S, Gong Y. et al. HIF-1-dependent heme synthesis promotes gemcitabine resistance in human non-small cell lung cancers via enhanced ABCB6 expression. Cell Mol Life Sci. 2022;79:343
307. Schmitz G, Langmann T. Transcriptional regulatory networks in lipid metabolism control ABCA1 expression. Biochim Biophys Acta. 2005;1735:1-19
308. He X, Wang J, Wei W, Shi M, Xin B, Zhang T. et al. Hypoxia regulates ABCG2 activity through the activivation of ERK1/2/HIF-1α and contributes to chemoresistance in pancreatic cancer cells. Cancer Biol Ther. 2016;17:188-98
309. He M, Wu H, Jiang Q, Liu Y, Han L, Yan Y. et al. Hypoxia-inducible factor-2α directly promotes BCRP expression and mediates the resistance of ovarian cancer stem cells to adriamycin. Mol Oncol. 2019;13:403-21
310. Woo SK, Kwon MS, Geng Z, Chen Z, Ivanov A, Bhatta S. et al. Sequential activation of hypoxia-inducible factor 1 and specificity protein 1 is required for hypoxia-induced transcriptional stimulation of Abcc8. J Cereb Blood Flow Metab. 2012;32:525-36
311. Yang SY, Song BQ, Dai SL, Yang KX, Jin Z, Shi KW. Effects of hypoxia-inducible factor-1α silencing on drug resistance of human pancreatic cancer cell line Patu8988/5-Fu. Hepatogastroenterology. 2014;61:2395-401
312. Hassan S, Peluso J, Chalhoub S, Idoux Gillet Y, Benkirane-Jessel N, Rochel N. et al. Quercetin potentializes the respective cytotoxic activity of gemcitabine or doxorubicin on 3D culture of AsPC-1 or HepG2 cells, through the inhibition of HIF-1α and MDR1. PLoS One. 2020;15:e0240676
313. Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev. 2010;20:51-6
314. Akakura N, Kobayashi M, Horiuchi I, Suzuki A, Wang J, Chen J. et al. Constitutive expression of hypoxia-inducible factor-1alpha renders pancreatic cancer cells resistant to apoptosis induced by hypoxia and nutrient deprivation. Cancer Res. 2001;61:6548-54
315. Chaika NV, Gebregiworgis T, Lewallen ME, Purohit V, Radhakrishnan P, Liu X. et al. MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proc Natl Acad Sci U S A. 2012;109:13787-92
316. Aubert S, Fauquette V, Hémon B, Lepoivre R, Briez N, Bernard D. et al. MUC1, a new hypoxia inducible factor target gene, is an actor in clear renal cell carcinoma tumor progression. Cancer Res. 2009;69:5707-15
317. Shukla SK, Purohit V, Mehla K, Gunda V, Chaika NV, Vernucci E. et al. MUC1 and HIF-1alpha Signaling Crosstalk Induces Anabolic Glucose Metabolism to Impart Gemcitabine Resistance to Pancreatic Cancer. Cancer Cell. 2017;32:71-87.e7
318. Nordh S, Ansari D, Andersson R. hENT1 expression is predictive of gemcitabine outcome in pancreatic cancer: a systematic review. World J Gastroenterol. 2014;20:8482-90
319. Bird NT, Elmasry M, Jones R, Psarelli E, Dodd J, Malik H. et al. Immunohistochemical hENT1 expression as a prognostic biomarker in patients with resected pancreatic ductal adenocarcinoma undergoing adjuvant gemcitabine-based chemotherapy. Br J Surg. 2017;104:328-36
320. Jordheim LP, Dumontet C. Do hENT1 and RRM1 predict the clinical benefit of gemcitabine in pancreatic cancer? Biomark Med. 2013;7:663-71
