10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(14):6179-6196. doi:10.7150/ijbs.110146 This issue Cite
Review
Circular RNA-Encoded Proteins in Disease Pathogenesis
1. Shenzhen Key Laboratory of Precision Diagnosis and Treatment of Depression, The Brain Cognition and Brain Disease Institute of Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen, Guangdong 518055, P.R. China.
2. Department of Mental Health and Public Health, Faculty of Life and Health Sciences, Shenzhen University of Advanced Technology, Shenzhen, Guangdong 518055, P.R. China.
3. University of Chinese Academy of Sciences, Beijing 100049, P. R. China.
4. Department of Neurology in Affiliated Zhongda Hospital and Jiangsu Provincial Medical Key Discipline, School of Medicine, Institute of Neuropsychiatry, Key Laboratory of Developmental Genes and Human Disease of Ministry of Education, Southeast University, Nanjing, Jiangsu 210009, P. R. China.
Received 2025-1-8; Accepted 2025-7-10; Published 2025-9-29
Abstract
Circular RNAs (circRNAs) are structurally stable and covalently-linked ring RNA molecules. Due to backsplicing, they are directly joined by phosphodiester linkage, which confers much greater stability relative to their linear mRNA counterparts. Recent studies indicate that circRNAs also encode proteins involved in mechanisms associated with the pathogenesis of various diseases, offering new treatment insights. This review briefly summarizes the history, characteristics, and functions of circRNAs; the formation and translation mechanisms of circRNA-encoded proteins; and computational and experimental techniques for identifying/predicting the protein-encoding potential of circRNAs. We also summarized their role in disease pathogenesis, which includes how they could be targeted and harnessed as novel therapeutic options for disease treatment. Finally, we stated some current limitations to studies on circRNA-encoded proteins and concluded with a discussion of future research directions to facilitate effective clinical translation.
Keywords: Circular RNAs, Circular RNA-encoded proteins or translatable circular RNAs, pathogenesis, molecular mechanisms, biomarkers, clinical relevance
Introduction
CircRNAs are an intricate class of endogenous, single-stranded (ss) RNA, formed predominantly by the backsplicing of precursor mRNAs (pre-mRNAs) [1, 2]. The formation of circRNAs is aided by the complementarity of the RNA sequences at both ends of the RNA transcript [3, 4]. Through the process of backsplicing, the downstream 5' splice site (splice donor) of the RNA molecule is joined via a phosphodiester bond to the upstream 3' splice site (splice acceptor), resulting in a covalently closed circRNA molecule [5].
The discovery of circRNAs dates to the mid-1970s when they were described as defective interfering (DI) particles with an uncharacteristic ability to form circular structures, indicating that DI-RNAs possess complementary ends [6]. That same year, circRNAs were also reported as viroids, which are single-stranded, highly thermostable infectious RNA molecules that affect higher plants [4]. The circular form of ssRNA was initially observed in the cytoplasm of eukaryotic cells, although its functions were largely unknown [3]. Within the next decade, yeasts such as Saccharomyces cerevisiae were found to contain mitochondrial circRNAs [7]. The circRNA genome was first observed in an animal virus: hepatitis delta (δ) [8]. In the 1990s, exonic circRNAs were reported as stable, mis-spliced products localized in the cytoplasmic partition in human eukaryotic cells, processed from nuclear precursor mRNAs [9]. Subsequently, studies began to emerge on the functional roles of some circRNAs and their abundance in mostly eukaryotic cells [10-12]. However, it is very crucial to note that extensive evidence has been provided that the majority of circular RNAs are indeed non-functional and are mainly byproducts of splicing errors [13]. Therefore, while some circular RNAs have been proven to possess functional roles, a great majority of them are likely non-functional splicing byproducts and may be detrimental [13].
The circular structure of circRNAs confers an unusual stability compared to their linear counterparts [14, 15]. As a result, circRNAs can resist degradation upon reaction with ribonucleases, particularly exonucleases [16]. Further studies have shown that circRNAs are highly tissue-specific and significantly enriched in the brain [11]. Despite being produced in the nucleus, circRNAs are mostly localized in the cytoplasm. A recent study showed that nuclear export of circRNAs from the nucleus to the cytoplasm is aided by nuclear Ran-GTP, which enhances the interaction of IGF2BP1 with circRNA, recruiting exportin-2 for the export of circRNAs from the nucleus to the cytoplasm [17]. During neurogenesis, additional factors necessary for efficient circRNA translocation have been identified. CircRNAs with adenosine (A)-rich motifs were retained in the nucleus through interaction with PABPC and remained trapped by TPR protein - this action was reversed following neuronal differentiation [18].
The development of high-throughput RNA-sequencing technology was a crucial breakthrough for the identification and quantification of novel circRNAs [19]. However, due to their complexity, circRNAs remain difficult to characterize. Modern methods rely on RNA-sequencing methodologies for circRNA characterization, which can contribute to false reads and result in the loss of the majority of circRNAs because of their low abundance [20].
At present, some widely known functions of non-coding circular RNAs include the following: acting as sponges for miRNAs [2], thereby regulating gene expression; interacting with RNA binding proteins (RBPs) [21]; and acting as transcriptional modulators by binding to and regulating RNA polymerase II (Pol II) [12, 22]. Most circRNAs do not contain ORFs or harbor specific IRES sequences, thus, would rarely undergo translation, and have been classified as untranslatable non-coding RNAs. For instance, studies employing ribosome profiling, standard proteomics and extensive analysis across species have shown that even among AUG circRNAs, which are the most conserved and abundant circRNA group, no indication of translation was recorded, thus suggesting that circRNAs are majorly non-coding [23]. However, recent works have revealed that some circRNAs actually do encode proteins implicated in notable disease-related pathways, such as the Wnt/β-catenin pathway, hedgehog signaling pathway, and MAPK/ERK pathway [24]. Thus, circRNA-encoded proteins have garnered much attention as potential therapeutic targets with available treatment options such as gene therapy approaches, small molecule inhibitors and antibody-based therapies. This review aims to explicitly shed more light on the clinical relevance of circRNA-encoded proteins, reported to play a significant role in the pathogenesis of various diseases ranging from cancer to neurological and cardiac diseases, and suggests that targeting these proteins would provide a fresh outlook and novel perspective towards providing long-lasting treatment for these diseases.
Therefore, this review focuses on known, translatable circRNAs and highlights their role and involvement in the pathogenesis of diseases in relation to protein-protein interactions, while discussing the ongoing challenges associated with profiling circRNA-encoded proteins.
Formation mechanisms of circRNAs
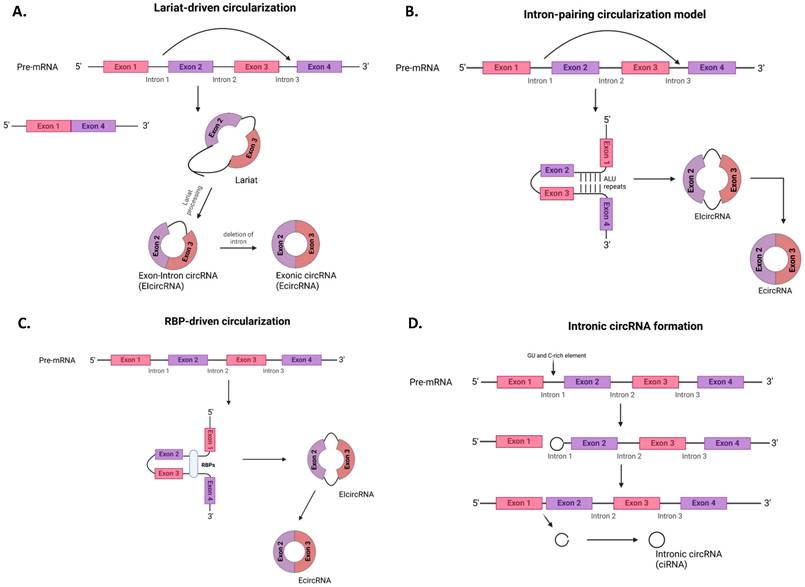
There are three major types of circular RNAs formed by different molecular mechanisms: exonic circular RNAs (EcircRNAs), exon-intron circular RNAs (EIcircRNAs), and intronic circular RNAs (ciRNAs) (Fig. 1) [12, 25, 26].
Lariat-driven model of circularization (exon-skipping)
EIcircRNAs are formed through the lariat-driven model of circularization commonly known as the exon-skipping method (Fig. 1a) [27]. Here, a linear mRNA with one or more skipped exons formed through traditional/canonical splicing gives rise to another RNA molecule called a lariat that contains the skipped exons. Subsequent lariat processing, followed by backsplicing, results in circRNA formation.
Intron-pairing circularization model
EcircRNAs and EIcircRNAs can be formed by the intron-pairing circularization model through complementarity, which is driven by the Alu repeats between adjacent introns located at the 5' and 3' splice sites of pre-mRNAs (Fig. 1b) [27].
RBP-driven circularization model
The formation of both EIcircRNAs and EcircRNAs can also be driven by the RBP-driven circularization model, which involves the linkage of the splice donor and splice acceptor of pre-mRNA by RBPs such as HNRNPL, MBNL1, FUS, etc. (Fig. 1c) [5, 28].
Intronic circular RNA (ciRNAs) formation
Finally, ciRNAs formation is aided by the localization of specific sequences, such as a 7 nt GU-rich element near the 5' splice site and an 11 nt C-rich element close to the branch site of the intron lariat (Fig. 1d) [5].
Formation mechanisms of circRNA. The different mechanisms for the formation of circular RNAs is shown above; a) Lariat-driven circularization model b) Intron-pairing circularization model c) RBP-driven circularization model d) Intronic circRNA formation. RBP, RNA-binding proteins. (Created in https://BioRender.com)
Comparison of translation mechanisms between circRNAs and mRNAs
CircRNAs were initially thought to be untranslatable or non-coding RNAs due to the absence of the 3' poly A tail and 5' N7 methylguanosine (m7G) cap required for canonical translation of mRNAs into their respective polypeptides. However, recent observations have debunked that idea. Some endogenous circRNAs possess an open reading frame (ORF) together with start and stop codons, which accentuates a broader potential for translation than previously recognized [29]. Thus, some circRNAs are indeed translatable and can undergo translation via mechanisms different from the canonical cap-dependent translational mechanism. Importantly, circRNAs have been identified in heavy polysomic fractions, as validated by ribosome fragment analysis, transcriptome-wide ribosome profiling, and polysome profiling data [30].
