10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2025; 21(14):6252-6269. doi:10.7150/ijbs.120058 This issue Cite
Review
Unveiling the dynamics and therapeutic potential of m6A methyltransferases and demethylases in liver diseases
1. Inflammation and Immune Mediated Diseases Laboratory of Anhui Province, Anhui Institute of Innovative Drugs, The Third Affiliated Hospital, School of Pharmacy, Anhui Medical University, Hefei 230032, China.
2. The Key Laboratory of Anti-inflammatory and Immune Medicines, Anhui Medical University, Ministry of Education, Hefei 230032, China.
3. Institute for Liver Diseases of Anhui Medical University, ILD-AMU, Anhui Medical University, Hefei 230032, China.
4. Department of Pharmacology and Laboratory of Cerebrovascular Pharmacology, School of Pharmaceutical Sciences, Soochow University, Suzhou 215123, China.
*These authors contributed equally to this work.
Received 2025-6-21; Accepted 2025-9-8; Published 2025-10-1
Abstract
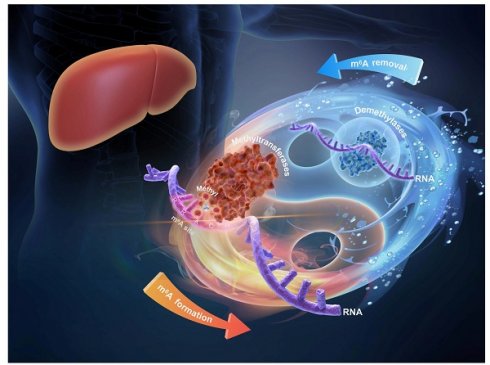
N6-methyladenosine (m6A), a well-known adenosine modification with newly recognized epigenetic functions, reportedly participates in the development of diverse liver diseases. Methyltransferases and demethylases, commonly referred to as “writers” and “erasers”, respectively, play crucial roles in maintaining the balance of m6A modification. In liver disease research specifically, the functioning of these enzymes has piqued significant interest, revealing new perspectives on molecular pathogenic mechanisms. Writer proteins collaborate with co-factors to install m6A modification on RNA, while eraser proteins, exemplified by Fto and Alkbh5, remove modifications via different mechanisms. In liver diseases, the two are not simply antagonistic, but rather act jointly to affect disease progression. By focusing this review on the mechanisms of methyltransferases and demethylases in various liver diseases, we seek to enhance comprehension of m6A modification's role and support the advancement of related research and treatment strategies.
Keywords: m6A modification, liver diseases, methyltransferase, demethylase
1. Introduction
Since the landmark discovery of pseudouridine as the first chemically modified nucleoside in the 1950s[1, 2]. In recent years, RNA modification biology has attracted renewed widespread attention due to the recognition of the prevalence and functional significance of internal mRNA modifications, particularly N6-methyladenosine (m6A)[3]. Spurred by advances in high sensitivity, high throughput sequencing methodologies for transcriptome wide mapping, N6-methyladenine modifications have now been documented across phylogenetically diverse organisms spanning prokaryotes to humans[4]. Among these modifications, m6A stands out due to its exceptional abundance and recognition as a pivotal post-transcriptional regulator[5]. This modification serves as an important regulatory marker in multiple RNA species, including mRNA, tRNA, rRNA, circRNA, miRNA, and lncRN[1, 6].
Spatiotemporally coordinated interactions among “writers” “erasers” and “readers” triads govern the m6A epitranscriptome. The “writers” complex spearheaded by Mettl3, Mettl14, and Mettl16 catalytic triumvirate within the Mettl methyltransferase family collaborates with “erasers” to establish reversible modification landscapes[7]. As shown in (Table 1), key contributions to the m6A methyltransferase complex include the following: Mettl3 interacts with S-adenosylmethionine (SAM) to achieve methyl transfer[3] and forms heterodimers with Mettl14 to enhance catalytic efficiency and promote substrate binding[8]; Mettl16 is involved in specific RNA modifications; Wilms tumor 1-associating protein (Wtap) assists in the localization and catalysis of Mettl3/Mettl14[9]; RNA-binding motif protein Rbm15 and Rbm15b recruits the complex to specific RNA sites[10]; Casitas B-lineage lymphoma transforming sequence-like 1 (Cbll1, also called E3 ubiquitin-protein ligase Hakai) maintains the stability of modifications[11]; Vir-like m6A methyltransferase associated (Virma), also designated as Kiaa1429, guides regional-specific methylation[12]; and zinc finger CCCH-type containing 13 (Zc3h13, also known as Flacc) maintains the nuclear localization of the related complex[13].
Among eraser proteins, fat mass and obesity-associated protein (Fto) and AlkB homolog 5 (Alkbh5) demonstrate particularly distinctive characteristics[14]. Fto, the first demethylase to be discovered, exhibits different substrate preferences in the nucleus and cytoplasm and removes methyl groups via oxidation reaction[15, 16]. Alkbh5 is an endogenous demethylase that mainly mediates the demethylation of the 3'-untranslated region (UTR) of specific transcripts[17, 18].
The impact of m6A modification on gene expression also requires the involvement of reader proteins, particularly members of the insulin-like growth factor 2 mRNA-binding protein (Igf2bp) and YTH domain family[19, 20]. Reader proteins, by interacting with other molecules, decipher the information carried by m6A modifications to regulate the metabolic processes of mRNAs, such as splicing, nuclear export, translocation, and stability[21]. Critically, disruption of the dynamic equilibrium between m6A methyltransferases and demethylases induces reader protein dysregulation, thereby driving disease pathogenesis. This mechanistic cascade has been validated by the following studies: The opposing actions of Mettl14 mediated m6A methylation and Alkbh5-dependent demethylation on Tgf-β1 mRNA establish a dynamic regulatory switch that differentially controls hepatic stellate cell activation and profibrotic signaling[22, 23]. Alcoholic hepatitis exhibits Mettl3-mediated m6A hypermethylation and Fto deficient demethylation of Il-17r mRNA, driving its pathological overexpression and inflammation amplification[24].
Recognizing the critical role of m6A modification in liver diseases, our group has systematically investigated m6A-related proteins. We demonstrated how m6A readers regulate gene expression and disease progression[25], highlighted m6A's importance in liver pathophysiology[26], and revealed that Wtap mediates m6A modification of circDcbld2 and interacts with Igf2bp2, uncovering a key mechanism in hepatic fibrosis[27]. However, comprehensive reviews elucidating the dynamic regulatory mechanisms of m6A methyltransferases and demethylases in hepatic disorders remain notably lacking.
The role of m6A methyltransferases and demethyltransferases in liver diseases
| Type | Diseases | m6A regulators | Target genes | Functions |
|---|---|---|---|---|
| Core catalytic writer | ALI | Mettl14↑ | Ccl2, Ccl5↓ | Suppress inflammatory response and ameliorate hepatic damage |
| Core catalytic writer | ALI | Mettl3↑ | Pck1↑ | Promote gluconeogenesis and reduces lactate accumulation |
| Core catalytic writer | NAFLD | Mettl3↓ | Cd36↓ | Drive NAFLD to NASH progression |
| Core catalytic writer | NAFLD | Mettl3↑ | Rubicon↑ | Suppress autophagy impairs lipid droplet clearance |
| Core catalytic writer | NAFLD | Mettl14↓ | Nlrp3↑ | Promote hepatic inflammation and exacerbates liver injury |
| Core catalytic writer | NAFLD | Mettl16↑ | Cidea↑ | Promote hepatic lipid accumulation and metabolic dysregulation |
| Accessory factor | NAFLD | Rbm15↑ | Rock1↓ | Suppress lipid synthesis and inflammation |
| Core catalytic writer | HF | Wtap↓ | Ptch1↑ | Suppress aberrant fibrotic progression |
| Core catalytic writer | HCC | Mettl3↑ | State3↑ | Enhance nuclear translocation and evasion of tumor cells |
| Core catalytic writer | HCC | Mettl14↑ | Usp48↑ | Reduce glycolytic activity and malignancy in HCC |
| Accessory factor | HCC | Kiaa1429↑ | Gata3↓ | Enhance the migratory and invasive capacities of HCC |
| Core catalytic writer | HCC | Mettl3↑ | Egfr↑ | Lenvatinib treatment resistance |
| Accessory factor | HCC | Cbll1/Hakai↓ | Ajuba↓ | Promote the growth of HCC cells and tumors |
| Accessory factor | — | Zcchc4 | — | Site specific m6A methylation of 28S rRNA |
| Accessory factor | — | Zc3h13 | — | Interact with Wtap and bind to Rbm15 and Rbm15b |
| Accessory factor | — | Znf217 | — | Mediate m6A RNA methylation through targeted DNA binding |
| m6A eraser | NAFLD | Fto↑ | Pparγ↑ | Suppress of hepatic steatosis |
| m6A eraser | NAFLD | Alkbh5↑ | Linc01468 | Promote hepatic inflammation and exacerbates liver injury |
| m6A eraser | ALD | Fto↑ | Il-17ra↑ | Recruit immune cells and exacerbate hepatic inflammation |
| m6A eraser | HF | Fto↑ | Becn1↓ | Suppress of autophagy mitigates ferroptosis |
| m6A eraser | HCC | Fto↑ | Sox2, Klf4↑ | Maintain of cancer stem cell properties |
| m6A eraser | HCC | Alkbh5↑ | Tirap↑ | Reduce the radiosensitivity of hepatocellular carcinoma cells |
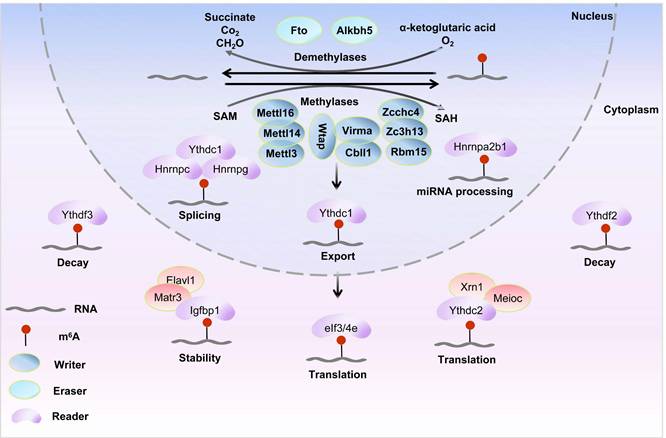
Reversible m6A modification and molecular functions on mRNA. The m6A methylation is catalyzed by the writer complex including Mettl3, Mettl14, Mettl16, Wtap, Virma, Rbm15/15b, Cbll1, Kiaa1429, and Zc3h13. The m6A modification is erased by demethylases including Fto and Alkbh5. Methylases and demethylases achieve the reversible regulation of m6A modification via dynamic equilibrium.
This review systematically synthesizes recent advances in m6A modification patterns across major liver diseases, including acute liver injury (ALI), viral hepatitis, nonalcoholic fatty liver disease(NAFLD), hepatic fibrosis (HF), and hepatocellular carcinoma (HCC), with the ultimate goal of developing mechanism based therapeutic strategies to improve clinical outcomes.
2. Formation and removal of THE m6A modification
As shown in (Figure 1), the m6A modification, which is the most common internal epigenetic mark in mRNA, is added by m6A methyltransferases or writers and is removed by demethylases or erasers (e.g., Fto and Alkbh5). These modifications can be recognized by m6A reader proteins, thereby participating in the post-transcriptional regulation of RNA[28].
2.1 m6A methyltransferases
m6A modification, catalyzed by specific methyltransferases, plays extensive roles in regulating gene expression and RNA metabolism. Precise control of this modification is crucial for maintaining its homeostasis, and in-depth investigation of these enzymes will facilitate the elucidation of RNA epigenetic regulatory mechanisms.
2.1.1 m6A methyltransferases of the Mettl family
The Mettl family proteins (including Mettl3, Mettl14, Mettl5, and Mettl16) constitute the core catalytic machinery for m6A deposition[29]. As introduced in Section 1, Mettl3 serves as the catalytic subunit that directly binds SAM to transfer methyl groups to RNA substrates, while Mettl14 acts as an allosteric activator enhancing Mettl3's RNA-binding affinity and catalytic efficiency within the heterodimeric complex[30]. Structural studies reveal that the catalytic site of Mettl3 is the exclusive domain for SAM binding, confirming its role as the sole active center[31, 32]. Furthermore, Mettl14's intrinsically disordered C-terminal domain interacts with protein arginine methyltransferase 1 (Prmt1) to maintain complex integrity and promote RNA substrate engagement, thereby augmenting methylation activity[33, 34]. Notably, beyond its catalytic function, Mettl3 contains a CCCH-type zinc-binding motif essential for in vitro RNA methylation[32]. Mettl16 regulates Mat2a mRNA splicing via methylation of its hairpin structure and modulates U6 snRNA methylation to influence mRNA splicing[35], while Mettl5, stabilized by dimerization with Trmt112, mediates m6A modification at position A1832 of 18S rRNA[36, 37].
