Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(4):486-497. doi:10.7150/ijbs.3982 This issue Cite
Research Paper
A Comparative Study of Mammalian Diversification Pattern
1. Jiangsu Key Laboratory for Biodiversity and Biotechnology, College of Life Sciences, Nanjing Normal University, Nanjing 210046, China;
2. College of Life Sciences, Guangzhou University, Guangzhou 510006, China
Received 2011-12-19; Accepted 2012-3-11; Published 2012-3-14
Abstract
Although mammals have long been regarded as a successful radiation, the diversification pattern among the clades is still poorly known. Higher-level phylogenies are conflicting and comprehensive comparative analyses are still lacking. Using a recently published supermatrix encompassing nearly all extant mammalian families and a novel comparative likelihood approach (MEDUSA), the diversification pattern of mammalian groups was examined. Both order- and family-level phylogenetic analyses revealed the rapid radiation of Boreoeutheria and Euaustralidelphia in the early mammalian history. The observation of a diversification burst within Boreoeutheria at approximately 100 My supports the Long Fuse model in elucidating placental diversification progress, and the rapid radiation of Euaustralidelphia suggests an important role of biogeographic dispersal events in triggering early Australian marsupial rapid radiation. Diversification analyses based on family-level diversity tree revealed seven additional clades with exceptional diversification rate shifts, six of which represent accelerations in net diversification rate as compared to the background pattern. The shifts gave origin to the clades Muridae+Cricetidae, Bovidae+Moschidae+Cervidae, Simiiformes, Echimyidae, Odontoceti (excluding Physeteridae+Kogiidae+Platanistidae), Macropodidae, and Vespertilionidae. Moderate to high extinction rates from background and boreoeutherian diversification patterns indicate the important role of turnovers in shaping the heterogeneous taxonomic richness observed among extant mammalian groups. Furthermore, the present results emphasize the key role of extinction on erasing unusual diversification signals, and suggest that further studies are needed to clarify the historical radiation of some mammalian groups for which MEDUSA did not detect exceptional diversification rates.
Keywords: mammal, diversification pattern, radiation, MEDUSA
Introduction
The question as to why some groups of organisms diversify vastly greater numbers of species than other groups has long been a subject of interest and debate [1-5]. Evolutionary biologists focus on identifying exceptional diversification patterns and the associated attributes underlying the heterogeneous diversity patterns observed among clades [6]. One leading explanation on it holds that differences in net diversification rates (speciation rate minus extinction rate) among groups are associated with varying ecological opportunities, which could relax or intensify selective pressures acting upon specific ecological traits and diversifying process through abiotic and/or biotic factors [7-10]. However, this approach relies on the identification of unusual diversification patterns that cannot be explained by those general patterns observed at broader scales.
Mammals are an extremely species-rich group, with highly diverse eco-morphologies, physiologies, life histories and behaviors [11-14]. Over 5,000 mammalian species have adapted to nearly all habitats throughout the world [12]. Indeed, there is little doubt that mammals represent one of the most successful radiations among vertebrates [4, 11, 13-15]. However, similar to scaled amphibians, reptiles and songbirds [16, 17], biodiversity pattern within mammals is also highly heterogeneous, which implies differential speciation and extinction probabilities [12]. Some groups, such as rodents, bats, primates, carnivores, and shrews (family Soricidae) are extremely rich with regard to their ecology, morphology and taxonomy, whereas groups such as aardvark (order Tubulidentata), Monito del Monte (order Microbiotheria) and colugos (order Dermoptera) exhibit evolutionary stasis with spare lineages [11, 12, 14]. Although identifying and understanding such patterns is critical to clarifying progress of mammalian macroevolution, the diversification patterns of mammals remain poorly known due to a lack of robust chronogram utilizing large-scale taxonomic sampling schemes, as well as a lack of suitable statistical comparative methods [18-25].
Using a recently published phylogenetic supermatrix including nearly all extant mammalian families [26] and a novel comparative likelihood method, MEDUSA [Modeling Evolutionary Diversification Using Stepwise Akaike Information Criterion (AIC)] [4], we examined the mammalian diversification pattern to address four questions: (1) what is the background pattern of mammalian diversification; (2) which, if any, mammalian clades exhibit exceptionally fast or slow diversifications; and (3) whether there is evidence of increasing diversification among some ecologically and morphologically diverse mammalian groups, such as rodents, bats, primates, carnivores, and shrews; (4) what kind of caveat we should pay attention to when using MEDUSA.