321. Perera S, Jang GH, Wang Y, Kelly D, Allen M, Zhang A. et al. hENT1 Expression Predicts Response to Gemcitabine and Nab-Paclitaxel in Advanced Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2022;28:5115-20
322. Xi Y, Yuan P, Li T, Zhang M, Liu MF, Li B. hENT1 reverses chemoresistance by regulating glycolysis in pancreatic cancer. Cancer Lett. 2020;479:112-22
323. Zhou B, Lei JH, Wang Q, Qu TF, Cha LC, Zhan HX. et al. Cancer-associated fibroblast-secreted miR-421 promotes pancreatic cancer by regulating the SIRT3/H3K9Ac/HIF-1α axis. Kaohsiung J Med Sci. 2022;38:1080-92
324. Zhang J, Han L, Yu J, Li H, Li Q. miR-224 aggravates cancer-associated fibroblast-induced progression of non-small cell lung cancer by modulating a positive loop of the SIRT3/AMPK/mTOR/HIF-1α axis. Aging (Albany NY). 2021;13:10431-49
325. Wang Q, Wei S, Li L, Qiu J, Zhou S, Shi C. et al. TGR5 deficiency aggravates hepatic ischemic/reperfusion injury via inhibiting SIRT3/FOXO3/HIF-1ɑ pathway. Cell Death Discov. 2020;6:116
326. Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986-96
327. Spivak-Kroizman TR, Hostetter G, Posner R, Aziz M, Hu C, Demeure MJ. et al. Hypoxia triggers hedgehog-mediated tumor-stromal interactions in pancreatic cancer. Cancer Res. 2013;73:3235-47
328. Carballo GB, Honorato JR, de Lopes GPF, Spohr T. A highlight on Sonic hedgehog pathway. Cell Commun Signal. 2018;16:11
329. Bailey JM, Swanson BJ, Hamada T, Eggers JP, Singh PK, Caffery T. et al. Sonic hedgehog promotes desmoplasia in pancreatic cancer. Clin Cancer Res. 2008;14:5995-6004
330. Bailey JM, Mohr AM, Hollingsworth MA. Sonic hedgehog paracrine signaling regulates metastasis and lymphangiogenesis in pancreatic cancer. Oncogene. 2009;28:3513-25
331. Li X, Wang Z, Ma Q, Xu Q, Liu H, Duan W. et al. Sonic hedgehog paracrine signaling activates stromal cells to promote perineural invasion in pancreatic cancer. Clin Cancer Res. 2014;20:4326-38
332. Singh S, Srivastava SK, Bhardwaj A, Owen LB, Singh AP. CXCL12-CXCR4 signalling axis confers gemcitabine resistance to pancreatic cancer cells: a novel target for therapy. Br J Cancer. 2010;103:1671-9
333. Sleightholm RL, Neilsen BK, Li J, Steele MM, Singh RK, Hollingsworth MA. et al. Emerging roles of the CXCL12/CXCR4 axis in pancreatic cancer progression and therapy. Pharmacol Ther. 2017;179:158-70
334. Singh AP, Arora S, Bhardwaj A, Srivastava SK, Kadakia MP, Wang B. et al. CXCL12/CXCR4 protein signaling axis induces sonic hedgehog expression in pancreatic cancer cells via extracellular regulated kinase- and Akt kinase-mediated activation of nuclear factor κB: implications for bidirectional tumor-stromal interactions. J Biol Chem. 2012;287:39115-24
335. Arora S, Bhardwaj A, Singh S, Srivastava SK, McClellan S, Nirodi CS. et al. An undesired effect of chemotherapy: gemcitabine promotes pancreatic cancer cell invasiveness through reactive oxygen species-dependent, nuclear factor κB- and hypoxia-inducible factor 1α-mediated up-regulation of CXCR4. J Biol Chem. 2013;288:21197-207
336. Venning FA, Wullkopf L, Erler JT. Targeting ECM Disrupts Cancer Progression. Front Oncol. 2015;5:224
337. Molnar J, Fong KS, He QP, Hayashi K, Kim Y, Fong SF. et al. Structural and functional diversity of lysyl oxidase and the LOX-like proteins. Biochim Biophys Acta. 2003;1647:220-4