To ensure proper m6A modification, Mettl3 and Mettl14 bind to Wtap, forming the Mettl3/Mettl14/Wtap complex identified via arabidopsis screening[38]. Wtap is essential for directing Mettl3/Mettl14 to nuclear speckles and boosting their catalytic activity in vivo[39, 40]. The complex primarily methylates the 3'-UTR of mRNA near stop codons within the DRACH motif (D=A/U/G, R=A/G, H=A/C/U), regulating mRNA processes[41].
2.1.2 Other m6A methyltransferases
In the realm of m6A modification, other pivotal factors and proteins are also intricately involved in the m6A methyltransferase machinery and its regulatory network. Virma binds RNA-dependently to polyadenylation factors, including cleavage and polyadenylation specificity factor subunits Cpsf5 and Cpsf6[42], and recruits the Mettl3-Mettl14-Wtap methyltransferase complex to catalyze site-specific m6A methylation proximal to mRNA stop codons and within 3'-UTRs[43]. The Virma-Wtap complex enhances the catalytic activity of Mettl3-Mettl14 toward target RNAs, thereby modulating cellular m6A modification levels[44]. This complex recruits the methyltransferase machinery to DRACH motif-enriched RNA regions through its interaction with homologous RNA-binding proteins Rbm15 and Rbm15b[45, 46]. In Drosophila models, Hakai/Cbll1 mutants exhibit >50% reduction in mRNA m6A, highlighting its role in maintaining modification homeostasis[47]. Mechanistically, Hakai/Cbll1 interacts with Mettl3, Mettl16, and Virma via its E3 ubiquitin ligase activity to stabilize m6A modifications[48]. In mouse embryonic stem cells, Zc3h13 depletion reduces m6A levels and mislocalizes the Wtap-Virma-Hakai/Cbll1 complex to the cytoplasm, demonstrating its essential role in maintaining nuclear compartmentalization of the m6A methyltransferase machinery[49]. Additionally, Zc3h13 bridges the Rbm15-Wtap-Fl(2)d complex to the mRNA-binding factor Nito, facilitating efficient m6A deposition[50]. Zcchc4, as a recently reported potential RNA methyltransferase[51], is conserved in other multicellular model organisms, but absent in yeast. Structural analysis of Zcchc4 revealed a putative m6A methyltransferase domain with a conserved catalytic DPPF motif and a CCHC-ZNF domain. These two domains cooperate to ensure Zcchc4 effectively mediates m6A modification at the A4220 site of 28S rRNA[52, 53].
In summary, Mettl-family methyltransferases and their associated constitute the core machinery for m6A methylation. Their precisely regulated protein interactions dynamically modulate mRNA m6A levels, and mechanistic insights into this network provide critical insights into liver pathophysiology.
2.2 m6A demethylases
The m6A modification can be reversed via active demethylation by the m6A demethylases Fto or Alkbh5, illustrating that methylation-dependent processes are reversible and controllable. As the first RNA demethylase to be identified, Fto is demonstrably capable of catalyzing the removal of methyl groups from RNA[54, 55]. In contrast to Fto, Alkbh5 was identified through biochemical screening[56]. Fto and Alkbh5 exhibit distinct characteristics and functions during m6A modification, which may result in different regulatory patterns and biological effects.
2.2.1 Context-dependent RNA demethylation by Fto
Fto, belonging to the non-heme Fe(II)/α-ketoglutarate-dependent AlkB dioxygenase family, exhibits functional similarity with Abh1-3 (oxidative demethylation of N-methylated DNA/RNA bases) and Abh8 (hydroxylation of tRNA wobble uridine)[57-59]. Experimental evidence have demonstrated that Fto can demethylate m³T and m³U in single-stranded (ss) DNA and ssRNA in vitro[60], but its activity is lower than those of other family proteins. The latest crystal structure reveals that Fto prefers single-stranded nucleic acids (ssNAs) as substrates[61], suggesting that its function may be modulated by context-dependent factors such as substrate conformation, reflecting the dynamic distribution of ssNAs across cellular compartments and physiological or pathological conditions. In mRNA, where m6A is the most abundant modification (3-5 marks per transcript)[62]. Research findings indicate that Fto targets m6A in mRNA, N6,2'-O-dimethyladenosine (m6Am) at the 5'-cap, and m1A in tRNA, thereby regulating these RNA modifications[63, 64]. Given that mRNA exists in a dynamic state shaped by nuclear cytoplasmic transport and protein interactions, the accessibility of m6A to Fto is likely context-dependent[65]. Given that Fto's activity on substrates can be influenced by environmental factors, it is reasonable to infer that its effect on m6A might also be context dependent.
Indeed, Fto exhibits compartment specific substrate preferences, underscoring its environmental sensitivity[66]. In the nucleus, where m6A is abundant, Fto functions as an Fe(II)/α-ketoglutarate-dependent dioxygenase to demethylate m6A via oxidative hydroxylation, producing formic acid/methanol and restoring adenosine[67]. Conversely, in the cytoplasm, Fto preferentially targets 5'-cap m6Am, potentially regulating mRNA stability, translation efficiency, and decay pathways[68]. These differential actions in the nucleus and cytoplasm clearly illustrate how the cellular environment shapes the substrate selection and catalytic function of Fto.
2.2.2 Substrate and roles of Alkbh5
Alkbh5, the second identified RNA demethylase, has only one known substrate: m6A[56]. As one of nine members of the AlkB family of ferrous iron- and 2-oxoglutarate-dependent nucleic acid oxygenases (Naoxs), it reportedly catalyzes the demethylation of m6A in RNA. This finding has improved our understanding of its substrate recognition specificity[63]. Crystallographic studies reveal that Alkbh5, primarily mediates the demethylation of m6A in the 3'-UTR of specific transcripts[69]. The expression patterns of Alkbh5 and Fto exhibit significant across tissues. For instance, Alkbh5 is most highly expressed in the testes, while Fto is predominantly expressed in the brain[70]. This differential tissue expression may be one explanation for the distinct biochemical pathways through which these enzymes participate in m6A demethylation. Comprehensive characterization of Alkbh5's properties, functions, and tissue specificity is crucial for mechanistically understanding its regulation of RNA methylation in biological processes.
3. Biological function and interaction of methyltransferaseS and demethylaseS
In the field of epigenetics, m6A modification is one of the most prevalent and well-studied RNA modifications, demonstrating widespread involvement in all aspects of mRNA metabolism, including export, translation, stability, and splicing. Specifically, methyltransferases are responsible for adding methyl groups to mRNA to complete the writing, demethylases remove these methyl groups to achieve erasure, and readers recognize the m6A modification sites on mRNA.
3.1 Regulation of mRNA splicing
The maturation of pre-mRNA involves 5'-capping, 3'-polyadenylation, and splicing[71]. Writer proteins like Mettl3 mediate m6A methylation at specific sites on introns and exons, correlating with transcription start sites (TSS) and stop codons to regulate RNA stability and splicing[72, 73]. While m6A does not disrupt Watson-Crick base pairing, it reduces double-stranded RNA stability by 1.4 kcal/mol[74] while stabilizing surrounding structures[75] or promoting the folding of adjacent RNA sequences[76], thereby influencing spliceosome assembly[77]. These modifications can be recognized by readers such as Hnrnpg, which further regulate mRNA splicing and stability. Ythdc1 (also known as Dc1) is another important reader protein. Some studies have shown that Ythdc1 interacts with splicing regulators, including Src-associated in mitosis, 68kDa (Sam68)[78], splicing factor Sc35[79], and serine-arginine-rich splicing factors Srsf1 and Srsf3[80], suggesting its involvement in splicing regulation. However, others reports indicate that Ythdc1 selectively regulates mRNA splicing by promoting the binding of Srsf3 while inhibiting the activity of Srsf10[81]. Fto-mediated m6A demethylation reduces Rbm15 binding at splicing junctions (e.g., 5'-AG|GUAAGU/3'-CAG|G), disrupting recruitment of Srsf/Hnrnp family proteins and increasing exon skipping in prostate cancer[82]. Collectively, writers and erasers coordinately regulate RNA splicing via dynamic m6A modification, with their dysregulation linked to disease pathogenesis.
3.2 Regulation of mRNA nuclear export
In eukaryotes, mRNA nuclear export critical for cytoplasmic protein synthesis depends on dynamic regulation by m6A writer and eraser proteins. The Mettl3-Mettl14 heterodimer, forming the core of the writer complex with Wtap and Virma, establishes context-specific m6A methylation on pre-mRNA[83]. This complex deposits m6A modifications to with site and transcript-specific patterns. For example, in human embryonic stem cells, Smad2/3 transcription factors bind to Mettl3, Mettl14, and Wtap, promoting m6A modification of target transcripts[84]. These m6A marks act as "identity tags," altering mRNA secondary structure to modulate interactions with nuclear export machinery components, thereby influencing export efficiency.
Fto and Alkbh5, as key m6A erasers, collaborate with writer proteins to maintain dynamic m6A homeostasis and regulate mRNA nuclear export. While Fto's demethylation activity toward m6A remains controversial, it demonstrates strong catalytic efficiency toward m6Am, primarily influencing snRNA methylation[85]. In contrast, nuclear localized Alkbh5 is a validated m6A demethylase. In cancer cells, Alkbh5 upregulation reduces m6A modification on specific mRNAs (e.g., Nanog and Foxm1), modifying their biophysical properties and potentially impairing nuclear export[86, 87]. This reduction in m6A modification alters the properties of these mRNAs and may consequently affect their nuclear export.
Reader proteins facilitate mRNA nuclear export by directly or indirectly mediating m6A-modified mRNA interactions. The nuclear reader Ythdc1 binds m6A-modified mRNAs and recruits Srsf3[80], a critical adaptor in the nuclear export factor (Nxf)1-dependent export pathway, to drive nuclear export[88]. Another reader protein, the shuttling reader Fmrp (fragile X mental retardation protein) preferentially associates with m6A modified transcripts, collaborating with chromosome region maintenance 1 (Crm1)[89] or Nxf2 adaptors to modulate target mRNA export[90].
In summary, writer and eraser proteins ensure the timeliness and precision of gene expression through the dynamic regulation of RNA nuclear export.
3.3 Regulation of mRNA translation
Writer and eraser proteins precisely regulate mRNA translation efficiency through dynamic m6A modifications, maintaining protein synthesis homeostasis in cells. This regulation is crucial for normal physiological processes and also plays an important role in disease pathogenesis.
Following nuclear export, mRNA translation efficiency is modulated by m6A modifications regulated by writers (e.g., Mettl3 and Mettl16) and erasers (e.g., Fto). Mettl3 enhances the interaction between poly(A)-binding protein cytoplasmic 1 (Pabpc1)[91] and eukaryotic translation initiation factor (eIf)4g by binding with Pabpc1, stabilizing eIf4f complex assembly, promoting the connection between the mRNA 5'-cap structure and the 3'-tail, facilitating ribosome recycling from termination sites back to the 5' end for subsequent translation rounds, ultimately boosting translation efficiency[92]. Mettl16 interacts with translation initiation factors (eIf3a, eIf3b) and rRNA to promote the assembly of the 43S pre-initiation complex and drive 80S initiation complex formation, ensuring that translation smoothly enters the elongation phase and thereby enhancing protein synthesis[7]. However, Fto diminishes the promotional effect of reader proteins (e.g., Ythdc1) on translation initiation by erasing m6A modifications from mRNA, reducing the loading of the 43S complex on mRNA and hindering the efficient assembly of the 80S complex[66]. This demethylation process reduces mRNA translational efficiency, thus forming a dynamic balance between the accuracy and speed of protein synthesis.
3.4 Regulation of mRNA decay
Decay, the final step in mRNA metabolism in which mRNAs become unstable and are degraded, is characterized by a precise mechanism involving writer and eraser proteins. Writer proteins (e.g., Mettl3, Mettl16) catalyze m6A deposition in mRNA subtelomeric regions, enhancing reader protein binding and shielding mRNA from RNase-mediated degradation to extend half-life. Stable m6A modifications further promote R-loop formation, supporting homologous recombination and telomere stability. Loss of Mettl3 and Mettl16 diminishes these m6A marks, accelerating mRNA decay and compromising telomere integrity and cellular function[93].