Methods
Due to the dependence of MEDUSA on phylogenetic relationship and resolution [4], one large DNA supermatrix containing ~94% of current mammalian families [26] was downloaded from Treebase (Study ID: S11872) to infer major cladogenesis events in the early mammalian history and examine general diversification patterns. The supermatrix comprising 35,603 base pairs (bp) consists of 21 protein-coding segments and five untranslated regions (UTRs), and most of them have been employed in mammalian phylogenetic analyses [21, 23, 26-31]. A maximum likelihood phylogeny was reconstructed using RAxML V7.2.7 [32] with 500 bootstrap replicates on CIPRES Science Gateway V3.1 [33]. Given that the phylogenetic relationship within marsupials [26] conflicted with recent phylogenies employing the solid homoplasy-free retroposon insertion markers and nucleotide sequences [34-37], a basal position of Didelphimorphia within Marsupialia was constrained in present study. Estimates of divergence times relied on two different relaxed molecular clock methods: a Bayesian approach implemented as in Meredith et al. [26], with both independent-rates and auto-correlated rates models using 'mcmctree' as implemented in PAML V4.4d [38] with the following parameters setting: Shape (α)=1 and scale (β)=5.24, and a sampling number of 10,000 generations after a burn-in of 1,000 generations, as well as a penalty likelihood approach [39]. In the latter case, divergence times were calculated using 77 mammalian paleontological constraints [26] and additive penalty as implemented in r8s V1.7 [39] after pruning non-mammalian outgroups. The smoothing parameter was optimized using cross-validation procedures. The bootstrap trees obtained from RAxML were used to estimate the uncertainty associated with divergence times using r8s V1.7 [39] and r8s-bootkit (T. Eriksson, http://www.bergianska.se/index_forskning_soft.html).
Chronograms encompassing 169 taxa captured major splitting events in the early evolutionary history of mammals [26]. These trees were subsequently pruned down to order- and family-level diversity trees, in which extant species richness were assigned to the corresponding branches. Species richness assignments were derived from the Mammal Species of the World database 3rd edition (http://www.press.jhu.edu) (Figs. 1 and 2) [12]. To identify general diversification pattern and rate shifts among lineages, we used a recently developed comparative likelihood method (MEDUSA) that integrates phylogenetic information with taxonomic richness data to estimate all speciation (b) and extinction (d) rates [4]. MEDUSA is a stepwise procedure that fits a series of birth-death models to the diversity tree using a joint phylogenetic and taxonomic likelihood function developed by Rabosky et al. [2]. The algorithm first fits a single diversification model (one speciation rate and one extinction rate) to the entire dataset. Then, a series of breaks are added to the diversity tree, so that different parts of the tree fit the birth-death models with different speciation and extinction rates. MEDUSA compares all single-breakpoint models (two speciation rates, two extinction rates, and one shift node) with the overall model (single diversification model), and selects the best one. Then all possible two-breakpoint models (three speciation rates, three extinction rates, and two shift nodes) are fitted and compared with the best single-breakpoint model, and so on. This process will continue until the addition of rate parameters no longer substantially improves the overall AIC score. In this study, an improvement of at least 4 units in the AIC was used as a threshold for retaining rate shifts [40]. To locate exceptional rate shifts more precisely, particularly in the case of unresolved phylogenetic branches, analyses were conducted using both order- and family-level diversity trees [4]. Note that we also performed the diversification analyses constraining Xenarthra and Afrotheria/Xenarthra clade as basal lineage within Placentalia, respectively [22, 41, 42], to estimate the influences of shift nodes since these alternative topologies. Given the consistent shift points from diversity trees with different placental roots (Table 1), only the diversity tree directly inferred from RAxML V7.2.7 [32] was thus illustrated. All diversification analyses were performed in R using the Ape [43], Geiger [44], Laser [45] libraries. Perl and Python scripts were also developed for the assignments of species richness and tree nodes as well as for the manipulation of the phylogenetic trees using Bio-Phylo-0.43 modules [46] in CPAN and ETE-2.1 [47]. Since the two molecular clock approaches were consistent in the identifications of clades with unusual shifting and revealed similar diversification patterns, we only presented the diversification results using r8s V1.7 [39].