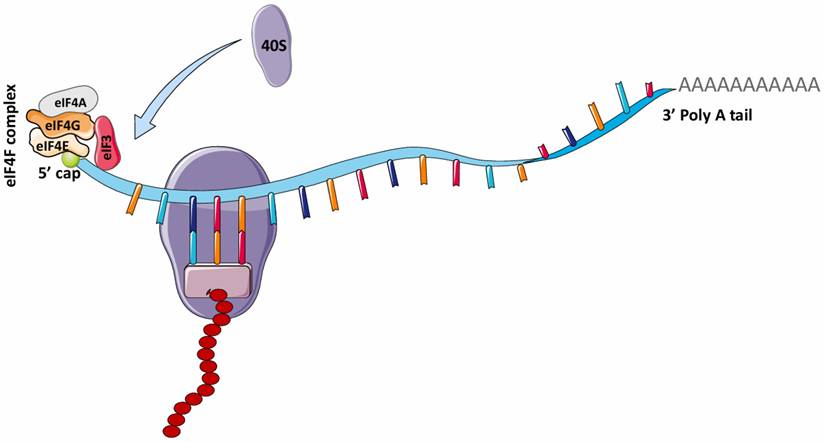
In 1975, it was reported that the addition of the 5' N7-methylguanosine (5' m7G) cap to eukaryotic mRNAs by the enzyme, guanine-7 methyltransferase is required to initiate protein synthesis (translation) in various eukaryotic cells, such as reovirus and vesicular stomatitis virus (VSV) mRNAs and reticulocyte mRNAs [31, 32]. This finding gave rise to the theory that the canonical cap-dependent translational mechanism is required for eukaryotic mRNA translation. For eukaryotic mRNAs, the traditional process of protein translation involves three major steps: initiation, elongation and termination. In mRNA translation initiation, the 5' m7G is recognized by eIF4F (eukaryotic initiation factor 4F), a protein complex comprising eIF4G (which binds eIF4E, eIF4A, and eIF3), eIF4A (an ATP-dependent RNA helicase), and eIF4E (Fig. 2) [33]. The binding of eIF4E to the 5' m7G structure of mRNA recruits these other factors to form a complex unit that enhances initiation of eukaryotic mRNA translation. During elongation, the ribosome simply travels along the mRNA and catalyzes the transfer and formation of peptide bonds between amino acids in the growing polypeptide chain. Upon termination, the polypeptide unit is released after recognition of the termination codon by the eukaryotic release factor (eRF).
However, this process is quite different for circRNAs and remains poorly understood. Research over the past decade has shown that circRNA can undergo translation through the independent ribosomal entry site (IRES) and non-IRES translation mechanisms. As neither process requires the presence of 5' or 3' ends for translation initiation, these are known as cap-independent translation mechanisms.
IRES translation mechanism
IRES are small, CU-rich, highly-structured nucleotide sequences of variable length that can recruit ribosomes (40S complex or ribosomal subunit) and translation initiation factors to facilitate cap-independent translation [34]. Several studies have confirmed that initiation of translation in circRNAs occurs through an IRES-mediated mechanism [35, 36]. In this form of translation, the binding of the ribosomal subunit to the IRES for translation is facilitated by the presence of eIF4G and eIF4A components on a specific domain of the IRES (Fig. 3) [33]. Also, it has been reported that IRES-like short elements consisting of AU-rich hexamer sequences, located in the 3' UTR of linear mRNAs, can also drive circRNA translation [29]. The IRES translation mechanism has been mainly observed in cells in severe stress or disease conditions [37]. Fan et al demonstrated that some trans-acting factors can function as RBPs by binding to IRES elements to initiate circRNA translation [29]. Chen et al maximized in vitro and in vivo translation of synthetic circRNAs by optimizing IRES elements, synthetic aptamers, 5' and 3' UTR, and vector topology, validating the role of IRES in circRNA translation initiation. Notably, increased in vitro circRNA translation was observed and may deliver more durable translation in vivo than mRNA protein expression [36]. When combined with IRES, other RBPs can aid in the initiation of circRNA translation. Zhong et al reported that the DEAD-box helicase 3 (DDX3) interacted with the IRES sequence of circEZH2 to drive EZH2-92aa protein translation. However, the involvement of other regulatory elements remains unknown [38].
mRNA translation in eukaryotes. The attachment of the eIF4F complex (eIF4G, eIF4A, eIF4E) and eIF3 complex to the 5' cap of mRNA is necessary for recruiting 40S ribosome for translation. This is known as the canonical cap-dependent translation. eIF4F, Eukaryotic initiation factor 4F; eIF4G, Eukaryotic initiation factor 4G; eIF4A, Eukaryotic initiation factor 4A; eIF3, Eukaryotic initiation factor 3. (Created in Servier Medical Art)
Various mechanisms of circRNA translation. CircRNA translation can proceed via different mechanisms depending on the internal features of the circRNA. In the presence of IRESs, IRES-like sequences such as m6A, or in their absence (for rolling circle translation), the respective eIFs recruit the 40S ribosome for circRNA translation. IRES, Internal ribosome entry site; eIF4G, Eukaryotic initiation factor 4G; eIF4A, Eukaryotic initiation factor 4A; eIF4G2, Eukaryotic initiation factor 4G2; m6A, N6-methyladenosine; YTHDF3, YTH N6-methyladenosine RNA Binding Protein 3. (Created in Servier Medical Art)
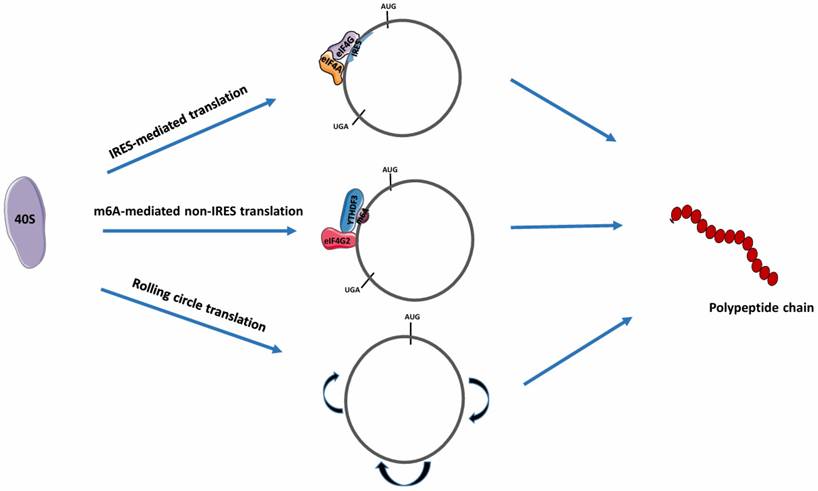
Non-IRES translation mechanism
This mechanism involves N6-methyl adenosine (m6A) mediated protein translation and rolling circle translation.
In m6A-mediated protein translation, the m6A sites in the 5' UTR are positioned on short nucleotide sequences. The binding of YTHDF3, an m6A -binding protein to circRNAs, enables recruitment of the eIF4G2 and ribosome complex, leading to the initiation of circRNA translation (Fig. 3) [39]. Tang et al reported that most circRNAs in male germ cells possess large ORFs with junction sequences containing significantly enriched m6A-modified start codons [40]. These junction sequences enable peptide synthesis by encoding peptides, validating the mechanism of m6A-dependent protein translation. Proteins besides YTHDF3 can also facilitate m6A modification during circRNA translation. Through m6A modification, facilitated by IGF2BP1, the translation of circMAP3K4 produces circMAP3K4-455aa, a 455 amino acid (aa) protein, which is associated with the progression of hepatocellular carcinoma [41].
For rolling circle translation, circRNAs with an infinite ORF (i.e., without stop codons) that only contain coding sequences can be efficiently translated into a large protein concatemer via a rolling circle amplification mechanism [42]. In this process, once translation is initiated, elongation proceeds indefinitely, and the ribosome does not have to continuously bind to the RNA (Fig. 3) [43]. Abe et al showed that synthetic circRNAs with an infinite ORF prepared in vitro can be translated in living human cells devoid of particular elements such as IRES, a 3' poly-A tail, or a 5' cap structure [44]. Notably, this process can produce a greater amount of protein compared to circRNAs with stop codons [29].
Although the protein-encoding potential of circRNAs containing IRESs has been deciphered, the fundamental principle of circRNA translation remains an open question. A thorough understanding of the mechanisms that drive circRNA translation is thus essential for developing efficient circRNA therapeutics capable of overcoming mRNA's fleeting nature and potentially exceeding the translation capabilities of mRNA [36].
Bioinformatic tools and experimental techniques for detecting translatable circRNAs
The identification and study of circRNA-encoded proteins have been facilitated by recent advances in computational/bioinformatic tools (Table 1). Although bioinformatics analysis is useful, experimental techniques are necessary to provide evidence of the protein-encoding ability of most circRNAs (Table 2).
Bioinformatic tools/databases and corresponding websites
Experimental techniques for detecting translatable circRNAs
| Technique | Description | References |
|---|---|---|
| RNA seq, m6A-seq, RNC-seq, YTHDF2 RIP-seq, ribosome profiling, ribosome immunoprecipitation, Ribosome fragment analysis, nanopore sequencing, ribosome affinity purification | To identify circular RNAs with encoding functions. | [30, 118] |
| Northern blotting | To validate circRNA expression and characterize the output of the overexpression of circRNAs using plasmids/vectors | [20, 119] |
| Sanger sequencing | To confirm the BSJ site of circRNAs | [120] |
| High-performance liquid chromatography-mass spectrometry (LC-MS), co-immunoprecipitation (co-IP), reciprocal immunoprecipitation, immunofluorescence staining | To provide direct evidence by identifying specific peptides | [38, 58, 121] |
| ELISA, western blotting | To detect putative peptides or proteins | [30, 122] |
| Dual luciferase reporter assay | For experimental evaluation of IRES/m6A activity of translatable circRNAs. | [114] |
| CCK-8, colony formation, EdU, Transwell and wound healing assays | For in vitro assessment of the influence of peptides on cell growth, migration, and invasion | [81] |
| Tagged RNA affinity purification (TRAP) assay and RNA immunoprecipitation (RIP) assay. | To identify/verify interaction between individual proteins and RNA. | [112] |
| si/shRNAs and CRISPR-Cas13 system. | To deplete circRNAs by targeting BSJs | [20, 122] |
| Plasmid and viral expression | For circRNA overexpression | [18, 45, 73] |
Functions and characteristics of proteins expressed from circRNAs
Our current understanding of the characteristics and functions of circRNA-encoded proteins has been broadened due to extensive evidence from recently published studies confirming their roles in various disease pathogenesis, such as cancer metastasis, cardiac and neurological diseases. The unusual stability of circRNAs, which results from their circular structure, allows for a longer expression of the proteins they encode [45, 46]. More so, circular RNAs are tissue-specific, which implies that the expression of the respective circRNA-encoded protein is exclusive to a particular tissue and can be targeted specifically [47, 48]. This suggests a prominent role of circular RNAs in affecting cellular physiology within specific tissues, serving as potential biomarkers for tissue-related diseases. Most importantly, most circRNAs originate from the nucleus but are transported to the cytoplasm, where they are subsequently translated into proteins that modulate various cellular pathways and physiology. In this section, we focus on a few of these circRNA-encoded proteins and their respective roles in disease progression.