Eraser proteins (e.g., Fto, Alkbh5) modulate mRNA stability by removing m6A modifications, influencing cancer progression and chemoresistance. Fto demethylates salt-inducible kinase 2 (Sik2) mRNA, reducing its binding to Igf2bp2 and promoting degradation, which inhibits autophagy and facilitates clear cell renal cell carcinoma proliferation and metastasis[94]. Alkbh5-mediated demethylation of Foxo1 mRNA enhances its stability, upregulating superoxide dismutase (Sod2) to lower Ros levels, thereby maintaining cancer stem cell traitsand conferring chemoresistance in triple-negative breast cancer; Alkbh5 depletion sensitizes cells to doxorubicin by decreasing Foxo1/Sod2 expression[95]. Dynamic m6A modification regulates RNA stability, tumorigenesis, and drug response, with m6A-binding proteins and demethylases directly linked to tumor cell growth and metastasis[96, 97]. Further studies reveal that m6A modification dynamic during ovarian development and aging are closely related to RNA stability and chromatin state[98]. This suggests a complex role for m6A modifications in modulating DNA epigenetics and cellular function. Therefore, the fine-tuned addition and removal of m6A modifications by writer and eraser proteins affect mRNA stability while also playing pivatol roles in cellular biological functions and tumor progression.
The coordinated actions of writer, eraser, and reader proteins are essential for dynamic m6A modification of RNA, governing processes ranging from RNA stability and splicing to nuclear export and translational efficiency. Dysregulation of these proteins to diverse diseases, emphasizing the importance of deciphering their functional networks. Further investigations into their functions in physiological and pathological contexts will deepen our understanding of RNA biology and inform novel therapeutic strategies for associated disorders.
4. Molecular mechanisms of methyltransferases and demethylases in the liver diseases
In recent years, the investigation of m6A RNA methylation has gained increasing attention in the field of liver diseases. Substantial evidence demonstrates that aberrant m6A modification is intimately associated with the pathogenesis and progression of various hepatic disorders, including ALI, NAFLD, liver cirrhosis, alcoholic liver disease (ALD), viral hepatitis, HF, and HCC. Elucidating the regulatory mechanisms of m6A modification will not only advance our understanding of disease pathogenesis but also potentially novel therapeutic targets for clinical intervention.
4.1 m6A modification in acute liver injury
The m6A modification plays a pivotal role in ALI, illustrating its dynamic regulation of liver diseases. Mettl3 employs m6A modification to regulate the expression of nuclear factor erythroid 2-related factor 2 (Nrf2), its downstream genes, and the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (Malat1). Mettl3 deficiency aggravates oxidative stress and hepatocyte damage, while pregnane X receptor (Pxr) activation upregulates Fto, reduces m6A modification of Malat1, restores Nrf2 activity, and enhances antioxidant capacity. These findings identify Mettl3, Fto, and Pxr as critical therapeutic targets for liver injury prevention and treatment[99]. Mettl14 exerts hepatoprotective effects in endoplasmic reticulum (ER) stress-induced ALI by suppressing the pro-apoptotic factor C/EBP homologous protein (Chop). Under ER stress conditions, inhibition of the HMG-CoA Reductase Degradation 1 (Hrd1) ubiquitination pathway stabilizes Mettl14 expression, thereby reducing hepatocyte apoptosis. Notably, while Mettl14-deficient mice develop severe liver injury under stress conditions, genetic ablation of Chop (Ddit3) reverses this phenotype, underscoring the importance of the Mettl14-Chop axis in ER stress response[100]. Mettl3 promotes gluconeogenesis via stabilizing Phosphoenolpyruvate Carboxykinase 1 (Pck1) mRNA, reducing lactate accumulation and improving liver function. Both Pck1 and Mettl3 knockout exacerbates ischemia/reperfusion injury, emphasizing the protective role of the Mettl3/m6A-Pck1 pathway[101]. Mettl3 deficiency in ALI upregulates sphingomyelinase Smpd3, causing ceramide accumulation and mitochondrial/ER stress-induced apoptosis. Targeting the Mettl3-sphingolipid metabolism axis via Smpd3 inhibition or sphingomyelin synthase 1 (Sgms1) upregulation alleviates liver injury[102]. In acetaminophen-induced ALI, Wtap collaborates with Mettl3 and Mettl14 to enhance m6A-modified antioxidant and anti-apoptotic genes expression, maintaining metabolic homeostasis and inhibiting Jnk hyperactivation. Wtap downregulation exacerbates hepatocyte injury, suggesting Wtap complex activation as a therapeutic strategy[103]. It is worth mentioning that Fto emerges as a critical regulator in age-related ALI, where its reduced expression enhances m6A modification of Acsl4 (acyl-CoA synthetase long-chain family member 4) and Tfrc (transferrin receptor 1), promoting ferroptosis and exacerbating lipid peroxidation and Ros accumulation. In ischemia-reperfusion models, Fto overexpression suppresses ferroptosis related molecules and mitigating tissue damage. Therapeutically, enhancing Fto activity via nicotinamide mononucleotide (NMN) represents a promising strategy to alleviate liver transplantation injury in elderly individuals[104].
While writers and erasers play crucial roles in ALI pathogenesis, the mechanisms of action of proteins such as Mettl3, Mettl14, and Fto in hepatocyte stress response, repair, and regeneration require elucidation. Specifically, the signaling pathway linking Pxr activation to Fto upregulates and affects the m6A modification of Malat1, and the upstream and downstream regulatory factors of the Mettl3 sphingolipid metabolism pathway in liver repair, require in-depth study. Exploration of these areas may guide the direction of future research on ALI.
4.2 m6A modification in viral hepatitis
The m6A modification plays a significant role in the interaction between chronic hepatitis B (CHB) and Covid-19, revealing its dynamic regulatory role in liver diseases. Clinical observations indicates that patients with CHB have an increased risk of hospitalization after contracting Covid-19. Furthermore, the immune disorders and liver injury induced by Covid-19 are exacerbated under abnormal m6A regulation[105]. Notably, Mettl3 enhances antiviral genes expression to activate the immune response; however, this can lead to uncontrolled inflammation[106]. Rbm15 contributes to Covid-19-associated hepatitis by binding SARS-CoV-2 RNA to modulate viral replication while simultaneously influencing host immune responses, particularly through regulation of key cytokines (Il-6, Tnf-α) and subsequent inflammatory cascades[107]. By contrast, Fto weakens RNA stability and interferes with antiviral immunity. These findings suggest that intervention strategies targeting m6A regulation could help to balance the antiviral and anti-inflammatory responses.
In hepatitis B virus (HBV)-related acute-on-chronic liver failure (ACLF), HBV infection upregulates Mettl3 and the miRNA miR-146a-5p, increases the m6A modification level, and exacerbates apoptosis, inflammation, and viral replication. Inhibiting Mettl3 can reduce hepatocyte damage and inhibit the maturation of miR-146a-5p to improve liver injury, highlighting Mettl3 as a key therapeutic target for ACLF[108]. Equally noteworthy is that, in HBV-induced acute liver failure (ALF) features dysregulated Mettl3, Igf2bp2, and Igf2bp3 expression that promotes immune cell infiltration (e.g. CD8+T cells and T helper 17 cells), with consequent Th17/Treg imbalance worsening hepatic injury. This has spurred development of m6A-based diagnostic models for early ALF detection and immunotherapeutic approaches[109]. The HBV X protein (HBx) upregulates Rbm15 to enhance m6A modification of circRNA Fam210a, accelerating its decay while activating Ybx1 transcriptional activity to drive hepatocarcinogenesis[110]. Additionally, HBV stimulates Kiaa1429-mediated m6A modification to upregulate Ccr9, which stabilizes drug transporters ATP-binding cassette subfamily B member 1 (Abcb1) and subfamily C member 1 (Abcc1) expression, fostering HCC chemoresistance and poor prognosis[111]. HBX further engage the m6A complex (Mettl3/Mettl14), recruiting it to viral cDNA to enhance transcript modification. This regulatory mechanism stabilizes viral transcription while reducing the expression of the HBX protein, forming a negative feedback loop to maintain CHB infection[112].
Hepatitis C virus (HCV) exploits the m6A machinery (Wtap-Mettl3/14) to promote its life cycle and immune evasion. Wtap directs m6A complex positioning on HCV RNA, preventing retinoic acid-inducible gene I (Rigi) detection while boosting virion production[113, 114].
The m6A modification has a negative regulatory effect on the hepatitis D virus (HDV) life cycle. Research shows that Mettl3/Mettl14-mediated m6A modification exerts negative regulation on HDV by reducing genomic RNA and delta antigen levels, yet paradoxically increasing extracellular genome accumulation. This modification further impedes virion assembly through Ythdc1-mediated interference with HDV genome-delta antigen interactions[115]. The demethylase Alkbh5 is upregulated under hypoxic conditions, reducing the m6A modification of HBV RNA to prolong its stability and promote viral replication[116]. Alkbh5 also enhances the stem cell-like properties and immune evasion of HCC by stabilizing Snai2 transcripts[117]. Meanwhile, alterations in Fto expression during HIV/HCV co-infection have been closely related to metabolic disorders, insulin resistance, and patient treatment response[118]. These two demethylases play crucial roles in the progression of viral infections by modulating viral RNA stability and host transcriptional responses, reducing the recognition of viral RNA by sensors such as Rigi, and enhancing the immune evasion ability of the virus[119].
Although it is known that m6A modification affects the stability and translation efficiency of viral RNA, other molecular aspects of its regulation in hepatitis virus infection require further research. Specifically, the mechanisms by which hepatitis viruses use the host cell's m6A modification system to evade the recognition and clearance of the immune system, as well as how the host cell resists virus infection by regulating m6A modification, remain unclear.
4.3 m6A modification in NAFLD
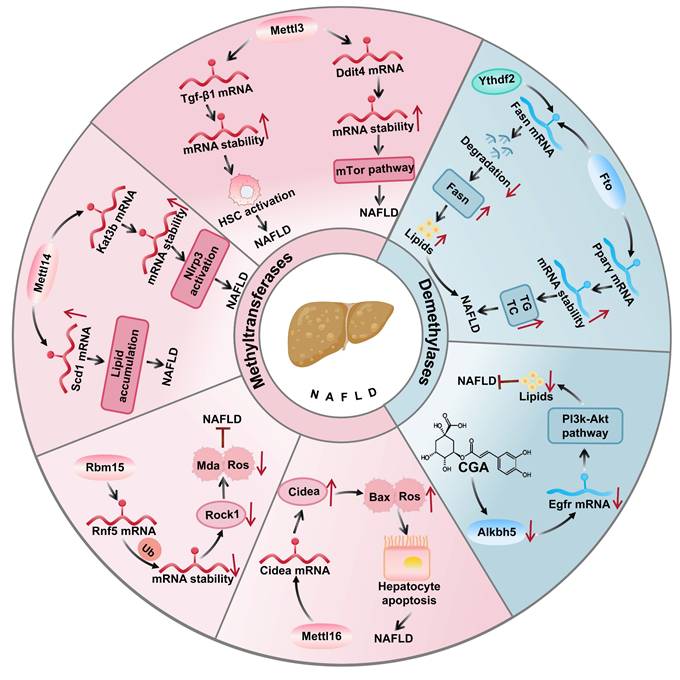
The role of m6A modification in NAFLD pathogenesis and therapeutic targets m6A modification crucially regulates NAFLD progression by modulating lipid metabolism, inflammatory responses, and cellular processes through m6A-related enzymes and target gene stability (Figure 2).
The main writer Mettl14 promotes inflammation by enhancing the stability of Nlrp3 inflammasome mRNA[120], while arsenite increases Nlrp3 m6A modification exacerbating NAFLD[121]. Lipopolysaccharide activates NF-κB p65 to transcribe Mettl3 and Mettl14, boosting Tgf-β1 mRNA m6A modification in the 5'-UTR for cap-independent translation and NAFLD progression[22]. Additionally m6A modification significantly influences in NAFLD and obesity, especially in the function of Mettl3 in myeloid cells[122]. Mettl3 in myeloid cells regulates Ddit4 mRNA stability via m6A to inhibit mTor/NF-κB signaling, counteracting NAFLD and obesity[123].
The autophagy mechanisms of m6A modification in NAFLD cannot be ignored. In NAFLD mouse models and free fatty acid-treated hepatocytes, elevated m6A modification levels correlate with Mettl3 upregulation[124]. In autophagic mechanisms, Mettl3 upregulation in NAFLD models modifies Rubicon mRNA, promoting its expression via Ythdf1, which blocks autophagosome-lysosome fusion and lipid clearance[125]. Mettl3-mediated Cyp2b6 m6A modification increases its expression, inhibiting insulin receptor substrate phosphorylation and exacerbating insulin resistance[126]. Further studies found that the overexpression of Mettl14 and Mettl3 promotes fatty acid synthesis and lipid accumulation by stabilizing the mRNAs of ATP citrate lyase (Acly) and stearoyl-CoA desaturase 1 (Scd1) and accelerating NAFLD to HCC[127], while adipose tissue Mettl3 and Mettl14 modify Adrb2/3, adipose triglyceride lipase (Atgl), and comparative gene identification 58 (Cgi58) transcripts to impair β-adrenergic signaling, reducing lipolysis and worsening obesity/NAFLD[128]. Mettl16 promotes NAFLD via m6A mediated cell death inducing DFFA-like effector A (Cidea) upregulation[129], whereas Rbm15 reduces NAFLD by m6A-methylating ring finger protein 5 (Rnf5) to ubiquitinate and degrade Rock1[130].