338. Vallet SD, Ricard-Blum S. Lysyl oxidases: from enzyme activity to extracellular matrix cross-links. Essays Biochem. 2019;63:349-64
339. Wang V, Davis DA, Yarchoan R. Identification of functional hypoxia inducible factor response elements in the human lysyl oxidase gene promoter. Biochem Biophys Res Commun. 2017;490:480-5
340. Cortes E, Lachowski D, Robinson B, Sarper M, Teppo JS, Thorpe SD. et al. Tamoxifen mechanically reprograms the tumor microenvironment via HIF-1A and reduces cancer cell survival. EMBO Rep. 2019 20
341. Yoshikawa Y, Takano O, Kato I, Takahashi Y, Shima F, Kataoka T. Ras inhibitors display an anti-metastatic effect by downregulation of lysyl oxidase through inhibition of the Ras-PI3K-Akt-HIF-1α pathway. Cancer Lett. 2017;410:82-91
342. Le Calvé B, Griveau A, Vindrieux D, Maréchal R, Wiel C, Svrcek M. et al. Lysyl oxidase family activity promotes resistance of pancreatic ductal adenocarcinoma to chemotherapy by limiting the intratumoral anticancer drug distribution. Oncotarget. 2016;7:32100-12
343. Jones L, Ghaneh P, Humphreys M, Neoptolemos JP. The matrix metalloproteinases and their inhibitors in the treatment of pancreatic cancer. Ann N Y Acad Sci. 1999;880:288-307
344. Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52-67
345. El Hajj EC, El Hajj MC, Ninh VK, Gardner JD. Inhibitor of lysyl oxidase improves cardiac function and the collagen/MMP profile in response to volume overload. Am J Physiol Heart Circ Physiol. 2018;315:H463-h73
346. Merico V, Imberti JF, Zanoni M, Boriani G, Garagna S, Imberti R. Inhibition of lysyl oxidase stimulates TGF-β signaling and metalloproteinases-2 and -9 expression and contributes to the disruption of ascending aorta in rats: protection by propylthiouracil. Heart Vessels. 2021;36:738-47
347. Kothapalli CR, Ramamurthi A. Lysyl oxidase enhances elastin synthesis and matrix formation by vascular smooth muscle cells. J Tissue Eng Regen Med. 2009;3:655-61
348. Ma LJ, Li YG, Huang L, Han M, Ma BJ, Sun BJ. et al. [Expression of LOX and MMP-2 in gastric cancer tissue and the effects of LOX and MMP-2 on tumor invasion and metastasis]. Zhonghua Zhong Liu Za Zhi. 2011;33:37-41
349. Ou J, Guan D, Yang Y. Non-contact co-culture with human vascular endothelial cells promotes epithelial-to-mesenchymal transition of cervical cancer SiHa cells by activating the NOTCH1/LOX/SNAIL pathway. Cell Mol Biol Lett. 2019;24:39
350. Shi XM, Zhao W, Yang YB, Lü BN. [Expression of LOX in Colorectal Cancer Tissues and Its Relationship with Progress and Prognosis]. Sichuan Da Xue Xue Bao Yi Xue Ban. 2017;48:566-9
351. Liu J, Ping W, Zu Y, Sun W. Correlations of lysyl oxidase with MMP2/MMP9 expression and its prognostic value in non-small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6040-7
352. Liu JL, Wei W, Tang W, Jiang Y, Yang HW, Li JT. Mechanism of lysyl oxidase (LOX) in breast cancer invasion and metastasis. Zhonghua Yi Xue Za Zhi. 2012;92:1379-83
353. Zheng Y, Wang X, Wang H, Yan W, Zhang Q, Chang X. Expression of the lysyl oxidase propeptide in hepatocellular carcinoma and its clinical relevance. Oncol Rep. 2014;31:1669-76