Results
Order-level diversification pattern
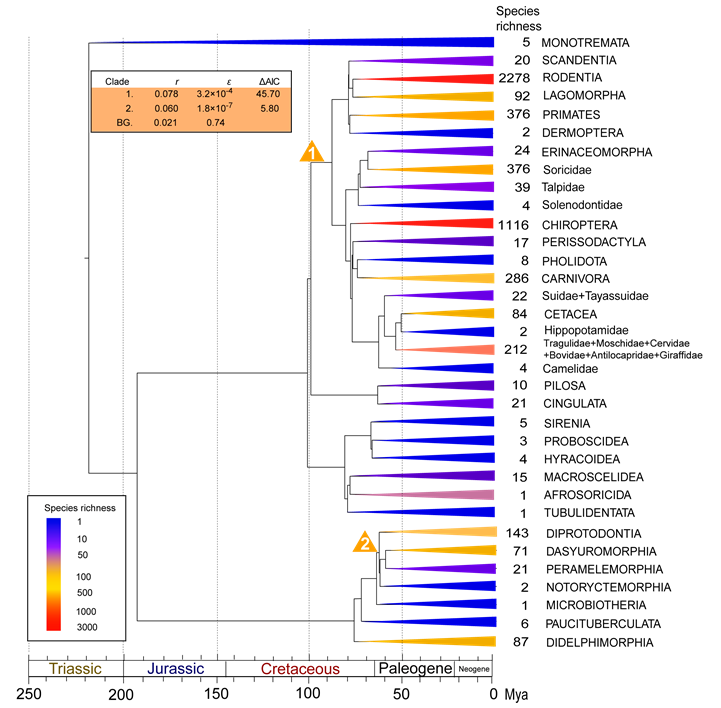
Timetrees were pruned down to order-level diversity trees with 34 branches, which summarized both phylogenetic and taxonomic information. A pure-birth (PB) model with one parameter (b) and a birth-death (BD) model with two parameters (b and d) were fitted to the diversity trees to test their suitability to explain the observed biodiversity. Likelihood scores were significantly lower for the PB model (b=0.05 sp/My, lnL=-301.03) than for the BD model (b=0.19 sp/My, d=0.14 sp/My, lnL=-249.61), indicating that the former model poorly explained the heterozygous biodiversity of extant mammalian orders (likelihood ratio test, Δ=102.84, P<0.001). However, the BD model showed a better match with recent species-rich stem lineages than with ancient species-poor clades. We subsequently tested whether any braches of the diversity trees exhibited exceptional diversification rate shifts using MEDUSA. The background diversification pattern was characterized with a low net diversification rate (r=0.021 sp/My) and a relatively high extinction rate (ε=d/b=0.74) (Fig. 1). Two large mammalian groups showed significant increases in net diversification rate throughout the mammalian history (Fig. 1), including the clade Boreoeutheria (Laurasiatheria+Euarchontoglires) (r=0.078 sp/My, rate shift 1 in Fig. 1), which encompasses over 90% of living mammals, and the clade Euaustralidelphia (r=0.060 sp/My, rate shift 2 in Fig. 1).