In the treatment of NAFLD, the demethylases Fto and Alkbh5 are targeted for their regulation of metabolism and inflammatory responses via the removal of m6A modifications. Fto upregulation enhances lipogenic gene expression and promotes lipid accumulation[131], while Alkbh5 affects proinflammatory genes[132]. Specifically, Fto catalyzes m6A demethylation altering the expression and splicing of lipid-related genes. As shown in (Figure 3), in the liver, Fto upregulation enhances sterol regulatory element binding protein 1c (Srebp1c) mediated lipogenesis which inhibiting fatty acid oxidation to exacerbate NAFLD[133]. By contrast, angiotensin-receptor blockers inhibit Fto to increase solute carrier family 7 member 11 (Slc7a11) m6A modification and suppress ferroptosis. Furthermore, chlorogenic acid (Cga) promotes autophagy and reduces lipid deposition by inhibiting the m6A demethylase activity of Alkbh5, revealing the potential application of m6A modification in the treatment of NAFLD[134].
The role of m6A writer and eraser proteins in NAFLD. In NAFLD, Mettl3 promotes disease progression by stabilizing Ddit4 mRNA to activate the mTor pathway and enhancing Tgf-β1 mRNA to drive liver fibrosis, while Mettl14 accelerates NAFLD by increasing Kat3b-mediated Nlrp3 expression. Conversely, Mettl3 inhibits hepatic lipid metabolism through Cd36 and Ccl2 regulation, Rbm15 reduces oxidative stress by destabilizing Rnf5 mRNA, and Fto attenuates NAFLD progression by suppressing Srebf1 mediated lipogenesis.
4.4 m6A modification in ALD
The progression of ALD encompasses multiple stages, from alcoholic fatty liver to cirrhosis and even hepatocellular carcinoma. In recent years, m6A, one of the most common types of mRNA modification, has been proven to play multifaceted regulatory roles in ALD development and progression. First, m6A modification affects the formation of alcoholic fatty liver by regulating the expression of genes related to lipid metabolism. Research shows that elevated m6A levels enhance the translation of lipogenic genes, promoting lipid synthesis and hepatic steatosis, while reduced the expression of the demethylase Fto may further exacerbate lipid accumulation[135]. Second, in alcoholic hepatitis, m6A modification exhibits a dual regulatory role in the liver inflammatory response. The m6A reader protein Ythdf2 promotes the degradation of inflammatory factors, thus alleviating inflammation[136]. However, stimulation with alcohol may change the m6A level, upregulating proinflammatory factors and exacerbating liver injury. Studies have shown that alcohol intake can trigger Kupffer cell pyroptosis and increase the release of proinflammatory factors such as Il-1β. Through regulation by the RNA-modifying enzyme Mettl3, m6A modification influences the inflammatory response and pyroptosis in Kupffer cells. Specifically, inhibiting Mettl3 was shown to relieve the inflammatory cascade reaction caused by Kupffer cell pyroptosis, thus reducing pathological damage in alcoholic steatohepatitis[137]. Fto, by reducing the m6A modification level of the Il-17a receptor gene Il-17ra, increasing its protein expression, thereby exacerbating the inflammatory response in the liver. Fto upregulation elevates Il-17-related inflammatory factors, whereas Fto inhibition enhances Il-17ra m6A modification, lowering its protein levels and attenuating inflammation[24]. In addition, at the stage of alcoholic HF, m6A modification affects fibrosis-related genes. For instance, m6A modification may affect the stability of key genes in the Tgf-β signaling pathway, thus promoting or inhibiting fibrosis[138]. Finally, during the progression of ALD to liver cancer, m6A modification influences the expression of oncogenes and tumor suppressor genes, with abnormalities promoting the proliferation and invasion of liver cancer cells, driving disease progression. Therefore, m6A modification affects different stages of ALD through multiple pathways, having a profound impact on the disease process.
In summary, m6A modification exerts multilevel regulatory effects on ALD pathogenesis, impacting key processes including lipid metabolism, inflammation, fibrosis, and carcinogenesis. Further investigation into the molecular mechanisms of m6A modification and its stage-specific roles in ALD may provide novel insights into pathogenesis machanisms. Such researchs could also uncover potential diagnostic biomarkers and therapeutic targets, paving the way for personalized treatment strategies.
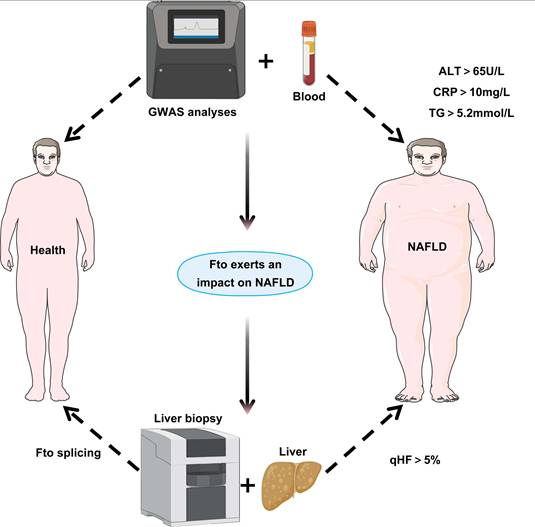
Clinical relevance of Fto in NAFLD. By collecting blood samples from healthy individuals and NAFLD patients, measuring key clinical parameters (ALT > 65 U/L, C-reactive protein (CRP) > 10 mg/L, Triglyceride (TG) > 5.2 mmol/L), and integrating GWAS data, this study combines hepatic biopsy for quantitative hepatic fat (qHF > 5%) assessment in NAFLD patients, followed by analysis of Fto splicing patterns in liver tissues to elucidate its impact on NAFLD development and progression.
Mechanisms of m6A “writers” and “eraser” in hepatic fibrosis. m6A writers (Mettl14/Mettl3/Wtap) promote fibrosis by destabilizing Lats2 to activate Yap-driven HSC activation and stabilizing Enpp1 to upregulate Hilpda-mediated oxidative stress. Conversely, Alkbh5 plays a dual regulatory role in hepatic fibrosis: while enhancing Drp1 expression to promote mitochondrial fission and oxidative stress, it simultaneously suppresses fibrosis by destabilizing Nova2 to inhibit Wnt/β-catenin signaling and stabilizing Ptch1 to block Gli1 activation, thereby comprehensively modulating HSC activation.
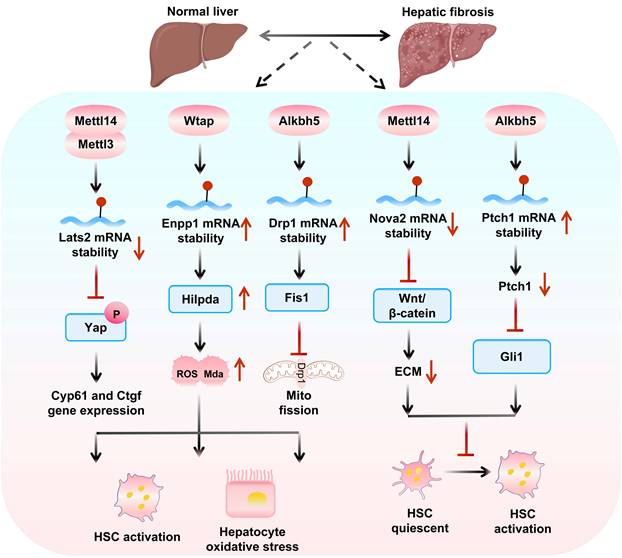
4.5 m6A modification in HF
In HF and related diseases, m6A modification regulates key gene expression, hepatic stellate cell (HSC) activation, and cell-cell interactions, highlighting its potential as a therapeutic target and providing new research directions (Figure 4).
The role of m6A modification-related enzymes in hepatic fibrosis and therapeutic implications Mettl14 exerts dual effects on HF progression via reader protein-dependent m6A mechanisms. On one hand, the Mettl14 and Ythdf1 axis reduces glutaminase 2 (Gls2) translation, creating an oxidative stress microenvironment that recruits Cx3cr1+/Ccr2+ monocytes. Crucially, Cx3cr1 activates the NF-κB pathway via Myd88, leading to transcriptional upregulation of profibrotic factors like S100a4, which drives HSC activation and fibrosis[139]. On the other hand, Mettl14 decreases m6A modification of Nova2 mRNA, enabling Ythdf2-mediated degradation and inhibit Nova2 activity[140], Conditional knockout of Mettl3, decreases m6A modification of Lats2 mRNA, thereby inhibiting the nuclear entry of Yap, downregulates profibrotic genes and causing HSCs to switch from an activated state to a quiescent state[141], blocking HF progression.
Sodium arsenite (NaAsO2) enhances m6A modification, promoting Mettl14 and Igf2bp2-mediated stability of Tgf-β1 mRNA; limiting this modification prevents NaAsO2-induced HSC activation[142]. Ectonucleotide pyrophosphatase and phosphodiesterase 1 (Enpp1), upregulated via Wtap-mediated m6A modification and Ythdf1-dependent translation[143], exacerbates fibrosis by promoting HSC lipid oxidation and proliferation. Conversely, N-acetyl-serine-aspartic acid-lysine-proline (AcSDKP) inhibits Hedgehog signaling by stabilizing patched 1 (Ptch1) mRNA via Wtap downregulation[144].
In the Fto/Unc51 like autophagy activating kinase 1 (Ulk1) axis, Fto promotes Ulk1-mediated autophagy and HSC activation, whereas Ythdc2-mediated Ulk1 regulation inhibits HF[145]. During ferroptosis, m6A modification stabilizes Becn1 mRNA to activate autophagy[94], a process enhanced by Ythdf1 and downregulated Fto. Dihydroartemisinin (DHA) inhibits HSC activation via Fto-upregulated autophagy and ferroptosis[146].
Alkbh5 exhibits context-dependent roles in HF. In Tgf-β-1-stimulated HSCs, Alkbh5 overexpression reduces Snail1 mRNA stability and suppresses profibrotic markers[23], while simultaneously activating Ptch1 to inhibit HSC activation[147] and blocking Drp1 mRNA m6A modification to restrain mitochondrial fission-dependent HSC proliferation/migration[148]. Notably, reduced Alkbh5 expression exacerbates HF progression, positioning it as a protective factor in fibrotic microenvironments. Given that the decreased expression of Alkbh5 exacerbates HF, research on the regulation of its demethylation activity is expected to open up new directions for the treatment of HF.
4.6 m6A modification in liver cirrhosis
Liver cirrhosis, characterized by extensive replacement of liver parenchyma with fibrous tissue and a significant decline in liver function, represents the terminal stage of liver diseases. Emerging evidences have shown that m6A-modified mRNAs play an important role in the pathogenesis and progression of liver cirrhosis. As a dynamic and reversible mode of epigenetic RNA regulation, m6A modification influences the key pathological processes of liver cirrhosis via regulation of gene expression, RNA stability, translation efficiency, and other mechanisms. As one of the main drivers of liver cirrhosis, the Tgf-β signaling pathway stabilizes its downstream Smad genes through m6A modification, thereby amplifying fibrogenic signal. This accelerates the activation of HSCs and the accumulation of extracellular matrix, promoting the development of fibrosis[149, 150]. In cirrhotic tissues, elevated expression of m6A writer enzymes Mettl3 and Mettl14 enhances the translation efficiency of fibrogenic genes, aggravating the fibrotic process and promoting the progression of liver cirrhosis[151]. Conversely, low expression of the m6A demethylase Fto leads to increased m6A modification of profibrotic genes, stabilizing their mRNAs and promoting the accumulation of fibrosis[152]. In addition, reduced levels of the m6A-binding protein Ythdf2 hinders the degradation of fibrotic and proinflammatory genes, resulting in the continuous expression of these genes and exacerbating the inflammatory and fibrotic responses. The m6A-mediated stabilization of proinflammatory cytokines Il-6 and Il-1β also perpetuates chronic inflammation, further driving cirrhosis progression[153]. Through these multifaceted mechanisms that regulates fibrotic and inflammatory gene expression, m6A modification significantly contributes to cirrhosis development. As an important epigenetic regulatory mechanism, m6A modification machinery provide new potential targets for the treatment of liver cirrhosis. Future therapies targeting m6A modification may potentially slow disease progression, opening new avenues for more effective treatment strategies.