354. Zhu S, Zhou Y, Wang L, Zhang J, Wu H, Xiong J. et al. Transcriptional upregulation of MT2-MMP in response to hypoxia is promoted by HIF-1α in cancer cells. Mol Carcinog. 2011;50:770-80
355. Zhu SK, Zhou Y, Cheng C, Zhong S, Wu HQ, Wang B. et al. Overexpression of membrane-type 2 matrix metalloproteinase induced by hypoxia-inducible factor-1α in pancreatic cancer: Implications for tumor progression and prognosis. Mol Clin Oncol. 2014;2:973-81
356. Ellenrieder V, Alber B, Lacher U, Hendler SF, Menke A, Boeck W. et al. Role of MT-MMPs and MMP-2 in pancreatic cancer progression. Int J Cancer. 2000;85:14-20
357. Zhao X, Gao S, Ren H, Sun W, Zhang H, Sun J. et al. Hypoxia-inducible factor-1 promotes pancreatic ductal adenocarcinoma invasion and metastasis by activating transcription of the actin-bundling protein fascin. Cancer Res. 2014;74:2455-64
358. Deng J, Guo Y, Hu X, Du J, Gu J, Kong L. et al. High Glucose Promotes Pancreatic Ductal Adenocarcinoma Gemcitabine Resistance and Invasion through Modulating ROS/MMP-3 Signaling Pathway. Oxid Med Cell Longev. 2022;2022:3243647
359. Dangi-Garimella S, Krantz SB, Barron MR, Shields MA, Heiferman MJ, Grippo PJ. et al. Three-dimensional collagen I promotes gemcitabine resistance in pancreatic cancer through MT1-MMP-mediated expression of HMGA2. Cancer Res. 2011;71:1019-28
360. Jin H, Zhao Y, Zhang S, Yang J, Zhang X, Ma S. Hyperthermia inhibits the motility of gemcitabine-resistant pancreatic cancer PANC-1 cells through the inhibition of epithelial-mesenchymal transition. Mol Med Rep. 2018;17:7274-80
361. Apte RS, Chen DS, Ferrara N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell. 2019;176:1248-64
362. Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R. et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732-44
363. Shi S, Xu J, Zhang B, Ji S, Xu W, Liu J. et al. VEGF Promotes Glycolysis in Pancreatic Cancer via HIF1α Up-Regulation. Curr Mol Med. 2016;16:394-403
364. Jang SI, Fang S, Baek YY, Lee DH, Na K, Lee SY. et al. Local Delivery of Gemcitabine Inhibits Pancreatic and Cholangiocarcinoma Tumor Growth by Promoting Epidermal Growth Factor Receptor Degradation. Int J Mol Sci. 2020 21
365. Bai X, Zhi X, Zhang Q, Liang F, Chen W, Liang C. et al. Inhibition of protein phosphatase 2A sensitizes pancreatic cancer to chemotherapy by increasing drug perfusion via HIF-1α-VEGF mediated angiogenesis. Cancer Lett. 2014;355:281-7
366. Raykov Z, Grekova SP, Bour G, Lehn JM, Giese NA, Nicolau C. et al. Myo-inositol trispyrophosphate-mediated hypoxia reversion controls pancreatic cancer in rodents and enhances gemcitabine efficacy. Int J Cancer. 2014;134:2572-82
367. Suzuki K, Takeuchi O, Suzuki Y, Kitagawa Y. Mechanisms of metformin's anti-tumor activity against gemcitabine-resistant pancreatic adenocarcinoma. Int J Oncol. 2019;54:764-72
368. Qian W, Li J, Chen K, Jiang Z, Cheng L, Zhou C. et al. Metformin suppresses tumor angiogenesis and enhances the chemosensitivity of gemcitabine in a genetically engineered mouse model of pancreatic cancer. Life Sci. 2018;208:253-61
369. Liu SL, Cai C, Yang ZY, Wu ZY, Wu XS, Wang XF. et al. DGCR5 is activated by PAX5 and promotes pancreatic cancer via targeting miR-3163/TOP2A and activating Wnt/β-catenin pathway. Int J Biol Sci. 2021;17:498-513