Roles of circRNA-encoded proteins in cancer progression
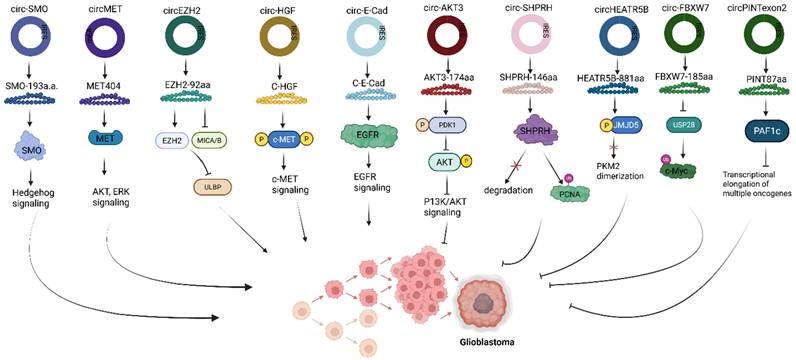
Circular RNA-encoded proteins can either function to inhibit or promote cancer tumor pathogenesis. For example, in glioblastoma, C-E-Cad, a 254 amino acid protein variant, encoded by the oncogenic circRNA, circ-E-Cad, is among the few circRNAs found to be upregulated in tumorigenic cells (Fig. 4) [49]. C-E-Cad activates the EGFR signaling pathway by binding to the EGFR CR2 domain, leading to glioblastoma tumorigenesis [49]. In another study, lending credence to the regulatory role of circRNA-encoded protein, highlights the significance of the Hedgehog pathway in driving tumorigenesis of many cancers, including glioblastoma. SMO-193aa, encoded by the circRNA circular SMO (circ-SMO), was shown to be crucial in the signaling activation of the Hedgehog pathway in glioblastoma [50]. Circ-SMO is produced from exons 3 - 6 of the SMO gene, translating into a novel protein SMO-193aa via the IRES-mediated circRNA translation mechanism (Fig. 4). Mechanistically, SMO-193aa, by sharing most of its predicted five-domain transmembrane sequence with SMO, increases SMO cholesterol modification by binding to SMO, thus inhibiting the action of Ptch1 protein [50]. Consequently, the Gli1 protein is activated for Shh downstream signaling. Fused in sarcoma (FUS), a downstream Gli1 protein target, is an RBP that has been shown to positively regulate the backsplicing of circ-SMO [51], promotes circ-SMO transcription. The network of Shh-Gli1-FUS-SMO-193aa forms a cycle that sustains HH signaling activation, resulting in glioblastoma progression [50].
Roles of translatable circRNAs in glioblastoma. Circ, Circular RNA; IRES, Internal ribosome entry site; SMO, G protein‑coupled‑like receptor smoothened; MET, Mesenchymal to epithelial transition; EZH2, Enhancer of Zeste Homolog 2; MICA/B, Major histocompatibility complex class I polypeptide-related sequence A/B; ULBP, UL16-binding protein; c-HGF, Cell-surface hepatocyte growth factor; c-MET, Cell-surface MET receptor; E-Cad, E-cadherin; EGFR, Epidermal growth factor receptor; PDK1, 3‑phosphoinositide‑dependent kinase 1; AKT, Protein kinase B; P13K/AKT, Phosphoinositide 3-Kinase/ Protein Kinase B; SHPRH, SNF2 histone linker PHD RING helicase; PCNA, proliferating cell nuclear antigen; HEATR5B, HEAT repeat containing 5B; JMJD5, Jumonji‑C domain‑containing protein 5; PKM2, Pyruvate Kinase M2; FBXW7, F‑box and WD repeat domain containing 7; USP28, ubiquitin specific peptidase 28; c-Myc, Cellular myelocytomatosis oncogene; PINT, p53‑induced transcript; PAF1c, Polymerase‑associated factor complex. Reproduced with permission from REF [24], CC BY 4.0 (https://creativecommons.org/licenses/by/4.0) (Created in https://BioRender.com)
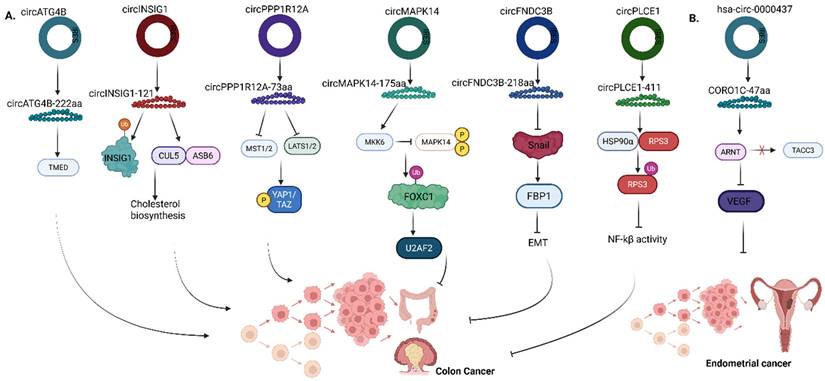
Autophagy is one of the key processes that induces chemoresistance, and enhances cancer cell survival. Previous studies have shown that the expression of a key autophagy-related factor known as ATG4B, significantly increased cancer chemoresistance in gastric, colorectal and colon cancers [52-54]. ATG4B acts by cleaving the MAP1LC3/LC3 complex to form LC3-I and is subsequently bound to phosphatidylethanolamine (PE) to generate lipidated LC3-II on phagophore membranes [55-57]. In colorectal cancer (CRC), the expression of circATG4B-222aa, encoded by circATG4B, was directly related to poor prognosis in patients with CRC who were treated with oxaliplatin. CircATG4B-222aa directly binds to TMED10, preventing TMED10 interaction with ATG4B, thus increasing autophagy, which triggers oxaliplatin resistance, thereby reducing the chemosensitivity of CRC cells (Fig. 5a) [56].
In gastric cancer, AXIN1-295aa, encoded by circAXIN1, functions as an oncogenic protein by interacting with adenomatous polyposis coli (APC) competitively (Fig. 6a) [58]. Functionally, APC forms a complex with glycogen synthase kinase 3 protein (GSK3), casein kinase 1 (CK1) and AXIN protein to recruit E3 ubiquitin ligase β-trcp to degrade β-catenin [59]. However, the competitive interaction with AXIN1-295aa leads to the dysregulation of the Wnt signaling pathway, thus allowing β-catenin to effectively translocate into the cellular nucleus, binding to the transcription factors (TCF/LEF) and activating downstream signaling, which promotes the progression and metastasis of gastric cancer cells [58].
In multiple myeloma (MM), Tang et al discovered that a novel peptide, circHNRNPU_603aa, which enhances the progression of MM, is encoded by the circular RNA, circHNRNPU (Fig. 6b) [60]. Analysis of the putative ORF of circHNRNPU on circRNA database showed that it contains an IRES sequence (from +201 to +374) and the ORF can potentially encode a 603-aa peptide, thus named, circHNRNPU_603aa. To determine if circHNRNPU_603aa promotes MM progression, colony formation assays, MTT and cell cycle assays were performed. The results of these assays indicated that circHNRNPU_603aa promoted MM cell proliferation and clonal expansion. Mechanistically, following the secretion of circHNRNPU, which encodes circHNRNPU_603aa in the bone marrows by MM cells, through the RNA-binding RGG-box region, circHNRNPU_603aa significantly regulates backsplicing of SKP2 exon and activates AMN1 to stabilize c-myc, thereby competitively inhibiting c-Myc ubiquitin (Fig. 6b) [60].
Roles of translatable circRNAs in the pathogenesis of (A) Colon cancer (B) Endometrial cancer. ATG4B, Autophagy Related 4B Cysteine Peptidase; TMED, Transmembrane Emp24 Domain-containing protein; INSIG1, Insulin Induced Gene 1; CUL5-ASB6 Complex, Cullin 5 - Ankyrin Repeat and SOCS Box containing protein 6; PPP1R12A - Protein Phosphatase 1 Regulatory Subunit 12A; MST1/2, Mammalian STE20-like protein kinase 1 and 2; LATS1/2, Large Tumor Suppressor Kinase 1 and 2; YAP1/TAZ, Yes-associated protein 1/Transcriptional coactivator with PDZ binding domain; MAPK14, Mitogen-activated protein kinase 14; MKK6, Mitogen-activated protein kinase kinase 6; FOXC1, Forkhead box C1; U2AF2, U2 small nuclear RNA auxiliary factor 2; FNDC3B, Fibronectin type III domain‑containing protein 3B; FBP1, Fructose-1,6-bisphosphatase; EMT, Epithelial‑mesenchymal transition; PLCE1, Phospholipase C Epsilon 1; HSP90α, heat‑shock protein 90α; RPS3, ribosomal protein S3; NF-Κb, Nuclear factor kappa-light-chain-enhancer of activated B cells; CORO1C - Coronin 1C; ARNT, Aryl hydrocarbon receptor nuclear translocator; TACC3, Transforming acidic coiled-coil containing protein 3; VEGF, Vascular endothelial growth factor. Reproduced with permission from REF [24], CC BY 4.0 (https://creativecommons.org/licenses/by/4.0) (Created in https://BioRender.com)
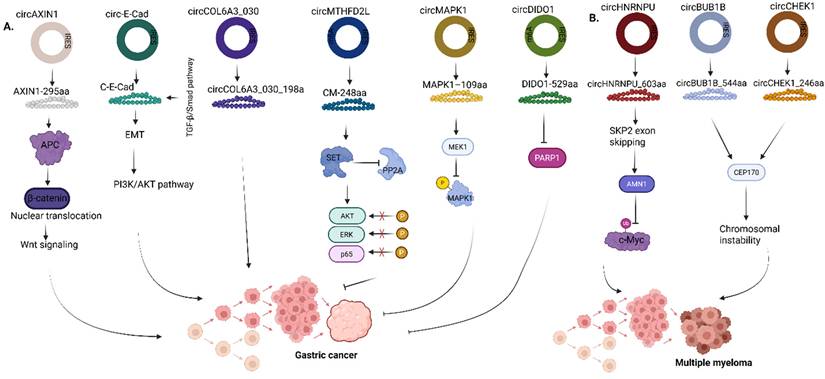
Roles of circRNA-encoded proteins in (A) Gastric cancer (B) Multiple myeloma. AXIN1, Axis inhibition protein 1; APC, Adenomatous polyposis coli; ERK, Extracellular signal-regulated kinase; MAPK1, Mitogen-activated protein kinase 1; MEK1, Mitogen-activated protein kinase kinase 1; DIDO1, Death inducer-obliterator 1; PARP1, Poly (ADP-Ribose) Polymerase 1; HNRNPU, Heterogeneous nuclear ribonucleoprotein U; SKP2, S-phase kinase-associated protein 2; AMN1, Aminopeptidase N 1; BUB1B, BUB1 mitotic checkpoint serine/threonine kinase B; CHEK1, Checkpoint kinase 1; CEP170, Centrosomal protein 170. Reproduced with permission from REF [24], CC BY 4.0 (https://creativecommons.org/licenses/by/4.0) (Created in https://BioRender.com)
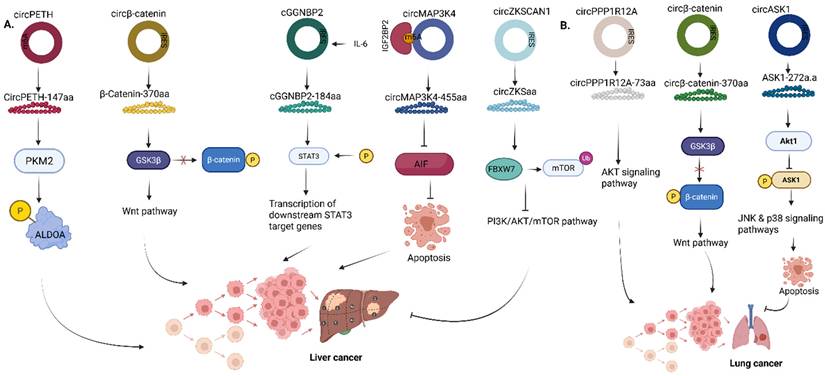
In liver cancer, the circRNA, circZKSCAN1, encodes a secretory protein located in the liver, circZKSaa, which is highly significant in the inhibition of proliferation of hepatocellular carcinoma (HCC) cells (Fig. 7a) [61]. The phosphoinositide-3-kinase (P13K)/AKT signaling pathway is an important pathway involved significantly in the regulation of various cellular processes, including cell metabolism, angiogenesis, cell proliferation and viability [62]. Irregularities in the P13K/AKT signaling pathway have been associated with the onset of different types of cancers, diabetes, inflammatory and autoimmune diseases, cardiovascular and neurological diseases [63-66]. circZKSaa overexpression inhibits HCC tumor metastasis by interacting with mTOR and also facilitating the interaction of FBXW7 with mammalian target of rapamycin (mTOR), which is one of the downstream members of the Akt pathway (Fig. 7a) [61].