4.7 m6A modification in HCC
As illustrated in (Figure 5), research on HCC has shown that m6A modification significantly impacts tumor progression and treatment response. Mettl3-mediated m6A modification stimulates Egfr mRNA translation, leading to lenvatinib resistance[154]. While Mettl3 is significantly upregulated in HCC and associated with shorter survival, with its knockout inhibiting tumorigenicity and metastasis. Mettl3 mediates Socs2 degradation, modulates Snail's epithelial-mesenchymal transition, and promotes cell proliferation/lipid production via modifying Rdm1 and stabilizing Linc00958[155, 156], forming a positive feedback loop with Stat3 by suppressing anti-tumor CD8+T cells through the Scap-cholesterol axis[157], promoting Stat3 mRNA translation, and accelerating metastasis as Stat3 upregulates Wtap to enhance Mettl3's nuclear function[158]. Additionally, Mettl3 promotes Tug1 upregulation to regulate Pd-l1/Cd47 and affect immune escape[159].
Mettl14-induced m6A modification stabilizes Usp48 and Sirt6, suppressing HCC glycolysis and malignancy[160], reduces circORC5 expression to inhibit gastric cancer, and enhances circSTX6 in HCC[161], while possibly downregulating Hnf3γ (which is negatively correlated with malignancy/survival) via m6A modification, with exogenous Hnf3γ promoting liver cancer cell differentiation and growth inhibition[162].
Mettl16, upregulated in HCC and associated with poor prognosis, binds to lncRNA Rab11b-as1 inducing its m6A modification and degradation, promoting cell proliferation/migration. Targeting the Mettl16-eIf3a/b axis may represent a novel anti-cancer strategy[7, 163]. Wtap-mediated m6A modification stabilizes lnc-OXAR via Igf2bp2, leading to oxaliplatin resistance in NASH-related HCC[164], promotes HCC progression via the HuR-est1-p21/p27 axis[165] and upregulates the expression of autophagy-related 5 (Atg5), promoting its translation during ferroptosis. HBX-interacting protein (Hbxip) enhances cisplatin resistance in liver cancer cells by upregulating Wtap and its m6A modification of poly (Adp-ribose) polymerase (Parp1)[166].
The role of m6A writer and eraser proteins in HCC. Methyltransferases: Mettl3 activates cholesterol synthesis via Scap mRNA translation to promote tumor growth; Wtap regulates P27 through Ets1 mRNA for HCC oncogenesis; Virma degrades Rnd3 mRNA to drive migration/invasion. Demethyltransferases: Fto generates Cxcl5 via Cebpa mRNA translation to facilitate HCC cell growth; Alkbh5 degrades Lypd1 mRNA to enhance migration, and activates tumor-associated macrophages (TAM) via Map3k8 mRNA, exacerbating migration/invasion through Jnk/Erk pathway and elevated Il10/Tgf-β, collectively demonstrating how m6A modifications influence HCC development and tumor microenvironment.
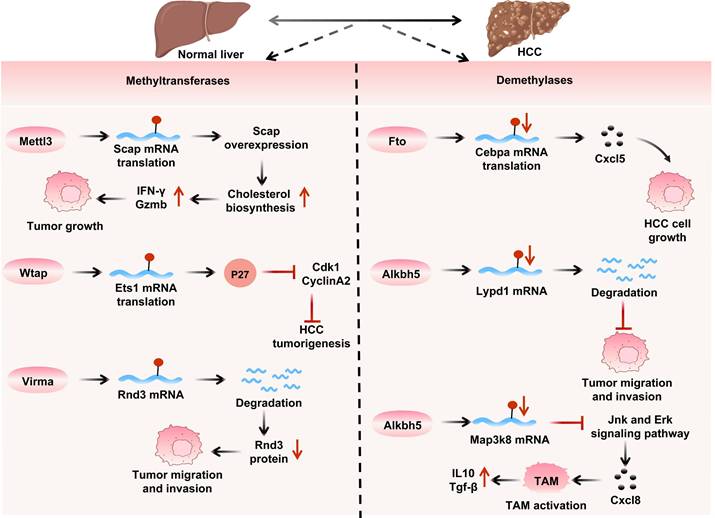
Regarding other related writers, Hakai interacts with Ajuba to enhance the proliferation of HCC cells[167]. HBX promotes the expression of Rbm15, increasing m6A levels on Fam210a mRNA and inducing its degradation, thereby improving the proliferation, stemness, and tumorigenicity of HCC cells. Kiaa1429 induces m6A methylation in the 3′-UTR of Gata Binding Protein 3 (Gata3) precursor mRNA, downregulating the expression of Gata3 and promoting the metastasis of HCC cells[168].
As m6A demethylases, Fto (highly expressed in HCC) is stabilized by Fto-interacting transcript 1 (Fto-It1) to upregulate Glut1/Pkm2/C-myc for glycolysis and proliferation[126]. Meanwhile FB23 and FB23-2 inhibitors increase Erbb3/Tubb4a m6A levels to inhibit Akt-Mtor signaling[169]. Additionally, adenosylmethionine decarboxylase 1 (Amd1) upregulates Sry-box transcription factor 2 (Sox2), Kruppel-like factor 4 (Klf4), and Nanog through Fto-mediated mRNA demethylation, maintaining tumor stemness[170].
Downregulation of Alkbh5 expression exerts a potential tumor and suppressive effect in HCC by inhibiting the transcription of Lypd1[171]. However, Alkbh5 has also been demonstrated to possess significant pro-tumorigenic functions: it enhances cancer stem cell properties and immune evasion by stabilizing Snai2[117], and its target, Linc02551, similarly drives HCC growth and metastasis[172]. Intriguingly, in radiation-induced hepatic stellate cells (HSCs), Alkbh5 activation promotes chemokine secretion via the Tirap/NF-κB signaling pathway[109], a mechanism potentially contributing to HCC radioresistance. This marked functional dichotomy observed across disease contexts highlights a critical unresolved question: how specific microenvironmental factors precisely regulate Alkbh5's substrate specificity and downstream signaling cascades. Future research elucidating the underlying mechanisms holds promise for developing context-specific therapeutic interventions targeting liver disease progression.
5. Investigation and advancement m6A of methylation-associated pharmaceuticals
In the treatment of liver diseases, novel chemical compounds influencing m6A modification have enabled m6A-based therapies, regulating activities of m6A-related enzymes. Fto inhibitors like Fb23 and Fb23-2 alter m6A levels of Erbb3 and signaling pathways, suppressing liver cancer cell proliferation and survival[173-175], while Dac51 inhibits Fto-mediated glycolysis, showing enhanced efficacy when combined with anti-PD-L1 blockers[176]. STM2457, a Mettl3 inhibitor, reduces Egfr's m6A modification to increase HCC cells sensitivity to lenvatinib[177], and UZH1A selectively binds to Mettl3 mRNA to exert antitumor effects in leukemia and osteosarcoma[178]. In NAFLD, STM2457 improves mitochondrial function and lipid oxidation[179]. ARBs inhibit Fto demethylation and promote Slc7a11 expression[180], and Cga/MV1035 inhibit Alkbh5 to enhance autophagy[181]. In ALI, NMN enhances Fto activity to alleviate liver transplantation injury[130].
Notably, traditional Chinese medicines show unique advantages: Resina Draconis extract induces HCC cell apoptosis by downregulating Mettl3[182]; Ling Gui Zhu Gan soup reduces liver fat degeneration by decreasing m6A methylation and Socs2 expression[183]; Gan Jiang Ling Zhu soup promotes Mettl14 and Ugt2a3 expression to alleviate NASH[184]; indole-3-lactic acid regulates Cyp8b1 via Fto/m6A/Ythdf2 to inhibit liver fat accumulation[185]; and cucurbitacin B covalently binds to Igf2bp1 to block m6A-modified mRNA recognition, offering new directions for liver cancer drug development[186].
6. Therapeutic implications and future directions
Emerging evidence have demonstrated that RNA m6A modification plays a pivotal role in the pathogenesis and progression of liver diseases. Elucidating the molecular mechanisms linking m6A modification to liver disease progression and may reveal promising therapeutic targets for drug development. Notably, targeting m6A regulators not only enhances treatment efficacy by modulating the liver disease microenvironment but also effectively overcomes drug resistance in clinical therapy, demonstrating promising application prospects.
Despite significant advancements in this field, only a limited number of m6A-targeting modulators have demonstrated ideal therapeutic efficacy in clinical applications for liver diseases. This limitation arises from multiple factors, including the lack of precision in current modulators for m6A modification regulation, often leading to off-target effects. For example, the Mettl3 inhibitor STM2457 may interfere with other RNA-modifying enzymes, disrupting snRNA methylation and causing global transcriptomic dysregulation[177]. Moreover, targeted interventions can induce compensatory feedback mechanisms, such as the upregulation of Fto and Alkbh5 following Mettl3 suppression, potentially counteracting therapeutic benefits[187]. Beyond specificity challenges, a key obstacle lies in the predominant focus on modulator activity optimization while overlooking critical pharmacokinetic properties, including absorption, distribution, metabolism, excretion, and lipophilicity. To address this, future research should prioritize the development of advanced delivery systems, such as TAT peptide-functionalized PLGA nanoparticles[156] or folate-modified exosome-liposome hybrid nanocarriers to enhance[188] tissue-specific drug delivery. Furthermore, the tissue-specific nature of m6A regulation introduces additional complexity. While hepatocyte specific Mettl3 upregulation exacerbates lipid accumulation in NAFLD[123], in contrast, it exerts anti-inflammatory effects in myeloid cells by stabilizing Ddit4 mRNA[125]. This stark functional dichotomy underscores the need for highly selective therapeutic strategies to avoid unintended systemic effects.
To address these challenges, future investigations should focus on three pivotal research directions: (1) Deciphering cell type-specific m6A epitranscriptomic landscapes in hepatic microenvironments through single-cell sequencing approaches; (2) Establishing physiologically relevant organoid models that faithfully recapitulate liver microenvironments for comprehensive pharmacodynamic evaluation of m6A modulators; (3) Engineering tissue-specific delivery platforms to enable precision therapeutic modulation. These strategic advancements will substantially facilitate the translation of m6A-targeted therapeutics from fundamental research to clinical implementation.
Abbreviations
Mettl1: Methyltransferase-like1; Mettl3: Methyltransferase-like3; Mettl5: Methyltransferase-like5; Mettl10: Methyltransferase-like10; Mettl14: Methyltransferase-like14; Mettl16: Methyltransferase-like16; Virma/Kiaa1429: Vir-like m6A methyltransferase associated; Wtap: Wilms tumor 1- associated protein; Zcchc4: Zinc Finger CCHC-type Containing 4; Znf217: Zinc Finger Protein 217; Hsp90: Heat Shock Protein 90; Stk33: Serine/Threonine Kinase 33; Clip1: Cytoplasmic Linker Protein 1; Trmt112: tRNA methyltransferase 112; Rbm15/Rbm15b: RNA binding motif protein 15/15B; Zfp217: Zinc Finger Protein 217; Zc3h13: Zinc Finger CCCH-type Containing 13; Pcif1: Phosphorylated CTD-interacting factor 1; Cbll1/Hakai: E3 ubiquitin protein ligase Hakai; Fto: Fat mass and obesity-associated; Alkbh5: AlkB homologue 5; ALI: Acute liver injury; NAFLD: non-alcoholic fatty liver disease; HF: liver fibrosis; HCC: hepatocellular carcinoma; Pck1: phosphoenolpyruvate carboxykinase 1; Fmrp: fragile X mental retardation protein; Crm1: chromosome region maintenance 1.
Acknowledgements
All authors express their sincere gratitude to Dr. Michelle Kahmeyer-Gabbe for her valuable assistance in improving the English language of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No.82570749, 82300722, 82570752, 82370630); the Anhui Provincial Natural Science Foundation (No.2308085QH248); the Research Fund of Anhui Institute of Translational Medicine (No.2021zhyx-B06, 2022zhyx-B07); the Fund of Traditional Chinese Medicine Institute of Anhui Dabie Mountain (No. TCMADM-2024-02).