Family-level diversification pattern
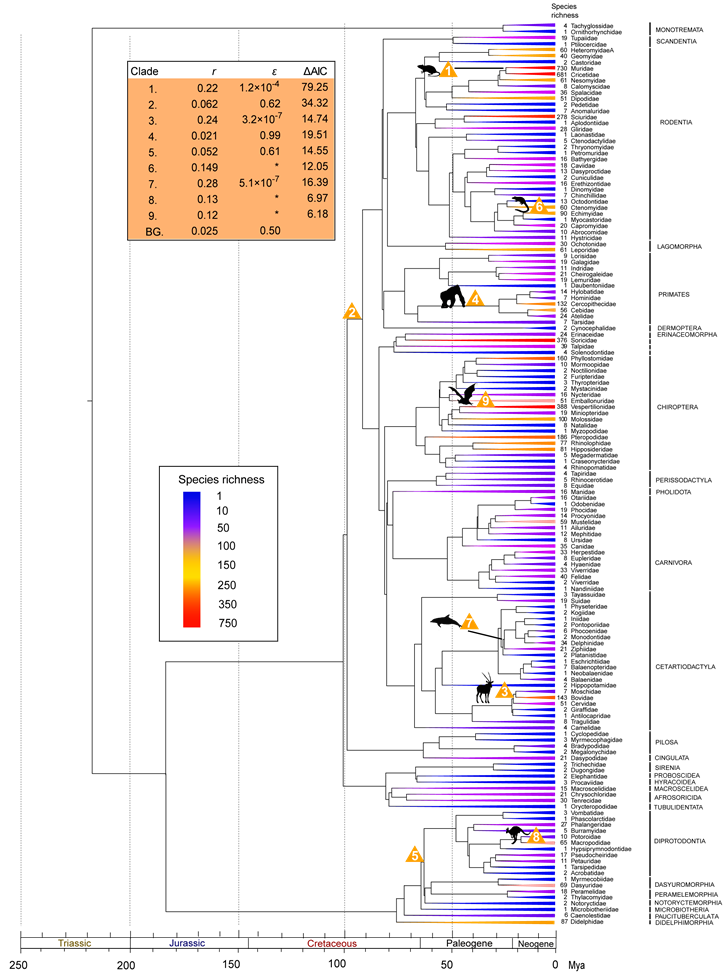
To avoid the impossibility of implementing the MEDUSA approach to unresolved part of the topology, diversification analyses were also performed using family-level diversity trees with 149 taxa. While the BD model (b=0.583 sp/My, d=0.548 sp/My, lnL=-1221.96) showed a substantially better fit to the data than the PB model (b=0.04 sp/My, lnL=-1360.31) (likelihood ratio test, Δ=276.70, P<0.001), it showed a poor match in the case of ancient lineages with poor species richness. These results indicate that neither simple diversification models was sufficient to explain the distinct diversification process associated with the macroevolution of mammals and their currently observed uneven biodiversity pattern among different clades.
The consistent clades with unusual shift rate indicates that different roots within Placentalia would not influence results from MEDUSA (Table 1). The background tempo of diversification inferred from the family-level tree revealed a low net diversification rate (r=0.025 sp/My), but with a moderate turnover rate where extinction rate was 50% of the speciation rate (Fig. 2). Seven additional clades with unusual rate shifts were identified as compared to the order-level diversification analysis, indicating that mammalian diversification was at least markedly shaped by nine shifting events (rate shifts 1-9 in Fig. 2). These additional lineages spread within Boreoeutheria and Marsupialia, including Muridae+Cricetidae (e.g., mouse, true hamsters, voles, lemmings, New World rats and mice), Bovidae+Moschidae+Cervidae (e.g., musk deers, deers, cattles, goats, sheep, and antelope), Simiiformes (New world and Old World monkeys, such as gibbons and great apes), Echimyidae (spiny rats), Odontoceti excluding Physeteridae+Kogiidae+Platanistidae (most toothed whales), Macropodidae (e.g., kangaroos and wallabies), and Vespertilionidae (evening bats) (Fig. 2).
Discussion
Molecular dating
Divergence times estimated by the auto-correlated rate model were more recent than those produced by the independent-rate model, whereas those estimated by the penalty likelihood approach were approximately intermediate between them (Table 2). Although the design concepts and algorithms of these relaxed molecular clock dating methods are entirely different [38, 39, 48], the divergence time estimates are still compatible with each other (Table 2). It highlights the importance in integrating the sufficient and appropriate fossil constraints spread in the phylogeny when estimating divergence times for taxonomic groups with lineage-specific rate variation rather than the methods themselves [26, 49]. Although the inferred ages in this study were elder than previous estimates using genomic data [50-52], the differences seem to be mainly due to the difference of the basal fossil calibrations. The fossil constraint of marsupial-placental split that occurred between ~120 My and ~140 My in those studies seems to be conflicted with a recent discovery of a stem eutherian from the Jurassic Period (~160 My) [53], which consequently caused the younger estimates [50-52]. By contrast, the present inferred ages of Theria in the Jurassic accord with this recently discovered fossil [53] (Table 2). The diversification analyses from MEDUSA indicate that the differences among these timetrees did not influence the general diversification pattern and the discrimination of clades with unusual diversification rates.