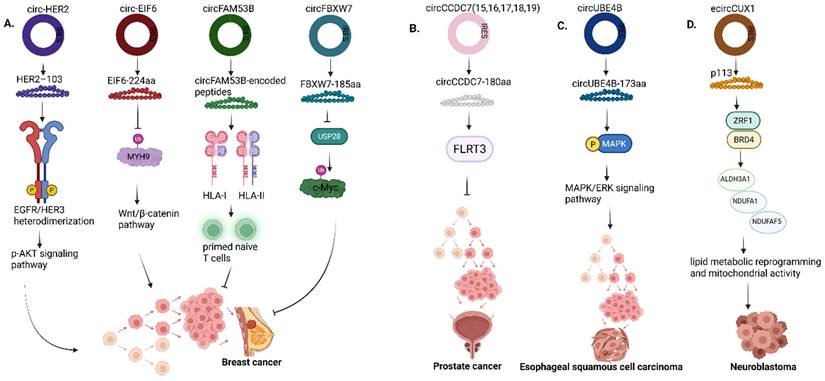
In breast cancer, EIF6-224aa encoded by circ-EIF6 was expressed endogenously in triple-negative breast cancer (TNBC) cells and tissues using a specific target antibody [67]. Although the KD of circ-EIF6 alleviated tumor cell progression of TNBC, the study showed that EIF6-224aa activates the Wnt/beta-catenin pathway via interaction with MYH9 protein (Fig. 8a). In the canonical Wnt pathway, the binding of the Wnt proteins to its receptors causes the release of β-catenin and its translocation into the nucleus, leading to the activation of downstream target genes such as CCND1, c-Myc, etc. involved in cell proliferation and migration. Conversely, the absence of Wnt proteins causes the ubiquitination of β-catenin by E3 ubiquitin ligase, and thus β-catenin cannot interact with TCF/LEF, which represses the expression of downstream target genes [59, 68]. Functionally, it was validated that EIF6-224aa overexpression decreased MYH9 ubiquitination, thereby shielding MYH9 from proteasomal degradation, and the expression of MYH9 correlates with the activation of the Wnt/beta-catenin pathway, as shown through the upregulation of downstream target genes such as CCND1, AXIN2, NKD1, etc, thereby leading to TNBC progression.
Finally, in prostate cancer, Wang et al showed that the circular RNA circCCDC7 (15,16,17,18,19) derived from exons 15 to 19 of the CCDC gene encodes a novel protein, circCCDC7-180aa, which functions as a tumor suppressor in prostate cancer by upregulating Fibronectin leucine-rich transmembrane protein 3 (FLRT3) (Fig. 8b) [69]. FLRT3 has already been linked to the regulation of endothelial cell survival stimulated by VEGF signaling, cell migration and tube formation in endothelial cells and neuronal cell outgrowth and morphogenesis [69-71].
Roles of circRNA-encoded proteins in neurological and cardiac diseases
Compared to the research on circRNA-encoded proteins in the pathogenesis of various types of cancers, few works have been done to decipher their roles in the pathogenesis of neurological and cardiac diseases due to several limitations. Mo et al discovered that a circular RNA, circAβ-a, which harbors the Aβ-coding region of the APP gene, encodes a novel Aβ-containing Aβ175 polypeptide (Fig. 9a) [72]. Aβ175 protein is processed into Aβ peptides, significant in Alzheimer's disease pathology. In this study, an alternative route for Aβ peptide production was confirmed because the Aβ175 contains β and γ-secretase cleavage sites; thus, there is a high probability of amyloid beta peptide production from Aβ175 through the cleavage of the β- and γ-secretase [72].
Roles of circRNA-encoded proteins in (A) Liver cancer (B) Lung cancer. PKM2, Pyruvate kinase M2; ALDOA, Aldolase A; GSK3β, Glycogen synthase kinase 3β; GGNBP2, Gamma-glutamyl-glycine beta-cytosine peptidase 2; STAT3, Signal transducer and activator of transcription 3; AIF, Apoptosis-inducing factor; MAP3K4, Mitogen-activated protein kinase kinase kinase 4; mTOR, Mechanistic target of rapamycin; ASK1, Apoptosis signal-regulating kinase 1. Reproduced with permission from REF.[24], CC BY 4.0 (https://creativecommons.org/licenses/by/4.0) (Created in https://BioRender.com)
Roles of circRNA-encoded proteins in (A) Breast cancer (B) Prostate cancer (C) Esophageal squamous cell carcinoma (D) Neuroblastoma. HER2, Human epidermal growth factor receptor 2; EGFR, Epidermal growth factor receptor; HER3, Human epidermal growth factor receptor 3; EIF6, Eukaryotic translation initiation factor 6; MYH9, Myosin Heavy Chain 9; FAM53B, Family with sequence similarity 53 member B; HLA I & II, Human leukocyte antigen I & II; CCDC7, Coiled-coil domain containing 7; FLRT3, Fibronectin leucine rich transmembrane protein 3; UBE4B, Ubiquitination factor E4B; MAPK, Mitogen-activated protein kinase; ERK, Extracellular signal-regulated kinases; ZRF1, Zuotin-related factor 1; BRD4, Bromodomain containing protein 4; ALDH3A1, Aldehyde dehydrogenase 3 family member A1; NDUFA1, NADH:Ubiquinone oxidoreductase subunit A1, NDUFAF5, NADH:Ubiquinone oxidoreductase complex assembly factor 5. Reproduced with permission from REF [24], CC BY 4.0 (https://creativecommons.org/licenses/by/4.0) (Created in https://BioRender.com)
Roles of circRNA-encoded proteins in (A) Alzheimer's disease (B) Major depressive disorder (C) Cardiac disease. Aβ, amyloid beta; FKBP8, FK506-binding protein 8; GR, Glucocorticoid receptor; HPA, Hypothalamic - pituitary - adrenal axis; Nlgn, Neuroligin; ING4, Inhibitor of growth family member 4; C8orf44-SGK3, Chromosome 8 open reading frame 44 - Serum/Glucocorticoid regulated kinase 3 fusion. Reproduced with permission from REF [24], CC BY 4.0 (https://creativecommons.org/licenses/by/4.0) (Created in https://BioRender.com)
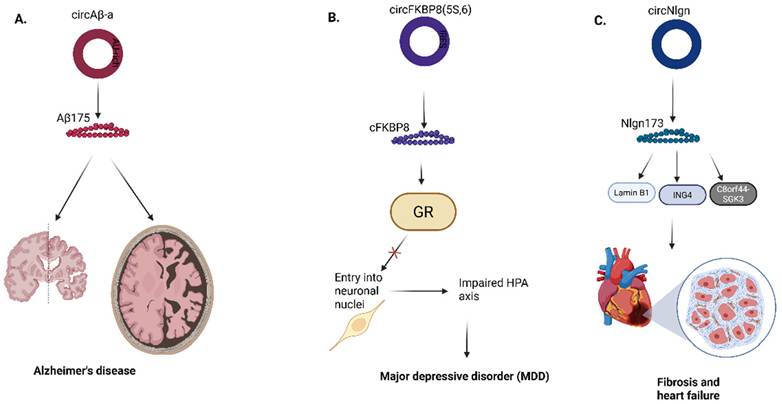
Recently, in the first study conducted on the role of circRNA-encoded proteins in the pathogenesis of major depressive disorder (MDD), Jiao et al showed that a circRNA termed circFKBP8(5S,6), which encodes a 127 aa protein termed cFKBP8, is significantly upregulated in stressed neuroblastoma cell lines and plasma neuronal-derived exosomes of human MDD patients. They further proved that cFKBP8 contributes to dysfunction of the hypothalamus-pituitary-adrenal (HPA) axis by preventing nuclear translocation of the glucocorticoid receptor (Fig. 9b) [73].
Lastly, in cardiac diseases, Du et al identified a circular RNA, circNlgn, produced by neuroglin that encodes a novel protein isoform, Nlgn 173, which promotes heart failure and cardiac remodeling (Fig. 9c) [74]. Nlgn173 contains a binding site for LaminB1, a protein located within the nuclear membrane, which aids in binding transcription factors [75]. The nuclear translocation of Nlgn173 is achieved via the interaction of LaminB1 with Nlgn173, where it functions as a transcription factor [74]. Past studies elucidated that SGK3 potentially interacts with and phosphorylates GSK-3β at serine-9, to form the pS9-GSK3β complex, which inactivates GSK3 activity and induces hypertrophic cardiomyopathy [76, 77]. On the other hand, ING4 can also induce cellular apoptosis and inhibit tumor cell growth through its interaction with p53 or BCL2 family proteins [78, 79].
Systematic circRNA-encoded protein discovery
The process of circRNA-encoded protein discovery in tumor diseases is similar to that for non-tumor diseases, such as cardiac diseases. This process involves the integration of both experimental and bioinformatics techniques for efficient circRNA identification. CircRNA-encoded proteins can regulate the pathogenesis of tumor diseases by either inhibiting or promoting tumor metastasis (Table 3, Figs. 4-8).
First, RNAs are extracted from tissue samples obtained from tumors and control patients. To efficiently identify and quantify circRNAs, high-throughput RNA sequencing is then carried out, and, various bioinformatics analysis tools, such as find_circ or circExplorer, are employed to identify unique circRNAs based on their characteristic back-splice junctions (BSJs). Subsequently, the already identified circRNAs are validated using techniques such as RT-PCR or Sanger sequencing. Because circRNAs exhibit differential levels of expression in tumors and normal tissues, comparing both tissues allows for the identification and confirmation of differentially expressed circular RNAs (DeCRs). Finally, the predominant cellular localization of circRNA is ascertained using fluorescent in situ hybridization (FISH) or subcellular fractionation assay to give insights into its protein-encoding ability.