Author contributions
Ya-Ning Chen, Sai Zhu and Li-Jiao Sun drafted the manuscript. Si-Jin Sun, Rui Zheng, Rong-Rong Zhou collected literature. Xiao Feng Li, Liang-Yun Li, Yu-Xin Zhao picture censorship. Cheng Huang, Xiao-Ming Meng, Lei Zhang, Xiong-Wen Lv, Hua Wang analysis conceptualization and supervision. Xin Chen and Jun Li provided some important guidance on the revised manuscript. Jun Li was involved in the critical revision of the manuscript. All authors read and approved the final paper.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sun H, Li K, Liu C, Yi C. Regulation and functions of non-m(6)A mRNA modifications. Nat Rev Mol Cell Biol. 2023;24:714-31
2. Huang S, Wylder AC, Pan T. Simultaneous nanopore profiling of mRNA m(6)A and pseudouridine reveals translation coordination. Nat Biotechnol. 2024;42:1831-5
3. Qi YN, Liu Z, Hong LL, Li P, Ling ZQ. Methyltransferase-like proteins in cancer biology and potential therapeutic targeting. J Hematol Oncol. 2023;16:89
4. Xu M, Xu HH, Lin Y, Sun X, Wang LJ, Fang ZP. et al. LECT2, a Ligand for Tie1, Plays a Crucial Role in Liver Fibrogenesis. Cell. 2019;178:1478-92 e20
5. Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E. et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022;50:D231-D5
6. Shi H, Wei J, He C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell. 2019;74:640-50
7. Su R, Dong L, Li Y, Gao M, He PC, Liu W. et al. METTL16 exerts an m(6)A-independent function to facilitate translation and tumorigenesis. Nat Cell Biol. 2022;24:205-16
8. Du W, Huang Y, Chen X, Deng Y, Sun Y, Yang H. et al. Discovery of a PROTAC degrader for METTL3-METTL14 complex. Cell Chem Biol. 2024;31:177-83 e17
9. Sun HL, Zhu AC, Gao Y, Terajima H, Fei Q, Liu S. et al. Stabilization of ERK-Phosphorylated METTL3 by USP5 Increases m(6)A Methylation. Mol Cell. 2020;80:633-47 e7
10. Chen L, Wang C, Sun H, Wang J, Liang Y, Wang Y. et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform. 2021;22:1706-28
11. Wei W, Zhang ZY, Shi B, Cai Y, Zhang HS, Sun CL. et al. METTL16 promotes glycolytic metabolism reprogramming and colorectal cancer progression. J Exp Clin Cancer Res. 2023;42:151
12. Xu Y, Chen Y, Yao Y, Xie H, Lu G, Du C. et al. VIRMA contributes to non-small cell lung cancer progression via N(6)-methyladenosine-dependent DAPK3 post-transcriptional modification. Cancer Lett. 2021;522:142-54
13. Liu Z, Gao L, Cheng L, Lv G, Sun B, Wang G. et al. The roles of N6-methyladenosine and its target regulatory noncoding RNAs in tumors: classification, mechanisms, and potential therapeutic implications. Exp Mol Med. 2023;55:487-501
14. Smolin EA, Buyan AI, Lyabin DN, Kulakovskiy IV, Eliseeva IA. RNA-Seq data of ALKBH5 and FTO double knockout HEK293T human cells. Data Brief. 2022;42:108187
15. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y. et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782
16. Faial T. FTO regulates LINE1 in early development. Nat Genet. 2022;54:921
17. Jiang Y, Wan YC, Gong M, Zhou SL, Qiu JN, Cheng WJ. RNA demethylase ALKBH5 promotes ovarian carcinogenesis in a simulated tumour microenvironment through stimulating NF-κB pathway. J Cell Mol Med. 2020;24:6137-48
18. Tang C, Klukovich R, Peng HY, Wang ZQ, Yu T, Zhang Y. et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. P Natl Acad Sci USA. 2018;115:E325-E33
19. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-95
20. Richter JD, Zhao X. The molecular biology of FMRP: new insights into fragile X syndrome. Nat Rev Neurosci. 2021;22:209-22
21. Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18:31-42
22. Feng Y, Dong H, Sun B, Hu Y, Yang Y, Jia Y. et al. METTL3/METTL14 Transactivation and m(6)A-Dependent TGF-beta1 Translation in Activated Kupffer Cells. Cell Mol Gastroenterol Hepatol. 2021;12:839-56
23. Zhou HY, Wang BQ, Chen MX, Wang YF, Jiang YF, Ma J. KDM4C represses liver fibrosis by regulating H3K9me3 methylation of ALKBH5 and m6A methylation of snail1 mRNA. J Dig Dis. 2024;25:298-309
24. Gan X, Dai Z, Ge C, Yin H, Wang Y, Tan J. et al. FTO promotes liver inflammation by suppressing m6A mRNA methylation of IL-17RA. Front Oncol. 2022;12:989353
25. Sun L, Chen X, Zhu S, Wang J, Diao S, Liu J. et al. Decoding m(6)A mRNA methylation by reader proteins in liver diseases. Genes Dis. 2024;11:711-26
26. Chen X, Zhu S, Li HD, Wang JN, Sun LJ, Xu JJ. et al. N(6)-methyladenosine-modified circIRF2, identified by YTHDF2, suppresses liver fibrosis via facilitating FOXO3 nuclear translocation. Int J Biol Macromol. 2023;248:125811
27. Zhu S, Chen X, Sun L, Li X, Chen Y, Li L. et al. N (6)-Methyladenosine modification of circDcbld2 in Kupffer cells promotes hepatic fibrosis via targeting miR-144-3p/Et-1 axis. Acta Pharm Sin B. 2025;15:296-313
28. Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20:608-24
29. Falnes PO. Closing in on human methylation-the versatile family of seven-beta-strand (METTL) methyltransferases. Nucleic Acids Res. 2024;52:11423-41
30. Jin X, Liu L, Liu D, Wu J, Wang C, Wang S. et al. Unveiling the methionine cycle: a key metabolic signature and NR4A2 as a methionine-responsive oncogene in esophageal squamous cell carcinoma. Cell Death Differ. 2024;31:558-73
31. Thomas CB, Scavetta RD, Gumport RI, Churchill ME. Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding. J Biol Chem. 2003;278:26094-101
32. Wang P, Doxtader KA, Nam Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol Cell. 2016;63:306-17
33. Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191-8
34. Wang Z, Pan Z, Adhikari S, Harada BT, Shen L, Yuan W. et al. m(6) A deposition is regulated by PRMT1-mediated arginine methylation of METTL14 in its disordered C-terminal region. EMBO J. 2021;40:e106309
35. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP. et al. The U6 snRNA m(6)A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell. 2017;169:824-35 e14
36. Peng H, Chen B, Wei W, Guo S, Han H, Yang C. et al. N(6)-methyladenosine (m(6)A) in 18S rRNA promotes fatty acid metabolism and oncogenic transformation. Nat Metab. 2022;4:1041-54
37. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47:7719-33
38. Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M. et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278-88
39. Su S, Li S, Deng T, Gao M, Yin Y, Wu B. et al. Cryo-EM structures of human m(6)A writer complexes. Cell Res. 2022;32:982-94
40. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-46
41. Jenjaroenpun P, Wongsurawat T, Wadley TD, Wassenaar TM, Liu J, Dai Q. et al. Decoding the epitranscriptional landscape from native RNA sequences. Nucleic Acids Res. 2021;49:e7
42. Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18:18-30
43. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z. et al. VIRMA mediates preferential m(6)A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10
44. Xu Y, Song M, Hong Z, Chen W, Zhang Q, Zhou J. et al. The N6-methyladenosine METTL3 regulates tumorigenesis and glycolysis by mediating m6A methylation of the tumor suppressor LATS1 in breast cancer. J Exp Clin Cancer Res. 2023;42:10
45. Horiuchi K, Kawamura T, Iwanari H, Ohashi R, Naito M, Kodama T. et al. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288:33292-302
46. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369-73
47. Zheng F, Du F, Qian H, Zhao J, Wang X, Yue J. et al. Expression and clinical prognostic value of m6A RNA methylation modification in breast cancer. Biomark Res. 2021;9:28
48. Bawankar P, Lence T, Paolantoni C, Haussmann IU, Kazlauskiene M, Jacob D. et al. Hakai is required for stabilization of core components of the m(6)A mRNA methylation machinery. Nat Commun. 2021;12:3778
49. Wen J, Lv R, Ma H, Shen H, He C, Wang J. et al. Zc3h13 Regulates Nuclear RNA m(6)A Methylation and Mouse Embryonic Stem Cell Self-Renewal. Mol Cell. 2018;69:1028-38 e6
50. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415-29
51. Doxtader KA, Wang P, Scarborough AM, Seo D, Conrad NK, Nam Y. Structural Basis for Regulation of METTL16, an S-Adenosylmethionine Homeostasis Factor. Mol Cell. 2018;71:1001-11 e4
52. Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R. et al. N(6-)Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15:88-94
53. Ren W, Lu J, Huang M, Gao L, Li D, Wang GG. et al. Structure and regulation of ZCCHC4 in m(6)A-methylation of 28S rRNA. Nat Commun. 2019;10:5042
54. Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ. et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4:1798
55. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-7
56. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
57. Wang C, Yang J, Song P, Zhang W, Lu Q, Yu Q. et al. FIONA1 is an RNA N(6)-methyladenosine methyltransferase affecting Arabidopsis photomorphogenesis and flowering. Genome Biol. 2022;23:40
58. Li P, Gao S, Wang L, Yu F, Li J, Wang C. et al. ABH2 couples regulation of ribosomal DNA transcription with DNA alkylation repair. Cell Rep. 2013;4:817-29
59. Sundheim O, Vagbo CB, Bjoras M, Sousa MM, Talstad V, Aas PA. et al. Human ABH3 structure and key residues for oxidative demethylation to reverse DNA/RNA damage. EMBO J. 2006;25:3389-97
60. Li H, Qiao S, Zhang H, Qiao Y, Liu J, Li Y. Highly sensitive and selective demethylase FTO detection using a DNAzyme-mediated CRISPR/Cas12a signal cascade amplification electrochemiluminescence biosensor with C-CN/PCN(V) heterojunction as emitter. Biosens Bioelectron. 2024;256:116276
61. Shishodia S, Demetriades M, Zhang D, Tam NY, Maheswaran P, Clunie-O'Connor C. et al. Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors. J Med Chem. 2021;64:16609-25
62. Liu N, Pan T. RNA epigenetics. Transl Res. 2015;165:28-35
63. Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767-72
64. Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP. et al. Reversible methylation of m(6)A(m) in the 5' cap controls mRNA stability. Nature. 2017;541:371-5
65. Shi Y, Lei Y, Chen M, Ma H, Shen T, Zhang Y. et al. A Demethylation-Switchable Aptamer Design Enables Lag-Free Monitoring of m(6)A Demethylase FTO with Energy Self-Sufficient and Structurally Integrated Features. J Am Chem Soc. 2024;146:34638-50
66. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC. et al. Differential m(6)A, m(6)A(m), and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell. 2018;71:973-85 e5
67. He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863-5
68. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y. et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172:90-105 e23
69. Bao J, Vitting-Seerup K, Waage J, Tang C, Ge Y, Porse BT. et al. UPF2-Dependent Nonsense-Mediated mRNA Decay Pathway Is Essential for Spermatogenesis by Selectively Eliminating Longer 3'UTR Transcripts. PLoS Genet. 2016;12:e1005863
70. Chokkalla AK, Jeong S, Mehta SL, Davis CK, Morris-Blanco KC, Bathula S. et al. Cerebroprotective Role of N(6)-Methyladenosine Demethylase FTO (Fat Mass and Obesity-Associated Protein) After Experimental Stroke. Stroke. 2023;54:245-54
71. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
72. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8:284-96