Radiations within Marsupialia
The estimated interfamily divergence times within Marsupialia are broadly compatible with recent molecular dating estimations [35-37, 54], further place the basal marsupial divergence in the Late Cretaceous (mean date ~77 My) (Table 2). Although current Australian and South American marsupials comprise only seven percent of the extent mammalian diversity, MEDUSA still located two different periods with rapid radiations occurring throughout their evolutionary history that could not be simply explained by the background diversification pattern (rate shift 1 in Fig. 1, and rate shifts 5 and 8 in Fig. 2).
The first period with exceptional acceleration in marsupial diversification occurring in the Early Paleocene is in the clade with exclusive extant Australian lineages that is recently referred as 'Euaustralidelphia' [34] (r=0.060 sp/My, rate shift 2 in Fig. 1; r=0.052 sp/My, rate shift 5 in Fig. 2). The relative short divergence time for following inter-order diverges prior to the Middle Eocene also pointed to sign of rapid diversification (Fig. 2). Given the exclusive marsupial lineages of the radiation, such diversification burst suggests an important role of biogeographic dispersal event from South America to Australia through Antarctica in triggering rapid radiation after occupation of the 'adaptive zones' (new habitats) due to the ecological release from competitive and selective pressures, and abundance of ecological opportunities [7-10]. However such radiation seems to have lasted in a short term because no constituents of extant families of Dasyuromorphia, Peramelemorphia, and Diprotodontia could be traced back prior to the Middle Eocene (Fig. 2). Although it is possible that the powerful extinction have masked the sign of long-term radiation, given the limited and discontinuous fossil of Australian marsupials in Tertiary in contrast with South American marsupial fossil deposits [13], the homogeneous vegetation type in Australia in the Paleocene (the limited differential ecological niches) [55], and no radical climatic changes causing the mass extinction prior to the Middle Eocene [58], the short-term radiation may be a more reasonable explanation. The other diversification bang of Marsupialia leaded to the tip of Macropodidae, the most species-rich group within Diprotodontia (r=0.13 sp/My, rate shift 8 in Fig. 2). Though MEDUSA is incapable of assigning the rate shift below the phylogenetic resolution and the shift point could be tipward along the branch when increasing the phylogenetic resolution, such detection still confirmed the rapid diversification within Macropodidae after the Early Miocene. Unlike the fungivore/omnivores of the potoroids, the most closely related sister group to Macropodidae, such progressive radiation should be associated with a series of digestive adaptations of the macropodids which enhance the nutritional yield of poor-quality food (such as large and sacculated stomachs) [11, 30, 55-57], the expansion of the arid zone and grasslands and replacement of rainforest in Australia at this period that created sufficient ecological niches for the grazing animals [55, 58], as well as the absence of placental ungulates since the geographic separation between Asia and Australia [13, 27, 57]. These results suggest that macroevolution of marsupials involve a complex interplay between external abiotic factors, such as climatic changes, motion of tectonic plates and variation of ecological niches, as well as the particular intrinsic adaptations to varied ecological niches.
Identified clades with unsual shift rate based on another two alternative topologies within Placentalia.