Translatable circRNAs and their roles in disease pathogenesis
| CircRNA | Protein | Disease | Translation mechanism | Protein interaction | References |
|---|---|---|---|---|---|
| circFAM53B | circFAM53B-encoded peptides (Upregulated) | Breast cancer | IRES-mediated | HLA-I and HLA-II molecules | [45] |
| circ-EIF6 | EIF6-224aa (Upregulated) | Triple Negative Breast cancer (TNBC) | IRES-mediated | MYH9 | [67] |
| circFBXW7 | FBXW7-185aa (Downregulated) | TNBC | IRES-mediated | USP28 | [123] |
| circ-HER2 | HER2-103 (Upregulated) | TNBC | IRES-mediated | EGFR/HER3 | [124] |
| circNlgn | Nlgn173 (Upregulated) | Cardiac disease | - | LaminB1, ING4 & C8orf44-SGK3 | [74] |
| circPLCE1 | circPLCE1‑411 (Downregulated) | Colorectal cancer | IRES-mediated | HSP90α/RPS3 complex | [107] |
| circPPP1R12A | circPPP1R12A-73aa (Upregulated) | Colorectal cancer | - | YAP1 | [125] |
| circFNDC3B | circFNDC3B-218aa (Downregulated) | Colorectal cancer | IRES-mediated | Snail | [121] |
| circINSIG1 | circINSIG1-121 (Upregulated) | Colorectal cancer | IRES-mediated | INSIG1 | [126] |
| circATG4B | circATG4B-222aa (Upregulated) | Colorectal cancer | IRES-mediated | TMED10 | [56] |
| circMAPK14 | circMAPK14‑175aa (Downregulated) | Colorectal cancer | IRES-mediated | MKK6 | [127] |
| hsa-circ-0000437 | CORO1C-47aa (Downregulated) | Endometrial cancer | IRES-mediated | ARNT | [122] |
| circAXIN1 | AXIN1-295aa (Upregulated) | Gastric cancer | IRES-mediated | APC | [58] |
| circMAPK1 | MAPK1-109aa (Downregulated) | Gastric cancer | IRES-mediated | MEK1 | [81] |
| circ-E-Cad | C-E-Cad (Upregulated) | Gastric cancer | IRES-mediated | p-AKT and p-PDK1 | [128] |
| circDIDO1 | DIDO1-529aa (Downregulated) | Gastric cancer | IRES-mediated/ m6A modification | PARP1 | [112] |
| circMTHFD2L | CM-248aa (Downregulated) | Gastric cancer | IRES-mediated and m6A modification | SET | [101] |
| circCOL6A3_030 | circCOL6A3_030_198aa (Upregulated) | Gastric cancer | IRES-mediated | - | [129] |
| circ-SMO | SMO-193a.a. (Upregulated) | Glioblastoma | IRES-mediated | SMO | [50] |
| circ-SHPRH | SHPRH-146aa (Downregulated) | Glioblastoma | IRES-mediated | SHPRH | [130] |
| circ-AKT3 | AKT3-174aa (Downregulated) | Glioblastoma | IRES-mediated | p-PDK1 | [131] |
| circEZH2 | EZH2-92aa (Upregulated) | Glioblastoma | IRES-mediated | MICA/B | [38] |
| circMET | MET404 (Upregulated) | Glioblastoma | m6A modification | MET | [30] |
| circ-E-Cad | C-E-Cad (Upregulated) | Glioblastoma | IRES-mediated | EGFR | [49] |
| circ-FBXW7 | FBXW7-185aa (Downregulated) | Glioblastoma | IRES-mediated | USP28 | [132] |
| circ-HGF | C-HGF (Upregulated) | Glioblastoma | IRES-mediated | c-MET | [133] |
| CircPINTexon2 | PINT87aa (Downregulated) | Glioblastoma | IRES-mediated | PAF1c | [134] |
| circHEATR5B | HEATR5B‑881aa (Downregulated) | Glioblastoma | IRES-mediated | JMJD5 | [114] |
| circPETH | CircPETH-147aa (Upregulated) | Hepatocellular carcinoma (HCC) | m6A modification | PKM2, ALDOA, HuR | [84] |
| circMAP3K4 | circMAP3K4‑455aa (Upregulated) | HCC | m6A modification | AIF | [41] |
| circZKSCAN1 | CircZKSaa (Downregulated) | HCC | IRES-mediated | mTOR | [61] |
| circβ-catenin | β-Catenin-370aa (Upregulated) | Liver cancer | IRES-mediated | GSK3β | [135] |
| cGGNBP2 | cGGNBP2-184aa (Upregulated) | Intrahepatic cholangiocarcinoma (ICC) | IRES-mediated | STAT3 | [136] |
| circASK1 | ASK1-272a.a (Downregulated) | Lung cancer | IRES-mediated | Akt1 | [85] |
| circPPP1R12A | circPPP1R12A-73aa (Upregulated) | Lung cancer | - | p-AKT | [137] |
| circβ‑catenin | circβ‑catenin‑370aa (Upregulated) | Non-small cell Lung cancer | IRES-mediated | GSK3β | [138] |
| circHNRNPU | circHNRNPU_603aa (Upregulated) | Multiple myeloma | IRES-mediated | AMN1 | [60] |
| circBUB1B | circBUB1B_544aa (Upregulated) | Multiple myeloma | IRES-mediated | CEP170 | [139] |
| circCHEK1 | circCHEK1_246aa (Upregulated) | Multiple Myeloma | IRES-mediated | CEP170 | [140] |
| circCCDC7(15,16,17,18,19) | circCCDC7-180aa (Downregulated) | Prostate cancer | IRES-mediated | FLRT3 | [69] |
| circAβ-a | Aβ175 (Upregulated) | Alzheimer's disease | IRES-like elements (AU-rich) | - | [72] |
| circUBE4B | circUBE4B‑173aa (Upregulated) | Esophageal squamous cell carcinoma | IRES-mediated | MAPK1 | [141] |
| ecircCUX1 | p113 (Upregulated) | Neuroblastoma | IRES-mediated | ZRF1 and BRD4 | [117] |
When studying circRNA-encoded proteins, it is necessary to predict the protein-encoding potential. The presence of an ORF sequence within the circRNA sequence indicates that a circRNA is indeed translatable [29]. Thus, circRNA sequences can be analyzed using online databases and tools containing a repository of information on circRNA supported by experimental evidence. In the case of IRES-mediated translation, the ORFs contain internal IRES sequences that initiate protein translation. Ribosome profiling can be used to confirm the association of circRNAs with ribosomes, suggesting translation into proteins, and further validated by LC-MS, Co-IP, or western blotting (by directing specific antibodies to precise peptide terminal sequences).
The effect of knockdown of circRNA or its encoded protein using siRNAs or CRISPR/Cas technologies in animal models or selected cell lines is assessed based on changes in cell migration, proliferation, and invasion [80]. Proper elucidation of the mechanism that drives the encoded protein to exert its tumor proliferative or inhibitory effect must be established. For example, Jiang et al found that MAPK1-109aa is significantly downregulated in patients with gastric cancer [81]. Thus, overexpression of circMAPK1, acting through its encoded protein, MAPK1-109aa, inhibits metastasis of gastric tumor cells. MAPK1-109aa acts as a tumor inhibitor by binding to MEK1 (mitogen-activated protein kinase kinase), thereby inhibiting phosphorylation of MAPK1 and its downstream effectors [81].
Circular RNA-encoded proteins as potential therapeutic targets
The discovery that circular RNA can actually encode various proteins that are implicated in mechanisms, pathways and signaling processes associated with various diseases, opened up a new window of opportunity towards their potential target for therapeutic intervention. This section explores some potential therapeutic targets associated with circRNA-encoded proteins and how they impact disease pathogenesis.
Glioblastoma is the most common form of brain cancer, and given that activated MET signaling/ MET alteration contributes significantly to glioblastoma [82], a thorough understanding of the MET signaling pathway is required to identify functional therapeutic targets to aid the development of effective treatment for this disease. Here, MET404, a 404-amino-acid encoded by the circular MET RNA, circMET, facilitates glioblastoma tumorigenesis, thus indicating its clinical relevance as a potential molecular target [30]. In mice, abrogation of MET signaling was achieved through the genetic alteration of circMET, which reduced MET404 expression. MET 404 promotes tumor cell proliferation by directly binding to the MET β subunit, leading to the outright activation of the MET receptor and downstream effectors (Fig. 4a). Previously, HGF was the only known MET ligand that binds to the SEMA domain on MET [30, 83]. This study revealed that MET 404 is also a MET ligand and binds to the (PSA+IPTI) domain on the MET receptor. Knockdown of the circMET gene was also shown to repress the expression of MET 404 in mice, while high expression of MET 404 further exacerbates glioblastoma tumorigenicity in vivo and in vitro. Thus, targeting MET 404 can lead to a potential treatment option for glioblastoma patients with MET hyperactivation. To confirm this, genetic ablation of circMET or the combined administration of MET404 antibody and onartuzumab, an FDA-approved traditional MET signaling inhibitor, significantly repressed MET signaling and improved cellular viability [30].
In a recent study, circPETH-147aa, encoded by circPETH (derived from exons 13-15 of the low-density lipoprotein receptor gene), was shown to enhance glycolysis and metastasis in HCC cells (Fig. 7a) [84]. A druggable MEG pocket of circPETH-147aa was identified and was confirmed to interact with PKM2, ALDOA and HuR. Overexpression of circPETH-147aa increased ALDOA phosphorylation by PKM2, increasing HuR-dependent SLC43A2 mRNA stability, which enhances cytotoxic CD8+ T cells exhaustion and abrogates anti-HCC immunity. High-throughput virtual screening identified a small-molecule compound, Norathyriol, which is a natural product that effectively showed a profound interaction with the MEG pocket of circPETH-147aa and was validated experimentally. Taken together, administration of Norathyriol alone or in combination with anti-PD1 antibodies effectively strengthened the activity of CD8+ T cells in a mouse model of lung metastasis and liver orthotopic intrahepatic metastasis [84].
Furthermore, Wang et al discovered that a novel protein encoded by circASK1, ASK1-272a.a, is involved in lung adenocarcinoma (LUDA) by showing that circASK1 originated from a MAPK signaling gene [85]. Given the reduced level of ASK1 in cells with acquired gefitinib resistance, it was chosen for further investigation. In in vitro experiments, KD of circASK1 made LUAD cells resistant to gefitinib treatment while circASK1 overexpression alleviated the gefitinib resistance of LUAD cells. Mechanistically, ASK1-272a.a activates ASK1 through competitive interaction with Akt1, thereby enhancing ASK1-mediated apoptosis through cellular susceptibility to gefitinib (Fig. 7b). Finally, ASK1-272a.a enhances the LUAD cells' sensitivity to gefitinib; therefore, ASK1-272a.a can act as a potential biomarker for tracking patients' response to gefitinib treatment.