73. Poh HX, Mirza AH, Pickering BF, Jaffrey SR. Alternative splicing of METTL3 explains apparently METTL3-independent m6A modifications in mRNA. PLoS Biol. 2022;20:e3001683
74. Kierzek E, Kierzek R. The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 2003;31:4472-80
75. Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L. et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707-19
76. Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. Structure and thermodynamics of N6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc. 2015;137:2107-15
77. Dai X, Ren T, Zhang Y, Nan N. Methylation multiplicity and its clinical values in cancer. Expert Rev Mol Med. 2021;23:e2
78. Luxton HJ, Simpson BS, Mills IG, Brindle NR, Ahmed Z, Stavrinides V. et al. The Oncogene Metadherin Interacts with the Known Splicing Proteins YTHDC1, Sam68 and T-STAR and Plays a Novel Role in Alternative mRNA Splicing. Cancers (Basel). 2019 11
79. Li K, Wang Z. Splicing factor SRSF2-centric gene regulation. Int J Biol Sci. 2021;17:1708-15
80. Roundtree IA, He C. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Trends Genet. 2016;32:320-1
81. Yang D, Zhao G, Zhang HM. m(6)A reader proteins: the executive factors in modulating viral replication and host immune response. Front Cell Infect Microbiol. 2023;13:1151069
82. Jiang T, Xiao Y, Zhou J, Luo Z, Yu L, Liao Q. et al. Arbutin alleviates fatty liver by inhibiting ferroptosis via FTO/SLC7A11 pathway. Redox Biol. 2023;68:102963
83. Li D, Cai L, Meng R, Feng Z, Xu Q. METTL3 Modulates Osteoclast Differentiation and Function by Controlling RNA Stability and Nuclear Export. Int J Mol Sci. 2020 21
84. Bertero A, Brown S, Madrigal P, Osnato A, Ortmann D, Yiangou L. et al. The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature. 2018;555:256-9
85. Seidler JF, Strasser K. Understanding nuclear mRNA export: Survival under stress. Mol Cell. 2024;84:3681-91
86. Lv D, Zhong C, Dixit D, Yang K, Wu Q, Godugu B. et al. EGFR promotes ALKBH5 nuclear retention to attenuate N6-methyladenosine and protect against ferroptosis in glioblastoma. Mol Cell. 2023;83:4334-51 e7
87. Liu L, Li H, Hu D, Wang Y, Shao W, Zhong J. et al. Insights into N6-methyladenosine and programmed cell death in cancer. Mol Cancer. 2022;21:32
88. Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG. et al. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat Commun. 2012;3:1006
89. Edens BM, Vissers C, Su J, Arumugam S, Xu Z, Shi H. et al. FMRP Modulates Neural Differentiation through m(6)A-Dependent mRNA Nuclear Export. Cell Rep. 2019;28:845-54 e5
90. Kerkow DE, Carmel AB, Menichelli E, Ambrus G, Hills RD Jr, Gerace L. et al. The structure of the NXF2/NXT1 heterodimeric complex reveals the combined specificity and versatility of the NTF2-like fold. J Mol Biol. 2012;415:649-65
91. Wei X, Huo Y, Pi J, Gao Y, Rao S, He M. et al. METTL3 preferentially enhances non-m(6)A translation of epigenetic factors and promotes tumourigenesis. Nat Cell Biol. 2022;24:1278-90
92. Zhai H, Qin W, Dong S, Yang X, Zhai X, Tong W. et al. PEDV N protein capture protein translation element PABPC1 and eIF4F to promote viral replication. Vet Microbiol. 2023;284:109844
93. Chen L, Zhang C, Ma W, Huang J, Zhao Y, Liu H. METTL3-mediated m6A modification stabilizes TERRA and maintains telomere stability. Nucleic Acids Res. 2022;50:11619-34
94. Shen M, Li Y, Wang Y, Shao J, Zhang F, Yin G. et al. N(6)-methyladenosine modification regulates ferroptosis through autophagy signaling pathway in hepatic stellate cells. Redox Biol. 2021;47:102151
95. Liu X, Li P, Huang Y, Li H, Liu X, Du Y. et al. M(6)A demethylase ALKBH5 regulates FOXO1 mRNA stability and chemoresistance in triple-negative breast cancer. Redox Biol. 2024;69:102993
96. Lan Q, Liu PY, Bell JL, Wang JY, Huttelmaier S, Zhang XD. et al. The Emerging Roles of RNA m(6)A Methylation and Demethylation as Critical Regulators of Tumorigenesis, Drug Sensitivity, and Resistance. Cancer Res. 2021;81:3431-40
97. Lan Q, Liu PY, Haase J, Bell JL, Huttelmaier S, Liu T. The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res. 2019;79:1285-92
98. Hu X, Lu J, Ding C, Li J, Zou Q, Xia W. et al. The N6-methyladenosine landscape of ovarian development and aging highlights the regulation by RNA stability and chromatin state. Aging Cell. 2025;24:e14376
99. Feng Y, Shen J, Lin Z, Chen Z, Zhou M, Ma X. PXR Activation Relieves Deoxynivalenol-Induced Liver Oxidative Stress Via Malat1 LncRNA m(6)A Demethylation. Adv Sci (Weinh). 2024;11:e2308742
100. Wei J, Harada BT, Lu D, Ma R, Gao B, Xu Y. et al. HRD1-mediated METTL14 degradation regulates m(6)A mRNA modification to suppress ER proteotoxic liver disease. Mol Cell. 2021;81:5052-65 e6
101. Yu S, Liu X, Xu Y, Pan L, Zhang Y, Li Y. et al. m 6 A-mediated gluconeogenic enzyme PCK1 upregulation protects against hepatic ischemia-reperfusion injury. Hepatology. 2025;81:94-110
102. Wang S, Chen S, Sun J, Han P, Xu B, Li X. et al. m(6)A modification-tuned sphingolipid metabolism regulates postnatal liver development in male mice. Nat Metab. 2023;5:842-60
103. Liu C, Li X, Gao M, Dong Y, Chen Z. Downregulation of hepatic METTL3 contributes to APAP-induced liver injury in mice. JHEP Rep. 2023;5:100766
104. Li R, Yan X, Xiao C, Wang T, Li X, Hu Z. et al. FTO deficiency in older livers exacerbates ferroptosis during ischaemia/reperfusion injury by upregulating ACSL4 and TFRC. Nat Commun. 2024;15:4760
105. Liu Z, Song L, Chen J, Zhou Y, Wang Y, Tang L. et al. Causal associations between chronic hepatitis B and COVID-19 in East Asian populations. Virol J. 2023;20:109
106. Qing X, Chen Q, Wang K. m6A Regulator-Mediated Methylation Modification Patterns and Characteristics in COVID-19 Patients. Front Public Health. 2022;10:914193
107. Meng Y, Zhang Q, Wang K, Zhang X, Yang R, Bi K. et al. RBM15-mediated N6-methyladenosine modification affects COVID-19 severity by regulating the expression of multitarget genes. Cell Death Dis. 2021;12:732
108. Cheng D, Wu C, Li Y, Liu Y, Mo J, Fu L. et al. METTL3 inhibition ameliorates liver damage in mouse with hepatitis B virus-associated acute-on-chronic liver failure by regulating miR-146a-5p maturation. Biochim Biophys Acta Gene Regul Mech. 2022;1865:194782
109. Li D, Liu Y, Zhou J, Chen Y, Yang C, Liu H. et al. m6A Regulator-mediated RNA Methylation Modulates Immune Microenvironment of Hepatitis B Virus-related Acute Liver Failure. Inflammation. 2023;46:1777-95
110. Yu J, Li W, Hou GJ, Sun DP, Yang Y, Yuan SX. et al. Circular RNA cFAM210A, degradable by HBx, inhibits HCC tumorigenesis by suppressing YBX1 transactivation. Exp Mol Med. 2023;55:2390-401
111. Lv Z, Liu L, You J, Zhou P, Su Y, Zhao K. et al. Small HBV surface antigen drives regorafenib resistance in HCC via KIAA1429-dependent m6A modification of CCR9. J Med Virol. 2024;96:e29894
112. Kim GW, Siddiqui A. Hepatitis B Virus X Protein Expression Is Tightly Regulated by N6-Methyladenosine Modification of Its mRNA. J Virol. 2022;96:e0165521
113. Sacco MT, Bland KM, Horner SM. WTAP Targets the METTL3 m(6)A-Methyltransferase Complex to Cytoplasmic Hepatitis C Virus RNA to Regulate Infection. J Virol. 2022;96:e0099722
114. Kim GW, Imam H, Khan M, Siddiqui A. N(6)-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J Biol Chem. 2020;295:13123-33
115. Kim GW, Moon JS, Gudima SO, Siddiqui A. N(6)-Methyladenine Modification of Hepatitis Delta Virus Regulates Its Virion Assembly by Recruiting YTHDF1. J Virol. 2022;96:e0112422
116. Tsukuda S, Harris JM, Magri A, Balfe P, Siddiqui A, Wing PAC. et al. The N6-methyladenosine demethylase ALKBH5 regulates the hypoxic HBV transcriptome. PLoS Pathog. 2024;20:e1011917
117. Meng Y, Shu Z, Wang X, Hong L, Wang B, Jiang J. et al. Hepatitis B Virus-Mediated m6A Demethylation Increases Hepatocellular Carcinoma Stemness and Immune Escape. Mol Cancer Res. 2024;22:642-55
118. Zhao L, Wang Y, Tian T, Rao X, Dong W, Zhang J. et al. Analysis of viral integration reveals new insights of oncogenic mechanism in HBV-infected intrahepatic cholangiocarcinoma and combined hepatocellular-cholangiocarcinoma. Hepatol Int. 2022;16:1339-52
119. Gong ZJ, De Meyer S, van Pelt J, Hertogs K, Depla E, Soumillion A. et al. Transfection of a rat hepatoma cell line with a construct expressing human liver annexin V confers susceptibility to hepatitis B virus infection. Hepatology. 1999;29:576-84
120. Li Y, Li J, Yu Q, Ji L, Peng B. METTL14 regulates microglia/macrophage polarization and NLRP3 inflammasome activation after ischemic stroke by the KAT3B-STING axis. Neurobiol Dis. 2023;185:106253
121. Qiu T, Wu C, Yao X, Han Q, Wang N, Yuan W. et al. AS3MT facilitates NLRP3 inflammasome activation by m(6)A modification during arsenic-induced hepatic insulin resistance. Cell Biol Toxicol. 2023;39:2165-81
122. Liao X, Cai D, Liu J, Hu H, You R, Pan Z. et al. Deletion of Mettl3 in mesenchymal stem cells promotes acute myeloid leukemia resistance to chemotherapy. Cell Death Dis. 2023;14:796
123. Qin Y, Li B, Arumugam S, Lu Q, Mankash SM, Li J. et al. m(6)A mRNA methylation-directed myeloid cell activation controls progression of NAFLD and obesity. Cell Rep. 2021;37:109968
124. Sun Y, Shen W, Hu S, Lyu Q, Wang Q, Wei T. et al. METTL3 promotes chemoresistance in small cell lung cancer by inducing mitophagy. J Exp Clin Cancer Res. 2023;42:65
125. Peng Z, Gong Y, Wang X, He W, Wu L, Zhang L. et al. METTL3-m(6)A-Rubicon axis inhibits autophagy in nonalcoholic fatty liver disease. Mol Ther. 2022;30:932-46
126. Li Y, Zhang D, Gao Y, Wang P, Wang Z, Zhang B. et al. METTL3 exacerbates insulin resistance in hepatocytes by regulating m(6)A modification of cytochrome P450 2B6. Nutr Metab (Lond). 2023;20:40
127. Luo P, Li S, Jing W, Tu J, Long X. N(6)-methyladenosine RNA modification in nonalcoholic fatty liver disease. Trends Endocrinol Metab. 2023;34:838-48
128. Kang Q, Zhu X, Ren D, Ky A, MacDougald OA, O'Rourke RW. et al. Adipose METTL14-Elicited N(6) -Methyladenosine Promotes Obesity, Insulin Resistance, and NAFLD Through Suppressing beta Adrenergic Signaling and Lipolysis. Adv Sci (Weinh). 2023;10:e2301645
129. Tang J, Zhao X, Wei W, Liu W, Fan H, Liu XP. et al. METTL16-mediated translation of CIDEA promotes non-alcoholic fatty liver disease progression via m6A-dependent manner. PeerJ. 2022;10:e14379
130. Li Y, Tian X, Yu Q, Bao T, Dai C, Jiang L. et al. Alleviation of hepatic insulin resistance and steatosis with NMN via improving endoplasmic reticulum-Mitochondria miscommunication in the liver of HFD mice. Biomed Pharmacother. 2024;175:116682
131. Yang Z, Yu GL, Zhu X, Peng TH, Lv YC. Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes Dis. 2022;9:51-61
132. Liu Y, Song R, Zhao L, Lu Z, Li Y, Zhan X. et al. m(6)A demethylase ALKBH5 is required for antibacterial innate defense by intrinsic motivation of neutrophil migration. Signal Transduct Target Ther. 2022;7:194
133. Tang Z, Sun C, Yan Y, Niu Z, Li Y, Xu X. et al. Aberrant elevation of FTO levels promotes liver steatosis by decreasing the m6A methylation and increasing the stability of SREBF1 and ChREBP mRNAs. J Mol Cell Biol. 2023 14