| Topology within Placentalia | Type of utilized diversity tree | Shift rate | Lineages/clades with shift rate |
|---|---|---|---|
| ((Afrotheria,Xenarthra), Boreoeutheria) | Order-level diversity tree | 1 | Boreoeutheria |
| 2 | Euaustralidelphia | ||
| Family-level diversity tree | 1 | Cricetidae, Muridae | |
| 2 | Boreoeutheria | ||
| 3 | Cervidae, Bovidae, Moschidae | ||
| 4 | Simiiformes | ||
| 5 | Euaustralidelphia | ||
| 6 | Echimyidae | ||
| 7 | Ziphiidae, Delphinidae, Monodontidae, Phocoenidae, Iniidae, Pontoporiidae | ||
| 8 | Macropodidae | ||
| 9 | Vespertilionidae | ||
| ((Afrotheria,Boreoeutheria), Xenarthra) | Order-level diversity tree | 1 | Boreoeutheria |
| 2 | Euaustralidelphia | ||
| Family-level diversity tree | 1 | Cricetidae, Muridae | |
| 2 | Boreoeutheria | ||
| 3 | Cervidae, Bovidae, Moschidae | ||
| 4 | Simiiformes | ||
| 5 | Euaustralidelphia | ||
| 6 | Echimyidae | ||
| 7 | Ziphiidae, Delphinidae, Monodontidae, Phocoenidae, Iniidae, Pontoporiidae | ||
| 8 | Macropodidae | ||
| 9 | Vespertilionidae |
Divergence times estimated for major groups using two molecular clock approaches. Asterisks denote estimates consistent with fossil constraints. Indentations in taxon names denote hierarchical relationships.
| Mcmctree | r8s | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Independent rates | Atuocorrelated rates | ||||||||||||||
| Taxon | mean | 95% CI | Mean | 95% CI | best tree | mean | SD | range | |||||||
| Mammalia | 218.3 | 210.2-225.1 | 221.4 | 216.2-231.4 | 218.5 | * | * | * | |||||||
| Monotremata | 30.0 | 24.2-37.2 | 34.7 | 22.6-59.6 | 25.5 | 25.8 | 0.88 | 25.2-28.8 | |||||||
| Theria | 188.2 | 176.5-198.6 | 201.3 | 191.9-208.7 | 182.0 | 182.7 | 2.25 | 176.9-192.0 | |||||||
| Marsupialia | 73.9 | 68.7-172.4 | 81.1 | 75.9-90.6 | 77.7 | 77.2 | 2.07 | 70.0-82.4 | |||||||
| Didelphimorphia | 33.8 | 28.2-36.5 | 37.1 | 32.2-40.2 | 34.2 | 32.1 | 1.21 | 30.3-39.9 | |||||||
| Paucituberculata | 8.9 | 6.4-10.5 | 10.6 | 7.7-15.2 | 10.2 | 10.9 | 0.72 | 9.3-12.1 | |||||||
| Diprotodontia | 45.2 | 42.4-47.9 | 48.7 | 46.7-51.2 | 50.1 | 52.6 | 1.99 | 44.0-55.7 | |||||||
| Dasyuromorphia | 26.3 | 22.9-30.3 | 29.1 | 21.2-35.3 | 27.4 | 27.5 | 0.98 | 24.7-30.4 | |||||||
| Peramelemorphia | 24.4 | 21.1-27.4 | 25.3 | 22.4-28.1 | 25.2 | 25.2 | 0.80 | 22.9-27.8 | |||||||
| Placentalia | 101.2 | 96.9-103.6 | 99.8 | 96.8-102.3 | 101..3 | 101.8 | 1.56 | 95.1-104.2 | |||||||
| Afrotheria | 78.5 | 74.0-82.9 | 80.1 | 77.2-83.5 | 82.6 | 81.6 | 1.53 | 75.3-83.9 | |||||||
| Sirenia | 30.2 | 26.9-31.8 | 31.6 | 30.9-32.9 | 31.4 | * | * | * | |||||||
| Proboscidea | 2.5 | 1.8-3.3 | 3.2 | 2.1-4.6 | 6.8 | * | * | * | |||||||
| Hyracoidea | 5.2 | 3.8-6.2 | 5.8 | 4.4-7.0 | 6.1 | * | * | * | |||||||
| Macroscelidea | 51.7 | 45.2-55.8 | 45.2 | 41.2-52.2 | 54.2 | 55.7 | 0.58 | 52.2-56.1 | |||||||