Vaccination utilizing the protein-encoding ability of circRNAs has also shown remarkable improvement towards targeting cancer tumors by harnessing and activating cytotoxic T cells. This was shown in a study in which the in vivo administration of peptide and circRNA vaccines consisting of circFam53B-219 peptide and circFAM53B, respectively, in mice bearing breast cancer tumors or melanoma, activated tumor-antigen specific cytotoxic T cells, resulting in increased tumor lysis and effective tumor management (Fig. 8a) [45].
Finally, MDD is a devastating mental health disease whose pathogenesis remains vague and is not completely understood. One of the theories that has been linked to the onset of MDD is the impairment of the hypothalamic-pituitary-adrenal HPA axis mediated by the glucocorticoid receptor (GR). Jiao et al. explained how this dysfunction occurs through investigation of the translational ability of circFKBP8(5S,6), an upregulated circular RNA in the peripheral plasma of MDD patients. They clearly showed through a series of in vitro, and in vivo experiments in mice and macaques, that circFKBP8(5S,6)-encoded protein, cFKBP8, inhibited GR entry into neuronal nuclei, associated with the dysregulation of HPA axis and depression (Fig. 9b). Thus, can serve as a reliable therapeutic target and diagnostic biomarker for the treatment of MDD [73].
Therefore, circRNA-encoded proteins implicated in disease pathogenesis can be potentially targeted through genetic-based alteration, use of antibodies and small-molecule inhibitors; they have also been proven to be effective biomarkers, beneficial for monitoring various disease conditions. Taken together, the above examples highlight the clinical relevance of circular RNA-encoded proteins as potential diagnostic biomarkers and therapeutic targets for disease treatment.
Limitations and controversies in circRNA-encoded protein research
Over the past one and a half decades, the field of circRNA biology has witnessed tremendous advancement regarding the detection and elucidation of their different physiological functions. Indeed, a significant amount of research has been carried out in this area. Despite this, we have to be aware of certain limitations and unsolved puzzles that still limit our understanding of this particular area of human biology.
Within the context of circRNA-encoded proteins implicated in CNS diseases, the sophisticated and heterogeneous nature of the CNS contributes to the varied expression of circRNAs in different brain regions, which can directly affect the accuracy of circRNA expression results. Also, clinical translation involves experimenting on live human brain tissues for proper validation of in vitro and in vivo experimental results to confirm these circRNAs as biomarkers. As it is unethical and nearly impossible to obtain live human brain tissues, especially from MDD patients, alternative techniques, such as the cerebral organoid technique, have been adopted [73, 86]. However, these approaches do not reflect real-life depressive-like patterns seen in humans. This presents a major challenge to research that needs to be addressed urgently.
A much more significant bottleneck associated with circRNA-encoded protein research is the experimental validation of circRNA-encoded peptides. Although circRNAs are more stable than linear mRNAs, they are still poorly expressed compared to their linear counterparts, thus contributing to false positives that inadvertently affect accurate detection and quantification of circRNAs. For instance, the use of RiboSeq for the detection of circRNA translation is challenging, and without the use of appropriate controls, results can be misleading [15]. Actually, a criterion for active translation depicted by a high-quality ribosome dataset is the presence of a strong triplet periodicity, otherwise known as phasing, along the regions/sequences where the ribosomes are bound [15]. Therefore, to ensure reliable circRNA translation results, circRNA Riboseq results showing reads mapped to the backsplicing site must, as a matter of fact, show strong phasing and must be compared stringently to control RNAs which do not undergo translation, such as miRNA, snoRNA [15]. Secondly, mass spectrometry is a widely adopted experimental technique for peptide identification; however, when it comes to unorthodox protein identification, such as those translated from circRNAs, rigorous analysis and checks are necessary to drastically minimize the risk of false discoveries. Therefore, the use of appropriate false discovery rate (FDR) control should be employed in these studies to efficiently validate circRNA translation [15].
Currently, there is still a huge debate amongst scientific researchers over the functional relevance of circRNA-encoded proteins. Despite the vast amount of published studies on circRNA-encoded proteins and their respective roles, a large skepticism still exists. The cap-independent mechanism of translation adopted by circRNAs, because of the absence of 5'cap, is highly inefficient due to sparse endogenous IRESs in the eukaryotic transcriptome [29], leading to low abundance of circRNA-encoded proteins; therefore, future research should aim to address this. There is a need to properly identify and validate all circRNAs that are indeed translatable using more sophisticated experimental and analytical platforms.
Another notable challenge with research on circRNA-encoded proteins is the detection of low-abundance circRNAs, including those that encode proteins. For example, there is evidence that trans-splicing byproducts of overexpression plasmids might have been misinterpreted as being translated from circRNAs [87]. The researchers discovered that deletion of the 3' splicing site required for circZNF609 formation or Alu elements necessary for backsplicing generated neither circZNF609 nor its corresponding protein in the former case. However, surprisingly, in the latter case, while circZNF609 was not produced, the translated protein was detected. Further concurrent deletion of both splicing site and Alu elements generated neither circZNF609 nor its protein product. Thus, establishing that the protein product might have been a result of by-products of linear splicing and not necessarily from circular RNA backsplicing [87]. Therefore, results from circRNA overexpression constructs should be interpreted with caution due to complications that can arise from overexpression systems. Most importantly, it should be noted that circRNAs possess similar sequences to their linear mRNA analogue, therefore conventional detection methods such as RT-qPCR may be faced with the challenge of non-specificity or insensitivity since genomic rearrangement does occur and the possibility for low-quality measurement exists.
In addition, interpretation of results from bulk circRNA RNA-seq of malignant tumor tissues compared to normal tissues, to determine differential circRNA expression, can also be misleading [88, 89]. Employing a broader and more in-depth technique, such as spatial analyses and single-cell analyses, is essential to correctly interpret transcriptomic results. For instance, studies employing spatial analyses have shown that the circRNA, ciRS-7 (CDR1as), which was previously thought to be abundant in tumors compared to normal tissues while correlating with poor prognosis in diseased patients, was actually not expressed in specific colon cancer cells but in stromal cells within the tumor microenvironment. Thus, the high expression from the bulk tissue analysis resulted from the stromal cell expression and not from the colon cancer cells themselves [89]. Generally, the expression of circRNAs, such as cirRS-7, is limited in proliferating cells, such as cancer cells but is more profound in quiescent cells, like muscle and stromal cells [88]. This may lead to misinterpretations when studying patient samples using bulk RNA analysis only without considering varying expressions within the different cell types [88].
Finally, the clinical translation of circRNA-encoded proteins is still in its early stages. Development of antibodies or small-molecule inhibitors for circRNA-encoded proteins has to resolve the issue of immune rejection, and a thorough understanding of the safety of the inhibitor drug for human administration is crucial. Nevertheless, off-target effects present a major bottleneck for gene-therapy treatment to target endogenous circRNAs.
Conclusion and future perspectives
Recent studies have highlighted the roles of circRNA-encoded proteins as potential clinical biomarkers for the diagnosis and treatment of several tumor and non-tumor diseases [19, 22]. Nevertheless, there are important gaps in knowledge that must be addressed in the future to improve the clinical relevance of circRNA-encoded proteins. At present, knowledge of the mechanism of circRNA translation within cells is still vague. How several molecular components interact within the cell to drive circRNA translation must be studied. Of course, this will require a great deal of time and effort. The special characteristics of circRNA that direct mediatory translational mechanisms also remain an open question.
Furthermore, the research involving circRNA-encoded proteins in central nervous system (CNS) diseases is not clinically valuable because of the lack of appropriate model organisms that reflect real-life phenotypic characteristics of the diseases. This presents a serious impediment to research in this field. Although research using organoids represents a crucial step forward, certain limitations exist when using such technologies, as addressed in the previous section.
Given the impressive potential of artificial intelligence (AI), the incorporation of advanced AI tools in circRNA research presents an important opportunity to deeply understand the functional characteristics of circRNAs and address open questions. Therefore, future research should aim to integrate AI, bioinformatics and omics technologies in circRNA studies [5].
In summary, circRNA-encoded proteins are potentially important clinical biomarkers and therapeutic targets for the treatment of tumors and non-tumor diseases. To actualize the potential of circRNA-encoded proteins, more research involving large cohorts of patients is needed to validate their expression levels in different diseases. Extensive elucidation of the functional mechanisms of all circRNA-encoded proteins should be prioritized to enable actualization of their therapeutic potential, either as specific biomarkers or as targets for small molecules. This review calls for a global and collective effort towards the development and refinement of robust strategies for the identification of all possible circRNA-encoded proteins to fully understand their relevance, as well as refining targeted therapeutic techniques while establishing a universal standard in adopting circular RNA-encoded proteins as unique biomarkers for disease diagnoses.
Acknowledgements
Sponsored by ANSO Scholarships for Young Talents.
Funding
This study was supported by grants from the China Science and Technology Innovation 2030 - Major Project (2022ZD0211701 and 2021ZD0200700) and the National Natural Science Key Foundation of China (82130042, 81830040 and 82471552), Shenzhen Science and Technology Serial Funds (GJHZ20210705141400002, KCXFZ20211020164543006, JCYJ20220818101615033, ZDSYS20220606100606014 and KQTD20221101093608028).
Alliance of International Science Organizations (ANSO) scholarship program.
Author contributions
Z. Z. - conceptualized this review and revised the manuscript. J. U - wrote the manuscript
Competing interests
The authors have declared that no competing interest exists.