134. Meng F, Song C, Liu J, Chen F, Zhu Y, Fang X. et al. Chlorogenic Acid Modulates Autophagy by Inhibiting the Activity of ALKBH5 Demethylase, Thereby Ameliorating Hepatic Steatosis. J Agric Food Chem. 2023;71:15073-86
135. Mizuno TM. Fat Mass and Obesity Associated (FTO) Gene and Hepatic Glucose and Lipid Metabolism. Nutrients. 2018 10
136. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M. et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626
137. Pan XS, Li BW, Wang LL, Li N, Lin HM, Zhang J. et al. Kupffer cell pyroptosis mediated by METTL3 contributes to the progression of alcoholic steatohepatitis. FASEB J. 2023;37:e22965
138. Ding H, Zhang X, Su Y, Jia C, Dai C. GNAS promotes inflammation-related hepatocellular carcinoma progression by promoting STAT3 activation. Cell Mol Biol Lett. 2020;25:8
139. Wang YF, Zhang WL, Li ZX, Liu Y, Tan J, Yin HZ. et al. METTL14 downregulation drives S100A4(+) monocyte-derived macrophages via MyD88/NF-kappaB pathway to promote MAFLD progression. Signal Transduct Target Ther. 2024;9:91
140. Hou X, Li Y, Song J, Peng L, Zhang W, Liu R. et al. METTL14 reverses liver fibrosis by inhibiting NOVA2 through an m6A-YTHDF2-dependent mechanism. Hepatol Commun. 2023 7
141. Li Y, Kang X, Zhou Z, Pan L, Chen H, Liang X. et al. The m(6)A methyltransferase Mettl3 deficiency attenuates hepatic stellate cell activation and liver fibrosis. Mol Ther. 2022;30:3714-28
142. Qiu T, Hou K, Zhang J, Wang N, Yao X, Yang G. et al. Sodium arsenite induces hepatic stellate cells activation by m(6)A modification of TGF-beta1 during liver fibrosis. Ecotoxicol Environ Saf. 2024;278:116435
143. Sun F, Wang J, Yang Y, Dong QQ, Jia L, Hu W. et al. Epitranscriptomic regulation of lipid oxidation and liver fibrosis via ENPP1 mRNA m(6)A modification. Cell Mol Life Sci. 2024;81:387
144. Wei A, Zhao F, Hao A, Liu B, Liu Z. N-acetyl-seryl-aspartyl-lysyl-proline (AcSDKP) mitigates the liver fibrosis via WTAP/m(6)A/Ptch1 axis through Hedgehog pathway. Gene. 2022;813:146125
145. Huang T, Zhang C, Ren J, Shuai Q, Li X, Li X. et al. FTO-mediated m (6)A demethylation of ULK1 mRNA promotes autophagy and activation of hepatic stellate cells in liver fibrosis. Acta Biochim Biophys Sin (Shanghai). 2024;56:1509-20
146. Shen M, Guo M, Li Y, Wang Y, Qiu Y, Shao J. et al. m(6)A methylation is required for dihydroartemisinin to alleviate liver fibrosis by inducing ferroptosis in hepatic stellate cells. Free Radic Biol Med. 2022;182:246-59
147. Yang JJ, Wang J, Yang Y, Yang Y, Li J, Lu D. et al. ALKBH5 ameliorated liver fibrosis and suppressed HSCs activation via triggering PTCH1 activation in an m(6)A dependent manner. Eur J Pharmacol. 2022;922:174900
148. Wang J, Yang Y, Sun F, Luo Y, Yang Y, Li J. et al. ALKBH5 attenuates mitochondrial fission and ameliorates liver fibrosis by reducing Drp1 methylation. Pharmacol Res. 2023;187:106608
149. Frangogiannis N. Transforming growth factor-beta in tissue fibrosis. J Exp Med. 2020;217:e20190103
150. Ilieva M, Uchida S. Epitranscriptomics in fibroblasts and fibrosis. Am J Physiol Cell Physiol. 2022;322:C1110-C6
151. Li T, Zhuang Y, Yang W, Xie Y, Shang W, Su S. et al. Silencing of METTL3 attenuates cardiac fibrosis induced by myocardial infarction via inhibiting the activation of cardiac fibroblasts. FASEB J. 2021;35:e21162
152. Mathiyalagan P, Adamiak M, Mayourian J, Sassi Y, Liang Y, Agarwal N. et al. FTO-Dependent N(6)-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation. 2019;139:518-32
153. Juanola A, Ma AT, de Wit K, Gananandan K, Roux O, Zaccherini G. et al. Novel prognostic biomarkers in decompensated cirrhosis: a systematic review and meta-analysis. Gut. 2023;73:156-65
154. Wang L, Yang Q, Zhou Q, Fang F, Lei K, Liu Z. et al. METTL3-m(6)A-EGFR-axis drives lenvatinib resistance in hepatocellular carcinoma. Cancer Lett. 2023;559:216122
155. Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of METTL3 as an Adverse Prognostic Biomarker in Hepatocellular Carcinoma. Dig Dis Sci. 2021;66:1110-26
156. Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13:117
157. Pan Y, Chen H, Zhang X, Liu W, Ding Y, Huang D. et al. METTL3 drives NAFLD-related hepatocellular carcinoma and is a therapeutic target for boosting immunotherapy. Cell Rep Med. 2023;4:101144
158. Liu B, Cao J, Wu B, Hao K, Wang X, Chen X. et al. METTL3 and STAT3 form a positive feedback loop to promote cell metastasis in hepatocellular carcinoma. Cell Commun Signal. 2023;21:121
159. Xi Q, Yang G, He X, Zhuang H, Li L, Lin B. et al. M(6)A-mediated upregulation of lncRNA TUG1 in liver cancer cells regulates the antitumor response of CD8(+) T cells and phagocytosis of macrophages. Adv Sci (Weinh). 2024;11:e2400695
160. Du L, Li Y, Kang M, Feng M, Ren Y, Dai H. et al. USP48 Is Upregulated by Mettl14 to Attenuate Hepatocellular Carcinoma via Regulating SIRT6 Stabilization. Cancer Res. 2021;81:3822-34
161. Lu J, Ru J, Chen Y, Ling Z, Liu H, Ding B. et al. N(6) -methyladenosine-modified circSTX6 promotes hepatocellular carcinoma progression by regulating the HNRNPD/ATF3 axis and encoding a 144 amino acid polypeptide. Clin Transl Med. 2023;13:e1451
162. Zhou T, Li S, Xiang D, Liu J, Sun W, Cui X. et al. m6A RNA methylation-mediated HNF3gamma reduction renders hepatocellular carcinoma dedifferentiation and sorafenib resistance. Signal Transduct Target Ther. 2020;5:296
163. Dai YZ, Liu YD, Li J, Chen MT, Huang M, Wang F. et al. METTL16 promotes hepatocellular carcinoma progression through downregulating RAB11B-AS1 in an m(6)A-dependent manner. Cell Mol Biol Lett. 2022;27:41
164. Lin Z, Huang Z, Qiu J, Shi Y, Zuo D, Qiu Z. et al. m(6)A-mediated lnc-OXAR promotes oxaliplatin resistance by enhancing Ku70 stability in non-alcoholic steatohepatitis-related hepatocellular carcinoma. J Exp Clin Cancer Res. 2024;43:206
165. Chen Y, Peng C, Chen J, Chen D, Yang B, He B. et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18:127
166. Fu XL, Guo SM, Ma JQ, Ma FY, Wang X, Tang YX. et al. HBXIP induces PARP1 via WTAP-mediated m(6)A modification and CEBPA-activated transcription in cisplatin resistance to hepatoma. Acta Pharmacol Sin. 2024;45:2405-19
167. Liu M, Jiang K, Lin G, Liu P, Yan Y, Ye T. et al. Ajuba inhibits hepatocellular carcinoma cell growth via targeting of beta-catenin and YAP signaling and is regulated by E3 ligase Hakai through neddylation. J Exp Clin Cancer Res. 2018;37:165
168. Shan M, Liu D, Sun L, Yang M, He M, Zhang Y. et al. KIAA1429 facilitates metastasis via m6A-YTHDC1-dependent RND3 down-regulation in hepatocellular carcinoma cells. Cancer Lett. 2024;584:216598
169. Jiang L, Liang R, Luo Q, Chen Z, Song G. Targeting FTO suppresses hepatocellular carcinoma by inhibiting ERBB3 and TUBB4A expression. Biochem Pharmacol. 2024;226:116375
170. Bian X, Shi D, Xing K, Zhou H, Lu L, Yu D. et al. AMD1 upregulates hepatocellular carcinoma cells stemness by FTO mediated mRNA demethylation. Clin Transl Med. 2021;11:e352
171. Chen Y, Zhao Y, Chen J, Peng C, Zhang Y, Tong R. et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1. Mol Cancer. 2020;19:123
172. Zhang H, Liu Y, Wang W, Liu F, Wang W, Su C. et al. ALKBH5-mediated m(6)A modification of lincRNA LINC02551 enhances the stability of DDX24 to promote hepatocellular carcinoma growth and metastasis. Cell Death Dis. 2022;13:926
173. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H. et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35:677-91 e10
174. Huang Y, Xia W, Dong Z, Yang CG. Chemical Inhibitors Targeting the Oncogenic m(6)A Modifying Proteins. Acc Chem Res. 2023;56:3010-22
175. Kiavue N, Cabel L, Melaabi S, Bataillon G, Callens C, Lerebours F. et al. ERBB3 mutations in cancer: biological aspects, prevalence and therapeutics. Oncogene. 2020;39:487-502
176. Liu Y, Liang G, Xu H, Dong W, Dong Z, Qiu Z. et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance. Cell Metab. 2021;33:1221-33 e11
177. Liu L, Zhao T, Zheng S, Tang D, Han H, Yang C. et al. METTL3 inhibitor STM2457 impairs tumor progression and enhances sensitivity to anlotinib in OSCC. Oral Dis. 2024;30:4243-54
178. Yanagi Y, Watanabe T, Hara Y, Sato Y, Kimura H, Murata T. EBV Exploits RNA m(6)A Modification to Promote Cell Survival and Progeny Virus Production During Lytic Cycle. Front Microbiol. 2022;13:870816
179. Gao Y, Wang P, Lu S, Ma W. [METTL3 inhibitor STM2457 improves metabolic dysfunction-associated fatty liver disease by regulating mitochondrial function in mice]. Nan Fang Yi Ke Da Xue Xue Bao. 2023;43:1689-96
180. Ren Y, Chen Y, Tang EH, Hu Y, Niu B, Liang H. et al. Arbidol attenuates liver fibrosis and activation of hepatic stellate cells by blocking TGF-beta1 signaling. Eur J Pharmacol. 2024;967:176367
181. Malacrida A, Rivara M, Di Domizio A, Cislaghi G, Miloso M, Zuliani V. et al. 3D proteome-wide scale screening and activity evaluation of a new ALKBH5 inhibitor in U87 glioblastoma cell line. Bioorg Med Chem. 2020;28:115300
182. Zhang L, Ke W, Zhao X, Lu Z. Resina Draconis extract exerts anti-HCC effects through METTL3-m6A-Survivin axis. Phytother Res. 2022;36:2542-57
183. Dang Y, Xu J, Yang Y, Li C, Zhang Q, Zhou W. et al. Ling-gui-zhu-gan decoction alleviates hepatic steatosis through SOCS2 modification by N6-methyladenosine. Biomed Pharmacother. 2020;127:109976
184. Wu J, Xian S, Zhang S, Yang Y, Pan J, Zhou W. et al. Gan-Jiang-Ling-Zhu decoction improves steatohepatitis induced by choline-deficient-high-fat-diet through the METTL14/N6-methyladenosine-mediated Ugt2a3 expression. J Ethnopharmacol. 2025;339:119153
185. Liu J, Liu Y, Huang C, He C, Yang T, Ren R. et al. Quercetin-Driven Akkermansia Muciniphila Alleviates Obesity by Modulating Bile Acid Metabolism via an ILA/m(6)A/CYP8B1 Signaling. Adv Sci (Weinh). 2025;12:e2412865
186. Liu Y, Guo Q, Yang H, Zhang XW, Feng N, Wang JK. et al. Allosteric Regulation of IGF2BP1 as a Novel Strategy for the Activation of Tumor Immune Microenvironment. ACS Cent Sci. 2022;8:1102-15
187. Li X, Ma S, Deng Y, Yi P, Yu J. Targeting the RNA m(6)A modification for cancer immunotherapy. Mol Cancer. 2022;21:76
188. Wu S, Yun J, Tang W, Familiari G, Relucenti M, Wu J. et al. Therapeutic m(6)A Eraser ALKBH5 mRNA-Loaded Exosome-Liposome Hybrid Nanoparticles Inhibit Progression of Colorectal Cancer in Preclinical Tumor Models. ACS Nano. 2023;17:11838-54
Author contact
![]() Corresponding authors: Jun Li, M.D., Ph.D. School of Pharmacy, Anhui Medical University, Hefei 230032, China. Tel/Fax: +86-0551-65161001. E-mail address: ljedu.cn. Xin Chen, M.D., Ph.D. The Third Affiliated Hospital, School of Pharmacy, Anhui Medical University, Hefei 230032, China. Tel/Fax: +86-0551-65167737. E-mail address: chenxinedu.cn.
Corresponding authors: Jun Li, M.D., Ph.D. School of Pharmacy, Anhui Medical University, Hefei 230032, China. Tel/Fax: +86-0551-65161001. E-mail address: ljedu.cn. Xin Chen, M.D., Ph.D. The Third Affiliated Hospital, School of Pharmacy, Anhui Medical University, Hefei 230032, China. Tel/Fax: +86-0551-65167737. E-mail address: chenxinedu.cn.

 Global reach, higher impact
Global reach, higher impact