| Afrosoricida | 69.9 | 65.0-74.76 | 70.1 | 67.2-74.3 | 70.9 | 72.6 | 1.32 | 67.1-75.2 | |||||||
| Xenarthra | 62.1 | 58.2-67.3 | 65.2 | 60.2-69.2 | 64.2 | 64.7 | 1.86 | 58.4-68.5 | |||||||
| Pilosa | 54.1 | 50.1-59.6 | 57.3 | 54.4-61.6 | 55.0 | 55.2 | 1.84 | 47.9-59.1 | |||||||
| Boreoeutheria | 91.9 | 81.6-86.9 | 99.1 | 96.1-102.2 | 92.6 | 93.2 | 1.59 | 85.1-97.2 | |||||||
| Laurasiatheria | 84.2 | 71.5-86.3 | 84.5 | 83.0-86.8 | 86.7 | 88.3 | 1.54 | 80.7-91.6 | |||||||
| Erinaceomorpha | 41.3 | 36.78-46.1 | 35.1 | 30.3-39.7 | 42.8 | 42.2 | 1.02 | 40.4-47.2 | |||||||
| Chiroptera | 65.4 | 63.3-68.1 | 68.4 | 66.2-68.7 | 67.6 | 70.5 | 1.72 | 63.2-72.5 | |||||||
| Perissodactyla | 55.8 | 55.1-57.5 | 56.5 | 55.1-58.5 | 55.5 | 55.8 | 0.24 | 55.1-57.2 | |||||||
| Pholidota | 20.5 | 16.7-25.6 | 26.2 | 19.8-33.8 | 25.6 | 25.2 | 1.45 | 20.9-29.8 | |||||||
| Carnivora | 51.2 | 47.6-54.3 | 58.5 | 56.4-61.9 | 58.4 | 59.5 | 2.43 | 48.3-62.3 | |||||||
| Cetartiodactyla | 65.8 | 63.4-68.2 | 69.8 | 67.3-72.9 | 69.8 | 69.4 | 1.89 | 61.4-72.5 | |||||||
| Euarchontoglires | 83.9 | 81.2-85.8 | 82.2 | 80.6-84.3 | 83.3 | 83.2 | 0.91 | 79.2-85.2 | |||||||
| Primates | 72.0 | 67.4-74.3 | 76.9 | 74.1-78.2 | 74.5 | 73.9 | 1.39 | 67.4-75.9 | |||||||
| Dermoptera | 7.6 | 5.3-9.2 | 7.2 | 4.7-10.2 | 10.1 | 10.5 | 0.59 | 8.2-11.1 | |||||||
| Scandentia | 57. | 50.9-61.8 | 56.4 | 51.7-60.3 | 57.2 | 54.6 | 1.03 | 51.2-58.4 | |||||||
| Rodentia | 72.6 | 70.3-72.5 | 68.8 | 67.4-69.6 | 68.9 | 68.5 | 0.32 | 67.3-69.2 | |||||||
| Lagomorpha | 53.2 | 48.2-51.5 | 48.2 | 47.5-52.1 | 49.6 | 49.0 | 0.91 | 48.3-53.0 | |||||||
Order-level diversity tree of mammals. Clades are colored as a function of the number of extant species within each order. Clades with unusual diversification rates are denoted with numbers, which indicate the order of which rate shifts were added by the stepwise AIC procedure. Yellow triangles indicate exceptional rates (as compared to background rates). Estimates of net diversification rates, relative extinction rates (ε=d/b) and improved AIC scores are shown on the table in the upper left corner. Asterisks indicate unresolved sub-clades for which relative extinction rates could not be calculated.
Family-level diversity tree of mammals. Clades are colored as a function of the number of extant species within each order. Clades with unusual diversification rates are denoted with numbers, which indicate the order of which rate shifts were added by the stepwise AIC procedure. Yellow triangles indicate exceptional rates (as compared to background rates). Estimates of net diversification rates, relative extinction rates (ε=d/b) and improved AIC scores are shown on the table in the upper left corner. Asterisks indicate unresolved sub-clades for which relative extinction rates could not be calculated.