References
1. Liu C-X, Chen L-L. Circular RNAs: Characterization, cellular roles, and applications. Cell. 2022;185:2016-34
2. Verduci L, Tarcitano E, Strano S, Yarden Y, Blandino G. CircRNAs: role in human diseases and potential use as biomarkers. Cell Death Dis. 2021;12:468
3. Hsu M-T, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339-40
4. Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A. 1976;73:3852-6
5. Pisignano G, Michael DC, Visal TH, Pirlog R, Ladomery M, Calin GA. Going circular: history, present, and future of circRNAs in cancer. Oncogene. 2023;42:2783-800
6. Kolakofsky D. Isolation and characterization of Sendai virus DI-RNAs. Cell. 1976;8:547-55
7. Arnberg AC, Ommen GJBV, Grivell LA, Bruggen EFJV, Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19:313-9
8. Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (δ) virus possesses a circular RNA. Nature. 1986;323:558-60
9. Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993;7:155-60
10. Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L. et al. Translation of CircRNAs. Molecular Cell. 2017;66:9-21.e7
11. You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G. et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603-10
12. Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H. et al. Circular Intronic Long Noncoding RNAs. Molecular Cell. 2013;51:792-806
13. Xu C, Zhang J. Mammalian circular RNAs result largely from splicing errors. Cell Rep. 2021;36:109439
14. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675-91
15. Hansen TB. Signal and noise in circRNA translation. Methods. 2021;196:68-73
16. Zhang X-O, Dong R, Zhang Y, Zhang J-L, Luo Z, Zhang J. et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016;26:1277-87
17. Ngo LH, Bert AG, Dredge BK, Williams T, Murphy V, Li W. et al. Nuclear export of circular RNA. Nature. 2024
18. Cao S-M, Wu H, Yuan G-H, Pan Y-H, Zhang J, Liu Y-X. et al. Altered nucleocytoplasmic export of adenosine-rich circRNAs by PABPC1 contributes to neuronal function. Molecular Cell. 2024;84:2304-19.e8
19. Yuan W, Zhang X, Cong H. Advances in the protein-encoding functions of circular RNAs associated with cancer (Review). Oncol Rep. 2023;50:160
20. Dodbele S, Mutlu N, Wilusz JE. Best practices to ensure robust investigation of circular RNAs: pitfalls and tips. EMBO Rep. 2021;22:e52072
21. Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L. et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609-20
22. Shi X, Liao S, Bi Z, Liu J, Li H, Feng C. Newly discovered circRNAs encoding proteins: recent progress. Front Genet. 2023;14:1264606
23. Stagsted LV, Nielsen KM, Daugaard I, Hansen TB. Noncoding AUG circRNAs constitute an abundant and conserved subclass of circles. Life Sci Alliance. 2019 Apr 26;2(3):e201900398
24. Zhang L, Gao H, Li X, Yu F, Li P. The important regulatory roles of circRNA-encoded proteins or peptides in cancer pathogenesis (Review). Int J Oncol. 2023;64:19
25. Li Z, Huang C, Bao C, Chen L, Lin M, Wang X. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256-64
26. Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L. Complementary Sequence-Mediated Exon Circularization. Cell. 2014;159:134-47
27. Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA (New York, NY). 2013;19:141-57
28. Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M. et al. circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell. 2014;56:55-66
29. Fan X, Yang Y, Chen C, Wang Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nature Communications. 2022;13:3751
30. Zhong J, Wu X, Gao Y, Chen J, Zhang M, Zhou H. et al. Circular RNA encoded MET variant promotes glioblastoma tumorigenesis. Nature Communications. 2023;14:4467
31. Monroy G, Spencer E, Hurwitz J. Purification of mRNA guanylyltransferase from vaccinia virions. J Biol Chem. 1978;253:4481-9
32. Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5'-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature. 1975;255:33-7
33. Hellen CUT. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2009;1789:558-70
34. Gritsenko AA, Weingarten-Gabbay S, Elias-Kirma S, Nir R, De Ridder D, Segal E. Sequence features of viral and human Internal Ribosome Entry Sites predictive of their activity. PLoS Comput Biol. 2017;13:e1005734
35. Chen C-K, Cheng R, Demeter J, Chen J, Weingarten-Gabbay S, Jiang L. et al. Structured elements drive extensive circular RNA translation. Molecular Cell. 2021;81:4300-18.e13
36. Chen R, Wang SK, Belk JA, Amaya L, Li Z, Cardenas A. et al. Engineering circular RNA for enhanced protein production. Nat Biotechnol. 2023;41:262-72
37. Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim Biophys Acta. 2009;1789:518-28
38. Zhong J, Yang X, Chen J, He K, Gao X, Wu X. et al. Circular EZH2-encoded EZH2-92aa mediates immune evasion in glioblastoma via inhibition of surface NKG2D ligands. Nature Communications. 2022;13:4795
39. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y. et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626-41
40. Tang C, Xie Y, Yu T, Liu N, Wang Z, Woolsey RJ. et al. m6A-dependent biogenesis of circular RNAs in male germ cells. Cell Res. 2020;30:211-28
41. Duan J-L, Chen W, Xie J-J, Zhang M-L, Nie R-C, Liang H. et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol Cancer. 2022;21:93
42. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30
43. Daubendiek SL, Ryan K, Kool ET. Rolling-Circle RNA Synthesis: Circular Oligonucleotides as Efficient Substrates for T7 RNA Polymerase. J Am Chem Soc. 1995;117:7818-9
44. Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H. et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep. 2015;5:16435
45. Huang D, Zhu X, Ye S, Zhang J, Liao J, Zhang N. et al. Tumour circular RNAs elicit anti-tumour immunity by encoding cryptic peptides. Nature. 2024;625:593-602
46. Wesselhoeft RA. Engineering circular RNA for potent and stable translation in eukaryotic cells. NATURE COMMUNICATIONS. 2018
47. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J. et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Briefings in Bioinformatics. 2016: bbw081.
48. Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18:680
49. Gao X, Xia X, Li F, Zhang M, Zhou H, Wu X. et al. Circular RNA-encoded oncogenic E-cadherin variant promotes glioblastoma tumorigenicity through activation of EGFR-STAT3 signalling. Nat Cell Biol. 2021;23:278-91
50. Wu X, Xiao S, Zhang M, Yang L, Zhong J, Li B. et al. A novel protein encoded by circular SMO RNA is essential for Hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol. 2021;22:33
51. Errichelli L, Dini Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D. et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nature Communications. 2017;8:14741
52. Lei Y, Tang L, Hu J, Wang S, Liu Y, Yang M. et al. Inhibition of MGMT-mediated autophagy suppression decreases cisplatin chemosensitivity in gastric cancer. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2020;125:109896
53. Li Y, Li C, Li D, Yang L, Jin J, Zhang B. lncRNA KCNQ1OT1 enhances the chemoresistance of oxaliplatin in colon cancer by targeting the miR-34a/ATG4B pathway. Onco Targets Ther. 2019;12:2649-60
54. Liu F, Ai F-Y, Zhang D-C, Tian L, Yang Z-Y, Liu S-J. LncRNA NEAT1 knockdown attenuates autophagy to elevate 5-FU sensitivity in colorectal cancer via targeting miR-34a. Cancer Med. 2020;9:1079-91
55. Cabrera S, Maciel M, Herrera I, Nava T, Vergara F, Gaxiola M. et al. Essential role for the ATG4B protease and autophagy in bleomycin-induced pulmonary fibrosis. Autophagy. 2015;11:670-84
56. Pan Z, Zheng J, Zhang J, Lin J, Lai J, Lyu Z. et al. A Novel Protein Encoded by Exosomal CircATG4B Induces Oxaliplatin Resistance in Colorectal Cancer by Promoting Autophagy. Advanced Science. 2022;9:2204513
57. Pengo N, Agrotis A, Prak K, Jones J, Ketteler R. A reversible phospho-switch mediated by ULK1 regulates the activity of autophagy protease ATG4B. Nature Communications. 2017;8:294
58. Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo X. et al. A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/β-catenin signaling pathway to promote gastric cancer progression. Mol Cancer. 2021;20:158
59. Krishnamurthy N, Kurzrock R. Targeting the Wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev. 2018;62:50-60
60. Tang X, Deng Z, Ding P, Qiang W, Lu Y, Gao S. et al. A novel protein encoded by circHNRNPU promotes multiple myeloma progression by regulating the bone marrow microenvironment and alternative splicing. J Exp Clin Cancer Res. 2022;41:85
61. Song R, Ma S, Xu J, Ren X, Guo P, Liu H. et al. A novel polypeptide encoded by the circular RNA ZKSCAN1 suppresses HCC via degradation of mTOR. Mol Cancer. 2023;22:16
62. Fadhal E. A Comprehensive Analysis of the PI3K/AKT Pathway: Unveiling Key Proteins and Therapeutic Targets for Cancer Treatment. Cancer Inform. 2023;22:11769351231194273
63. Goyal A, Agrawal A, Verma A, Dubey N. The PI3K-AKT pathway: A plausible therapeutic target in Parkinson's disease. Experimental and Molecular Pathology. 2023;129:104846
64. He X, Li Y, Deng B, Lin A, Zhang G, Ma M. et al. The PI3K/AKT signalling pathway in inflammation, cell death and glial scar formation after traumatic spinal cord injury: Mechanisms and therapeutic opportunities. Cell Prolif. 2022;55:e13275
65. He Y, Sun MM, Zhang GG, Yang J, Chen KS, Xu WW. et al. Targeting PI3K/Akt signal transduction for cancer therapy. Signal Transduction and Targeted Therapy. 2021;6:1-17
66. Nitulescu GM, Van De Venter M, Nitulescu G, Ungurianu A, Juzenas P, Peng Q. et al. The Akt pathway in oncology therapy and beyond (Review). Int J Oncol. 2018;53:2319-31
67. Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang H. et al. circ-EIF6 encodes EIF6-224aa to promote TNBC progression via stabilizing MYH9 and activating the Wnt/beta-catenin pathway. Molecular Therapy. 2022;30:415-30
68. Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X. et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduction and Targeted Therapy. 2022;7:1-23
69. Wang Q, Cheng B, Singh S, Tao Y, Xie Z, Qin F. et al. A protein-encoding CCDC7 circular RNA inhibits the progression of prostate cancer by up-regulating FLRT3. npj Precis Onc. 2024;8:11
70. Tsuji L, Yamashita T, Kubo T, Madura T, Tanaka H, Hosokawa K. et al. FLRT3, a cell surface molecule containing LRR repeats and a FNIII domain, promotes neurite outgrowth. Biochemical and Biophysical Research Communications. 2004;313:1086-91
71. Jauhiainen S, Laakkonen JP, Ketola K, Toivanen PI, Nieminen T, Ninchoji T. et al. Axon Guidance-Related Factor FLRT3 Regulates VEGF-Signaling and Endothelial Cell Function. Front Physiol. 2019;10:224
72. Mo D, Li X, Raabe CA, Rozhdestvensky TS, Skryabin BV, Brosius J. Circular RNA Encoded Amyloid Beta peptides-A Novel Putative Player in Alzheimer's Disease. Cells. 2020;9:2196
73. Jiao J, Xu D, Kong Y, Cao Y, Wang L, Hong Y. et al. circFKBP8(5S,6)-encoded protein as a novel endogenous regulator in major depressive disorder by inhibiting glucocorticoid receptor nucleus translocation. Science Bulletin. 2024: S2095927324004468.