370. Liu Y, Xu G, Li L. LncRNA GATA3-AS1-miR-30b-5p-Tex10 axis modulates tumorigenesis in pancreatic cancer. Oncol Rep. 2021 45
371. Tang Y, Song G, Liu H, Yang S, Yu X, Shi L. Silencing of Long Non-Coding RNA HOTAIR Alleviates Epithelial-Mesenchymal Transition in Pancreatic Cancer via the Wnt/β-Catenin Signaling Pathway. Cancer Manag Res. 2021;13:3247-57
372. Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao X. et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68-81
373. Ou ZL, Luo Z, Lu YB. Long non-coding RNA HULC as a diagnostic and prognostic marker of pancreatic cancer. World J Gastroenterol. 2019;25:6728-42
374. Shi C, Zhang H, Wang M, Tian R, Li X, Feng Y. et al. OPA Interacting Protein 5 Antisense RNA 1 Expedites Cell Migration and Invasion Through FOXM1/ Wnt/β-Catenin Pathway in Pancreatic Cancer. Dig Dis Sci. 2022;67:915-24
375. Lin C, Wang Y, Dong Y, Lai S, Wang L, Weng S. et al. N6-methyladenosine-mediated SH3BP5-AS1 upregulation promotes GEM chemoresistance in pancreatic cancer by activating the Wnt signaling pathway. Biol Direct. 2022;17:33
376. Zhang YY, Feng HM. MEG3 Suppresses Human Pancreatic Neuroendocrine Tumor Cells Growth and Metastasis by Down-Regulation of Mir-183. Cell Physiol Biochem. 2017;44:345-56
377. Wei YL, Hua J, Liu XY, Hua XM, Sun C, Bai JA. et al. LncNEN885 inhibits epithelial-mesenchymal transition by partially regulation of Wnt/β-catenin signalling in gastroenteropancreatic neuroendocrine neoplasms. Cancer Sci. 2018;109:3139-48
378. Wu C, Yang L, Qi X, Wang T, Li M, Xu K. Inhibition of long non-coding RNA HOTAIR enhances radiosensitivity via regulating autophagy in pancreatic cancer. Cancer Manag Res. 2018;10:5261-71
379. Zhang X, Zhao P, Wang C, Xin B. SNHG14 enhances gemcitabine resistance by sponging miR-101 to stimulate cell autophagy in pancreatic cancer. Biochem Biophys Res Commun. 2019;510:508-14
380. Liu C, Wang JO, Zhou WY, Chang XY, Zhang MM, Zhang Y. et al. Long non-coding RNA LINC01207 silencing suppresses AGR2 expression to facilitate autophagy and apoptosis of pancreatic cancer cells by sponging miR-143-5p. Mol Cell Endocrinol. 2019;493:110424
381. Zhang J, Gao S, Zhang Y, Yi H, Xu M, Xu J. et al. MiR-216a-5p inhibits tumorigenesis in Pancreatic Cancer by targeting TPT1/mTORC1 and is mediated by LINC01133. Int J Biol Sci. 2020;16:2612-27
Author contact
![]() Corresponding authors: Jingfeng Tang (Jingfeng_hutcom) and Cefan Zhou (cefanedu.cn), National "111" Center for Cellular Regulation and Molecular Pharmaceutics, Key Laboratory of Fermentation Engineering (Ministry of Education), Hubei University of Technology, Zip/Postal Code: 430068, 28 NanLi Road, Wuhan, Hubei, China Phone: +86 15872398415
Corresponding authors: Jingfeng Tang (Jingfeng_hutcom) and Cefan Zhou (cefanedu.cn), National "111" Center for Cellular Regulation and Molecular Pharmaceutics, Key Laboratory of Fermentation Engineering (Ministry of Education), Hubei University of Technology, Zip/Postal Code: 430068, 28 NanLi Road, Wuhan, Hubei, China Phone: +86 15872398415

 Global reach, higher impact
Global reach, higher impact