74. Du WW, Xu J, Yang W, Wu N, Li F, Zhou L. et al. A Neuroligin Isoform Translated by circNlgn Contributes to Cardiac Remodeling. Circ Res. 2021;129:568-82
75. Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560-4
76. Dai F, Yu L, He H, Chen Y, Yu J, Yang Y. et al. Human serum and glucocorticoid-inducible kinase-like kinase (SGKL) phosphorylates glycogen syntheses kinase 3 beta (GSK-3beta) at serine-9 through direct interaction. Biochemical and Biophysical Research Communications. 2002;293:1191-6
77. Woulfe KC, Gao E, Lal H, Harris D, Fan Q, Vagnozzi R. et al. Glycogen synthase kinase-3beta regulates post-myocardial infarction remodeling and stress-induced cardiomyocyte proliferation in vivo. Circ Res. 2010;106:1635-45
78. Li X, Zhang Q, Cai L, Wang Y, Wang Q, Huang X. et al. Inhibitor of growth 4 induces apoptosis in human lung adenocarcinoma cell line A549 via Bcl-2 family proteins and mitochondria apoptosis pathway. J Cancer Res Clin Oncol. 2009;135:829-35
79. Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S. et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373-8
80. Li S, Li X, Xue W, Zhang L, Yang L-Z, Cao S-M. et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat Methods. 2021;18:51-9
81. Jiang T, Xia Y, Lv J, Li B, Li Y, Wang S. et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol Cancer. 2021;20:66
82. Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061-8
83. Bradley CA, Salto-Tellez M, Laurent-Puig P, Bardelli A, Rolfo C, Tabernero J. et al. Targeting c-MET in gastrointestinal tumours: rationale, opportunities and challenges. Nat Rev Clin Oncol. 2017;14:562-76
84. Lan T, Gao F, Cai Y, Lv Y, Zhu J, Liu H. et al. The protein circPETH-147aa regulates metabolic reprogramming in hepatocellular carcinoma cells to remodel immunosuppressive microenvironment. Nature Communications. 2025;16:333
85. Wang T, Liu Z, She Y, Deng J, Zhong Y, Zhao M. et al. A novel protein encoded by circASK1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating ASK1-dependent apoptosis. Cancer Letters. 2021;520:321-31
86. Tang X-Y, Wu S, Wang D, Chu C, Hong Y, Tao M. et al. Human organoids in basic research and clinical applications. Signal Transduction and Targeted Therapy. 2022;7:168
87. Ho-Xuan H, Glazar P, Latini C, Heizler K, Haase J, Hett R. et al. Comprehensive analysis of translation from overexpressed circular RNAs reveals pervasive translation from linear transcripts. Nucleic Acids Res. 2020;48:10368-82
88. Garcia-Rodriguez JL, Korsgaard U, Ahmadov U, Jarlstad Olesen MT, Dietrich KG, Hansen EB. et al. Spatial Profiling of Circular RNAs in Cancer Reveals High Expression in Muscle and Stromal Cells. Cancer Res. 2023;83:3340-53
89. Kristensen LS, Ebbesen KK, Sokol M, Jakobsen T, Korsgaard U, Eriksen AC. et al. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. Nat Commun. 2020;11:4551
90. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
91. Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL. et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Research. 2010;38:e178
92. Zhang J, Chen S, Yang J, Zhao F. Accurate quantification of circular RNAs identifies extensive circular isoform switching events. Nature Communications. 2020;11:90
93. Gao Y, Zhao F. Computational Strategies for Exploring Circular RNAs. Trends in Genetics. 2018;34:389-400
94. Jakobi T, Dieterich C. Computational approaches for circular RNA analysis. WIREs RNA. 2019;10:e1528
95. Zhou Z, Zhang J, Zheng X, Pan Z, Zhao F, Gao Y. CIRI-Deep Enables Single-Cell and Spatial Transcriptomic Analysis of Circular RNAs with Deep Learning. Adv Sci (Weinh). 2024;11:e2308115
96. Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA (New York, NY). 2014;20:1666-70
97. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biology. 2016;13:34-42
98. Liu Y-C, Li J-R, Sun C-H, Andrews E, Chao R-F, Lin F-M. et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Research. 2016;44:D209-15
99. Huang W, Ling Y, Zhang S, Xia Q, Cao R, Fan X. et al. TransCirc: an interactive database for translatable circular RNAs based on multi-omics evidence. Nucleic Acids Research. 2021;49:D236-D42
100. Chen X, Han P, Zhou T, Guo X, Song X, Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985
101. Liu H, Fang D, Zhang C, Zhao Z, Liu Y, Zhao S. et al. Circular MTHFD2L RNA-encoded CM-248aa inhibits gastric cancer progression by targeting the SET-PP2A interaction. Molecular Therapy. 2023;31:1739-55
102. Liu M, Wang Q, Shen J, Yang BB, Ding X. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biology. 2019;16:899-905
103. Rombel IT, Sykes KF, Rayner S, Johnston SA. ORF-FINDER: a vector for high-throughput gene identification. Gene. 2002;282:33-41
104. Zhao J, Wu J, Xu T, Yang Q, He J, Song X. IRESfinder: Identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J Genet Genomics. 2018;45:403-6
105. Zhao J, Li Y, Wang C, Zhang H, Zhang H, Jiang B. et al. IRESbase: A Comprehensive Database of Experimentally Validated Internal Ribosome Entry Sites. Genomics Proteomics Bioinformatics. 2020;18:129-39
106. Mokrejs M, Vopálenský V, Kolenaty O, Masek T, Feketová Z, Sekyrová P. et al. IRESite: the database of experimentally verified IRES structures (www.iresite.org). Nucleic Acids Research. 2006;34:D125-30
107. Liang Z-x, Liu H-s, Xiong L, Yang X, Wang F-w, Zeng Z-w. et al. A novel NF-κB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol Cancer. 2021;20:103
108. Wang L, Park HJ, Dasari S, Wang S, Kocher J-P, Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Research. 2013;41:e74
109. Rey J, Murail S, de Vries S, Derreumaux P, Tuffery P. PEP-FOLD4: a pH-dependent force field for peptide structure prediction in aqueous solution. Nucleic Acids Research. 2023;51:W432-W7
110. Saladin A, Rey J, Thévenet P, Zacharias M, Moroy G, Tufféry P. PEP-SiteFinder: a tool for the blind identification of peptide binding sites on protein surfaces. Nucleic Acids Research. 2014;42:W221-6
111. Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845-58
112. Zhang Y, Jiang J, Zhang J, Shen H, Wang M, Guo Z. et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol Cancer. 2021;20:101
113. Pierce BG, Wiehe K, Hwang H, Kim B-H, Vreven T, Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771-3
114. Song J, Zheng J, Liu X, Dong W, Yang C, Wang D. et al. A novel protein encoded by ZCRB1-induced circHEATR5B suppresses aerobic glycolysis of GBM through phosphorylation of JMJD5. J Exp Clin Cancer Res. 2022;41:171
115. Yugandhar K, Gromiha MM. Protein-protein binding affinity prediction from amino acid sequence. Bioinformatics. 2014;30:3583-9
116. Sedan Y, Marcu O, Lyskov S, Schueler-Furman O. Peptiderive server: derive peptide inhibitors from protein-protein interactions. Nucleic Acids Research. 2016;44:W536-41
117. Yang F, Hu A, Guo Y, Wang J, Li D, Wang X. et al. p113 isoform encoded by CUX1 circular RNA drives tumor progression via facilitating ZRF1/BRD4 transactivation. Mol Cancer. 2021;20:123
118. Zhang S, Wang C, Wang Y, Zhang H, Xu C, Cheng Y. et al. A novel protein encoded by circRsrc1 regulates mitochondrial ribosome assembly and translation during spermatogenesis. BMC Biol. 2023;21:94
119. Tatomer DC, Liang D, Wilusz JE. Inducible Expression of Eukaryotic Circular RNAs from Plasmids. Methods Mol Biol. 2017;1648:143-54
120. Mo D, Li X, Raabe CA, Cui D, Vollmar J-F, Rozhdestvensky TS. et al. A universal approach to investigate circRNA protein coding function. Sci Rep. 2019;9:11684
121. Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang J. et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer. 2020;19:71
122. Li F, Cai Y, Deng S, Yang L, Liu N, Chang X. et al. A peptide CORO1C-47aa encoded by the circular noncoding RNA circ-0000437 functions as a negative regulator in endometrium tumor angiogenesis. Journal of Biological Chemistry. 2021;297:101182
123. Ye F, Gao G, Zou Y, Zheng S, Zhang L, Ou X. et al. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Molecular Therapy - Nucleic Acids. 2019;18:88-98
124. Li J, Ma M, Yang X, Zhang M, Luo J, Zhou H. et al. Circular HER2 RNA positive triple negative breast cancer is sensitive to Pertuzumab. Mol Cancer. 2020;19:142
125. Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B. et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18:47
126. Xiong L, Liu H-s, Zhou C, Yang X, Huang L, Jie H-q. et al. A novel protein encoded by circINSIG1 reprograms cholesterol metabolism by promoting the ubiquitin-dependent degradation of INSIG1 in colorectal cancer. Mol Cancer. 2023;22:72
127. Wang L, Zhou J, Zhang C, Chen R, Sun Q, Yang P. et al. A novel tumour suppressor protein encoded by circMAPK14 inhibits progression and metastasis of colorectal cancer by competitively binding to MKK6. Clin Transl Med. 2021;11:e613
128. Li F, Tang H, Zhao S, Gao X, Yang L, Xu J. Circ-E-Cad encodes a protein that promotes the proliferation and migration of gastric cancer via the TGF-β/Smad/C-E-Cad/PI3K/AKT pathway. Molecular Carcinogenesis. 2023;62:360-8
129. Geng X, Wang J, Zhang C, Zhou X, Jing J, Pan W. Circular RNA circCOL6A3_030 is involved in the metastasis of gastric cancer by encoding polypeptide: circCOL6A3_030 promotes metastasis. Bioengineered. 2021;12:8202-16
130. Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F. et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805-14
131. Xia X, Li X, Li F, Wu X, Zhang M, Zhou H. et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18:131
132. Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F. et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. JNCI: Journal of the National Cancer Institute. 2018;110:304-15
133. Saunders JT, Kumar S, Benavides-Serrato A, Holmes B, Benavides KE, Bashir MT. et al. Translation of circHGF RNA encodes an HGF protein variant promoting glioblastoma growth through stimulation of c-MET. J Neurooncol. 2023;163:207-18
134. Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P. et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nature Communications. 2018;9:4475
135. Liang W-C, Wong C-W, Liang P-P, Shi M, Cao Y, Rao S-T. et al. Translation of the circular RNA circβ-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20:84
136. Li H, Lan T, Liu H, Liu C, Dai J, Xu L. et al. IL-6-induced cGGNBP2 encodes a protein to promote cell growth and metastasis in intrahepatic cholangiocarcinoma. Hepatology. 2022;75:1402-19
137. Zhao W, Xue Y, Zhang Y, Zhu Y, Chen Z, Zhao X. A peptide translated from circPPP1R12A promotes the malignancy of non-small cell lung cancer cells through AKTsignaling pathway. Journal of Clinical Laboratory Analysis. 2022;36:e24644
138. Zhao W, Zhang Y, Zhu Y. Circular RNA circβ-catenin aggravates the malignant phenotype of non-small-cell lung cancer via encoding a peptide. Journal of Clinical Laboratory Analysis. 2021;35:e23900
139. Tang X, Guo M, Ding P, Deng Z, Ke M, Yuan Y. et al. BUB1B and circBUB1B_544aa aggravate multiple myeloma malignancy through evoking chromosomal instability. Signal Transduction and Targeted Therapy. 2021;6:361
140. Gu C, Wang W, Tang X, Xu T, Zhang Y, Guo M. et al. CHEK1 and circCHEK1_246aa evoke chromosomal instability and induce bone lesion formation in multiple myeloma. Mol Cancer. 2021;20:84
141. Lyu Y, Tan B, Li L, Liang R, Lei K, Wang K. et al. A novel protein encoded by circUBE4B promotes progression of esophageal squamous cell carcinoma by augmenting MAPK/ERK signaling. Cell Death Dis. 2023;14:346
Author contact
![]() Corresponding authors: Zhang Zhijun, email: janemengzhang163.com, Jude Uzoechina, email: uzoechinaac.cn.
Corresponding authors: Zhang Zhijun, email: janemengzhang163.com, Jude Uzoechina, email: uzoechinaac.cn.

 Global reach, higher impact
Global reach, higher impact