Exceptional rate shifts within Placentalia
The consistent identification of diversification bang of Boreoeutheria ~100 My ago from all the analyses (Table 1; r=0.078 sp/My, rate shift 1 in Fig. 1 and r=0.062 sp/My, rate shift 2 in Fig. 2) provides supports for the Long Fuse model of mammalian diversification [24, 59], as opposed to the delayed rise of present-day mammals hypothesis [60]. The time point of the radiation suggests an important role of environmental changes in releasing new ecological niches and triggering rapid diversification of Placentalia (e.g. Cretaceous Terrestrial Revolution, Cretaceous-Paleogene mass extinction, tectonic and climatic changes) [25, 26, 61-63]. These results also raise the question as to the causes underlying the successful radiation of Boreoeutheria instead of the whole Placentalia clade. In contrast with the mostly native distribution regions of historical and present afrotherians and xenarthrans [12, 13], two closest living relative of Boreoeutheria, boreoeutherian rapid radiation also appears to link with their early biogeographic events of expanding the habitat ranges and successfully colonizing into the unsaturated continental ecospace, which emphasizes the importance of accessing new geographical or ecological regions in boreoeutherian diversification and present-day uneven biodiversity [64]. Alternatively, it is possible that the absence of Afrotheria and Xenarthra in this diversification bang may be attributed to the historical extinction probabilities, masking their signal of early rapid radiation in the molecular phylogeny of the group [65-67], since unlike their sister group with spectacle biodiversity, the spare living lineages and tree shapes would fit the model that described diversification pattern of Boreoeutheria poorly. However, we argue that such assumption seems to be lacked of paleontological support because few early afrotherian and xenarthran fossil in the early diversification period was observed beyond their current distribution [7]. Compared with the background diversification pattern, the increased relative extinction rate of Boreoeutheria (ε=0.62; shift rate 2 in Fig. 2) suggests the turnover have also played an important role in shaping its disparity pattern of biodiversity, consistent with the notion which emphasizes the major role of frequent turnovers in mammalian diversification from paleontological spectacle [62, 63].
Within the diversification burst of Boreoeutheria, results based on family-level 'diversity tree' also pointed to six progressive exceptional diversifications involving with the recent diversifications of Cetartiodactyla, Chiroptera, Rodentia, and Primates (Table 1 and Fig. 2). The most significant one is the rapid radiation of the murids and cricetids (r=0.22 sp/My, rate shift 1 in Fig. 2), two most diverse and species-rich families of Mammalia with over 1,500 recognized species variously adapting to different habitats on most all landmass in the world except for Antarctica and New Zealand [12, 14]. Additionally, our study also located the other radiation within Rodentia leading to the Echimyidae, which is often referred as 'spiny rat' and occupies an amazing variety of habitats in the New World [12, 14] (r=0.149 sp/My, rate shift 6 in Fig. 2). When considering their most closely related sister groups, both diversification patterns reveal the extreme disparity in biodiversity and current distribution regions [12] (Fig. 2), which may imply the major role of biogeographic dispersals and expansions into novel and various habitats of Old and/or New World in achieving the spectacle taxonomic diversity in the short-term [7, 64, 68]. However, the in-depth studies on contributions of the rapid diversification are solely needed in the future. Two exceptional shift points were also detected in Cetartiodactyla (rate shifts 3 and 7 in Fig. 2). One substantial acceleration in net diversification rate of the clades Bovidae+Moschidae+Cervidae in the Late Oligocene (r=0.24 sp/My, rate shift 3 in Fig. 2) may have resulted from floral shifts associated with the expansion of the arid zone and grasslands (thus creating new ecological opportunities) due to global cooling and drying trend in the Oligocene [58, 69], the biogeographic scenario involving with expansion into different continents in the Miocene [70-72], as well as a series of digestive traits adapting the nutritional yield from poor-quality food [14]. Note that although the moschids are entirely Asian in their present distribution, the extensive fossil deposits from Eurasia and America indicate a rich diversity and cross-continental dispersal events in history [70, 73]. The other diversification burst within Cetartiodactyla was observed in a subclade of toothed whales including Ziphiidae, Delphinidae, Monodontidae, Phocoenidae, Iniidae, and Pontoporiidae in the Oligocene (r=0.28 sp/My, rate shift 7 in Fig. 2) instead of the crown whales that imply adaptive radiation of the crown group after the transition to new adaptive zone [10, 74]. This supports the major role of early abiotic drivers in shaping current biodiversity pattern of whales (e.g., tectonic-driven rearrangement of barriers, changes of mohaline circulation, and water temperature in the Oligocene) [75, 76], which was not detected using lineage-through-time plots (LTT plots) [61]. This shift node is also not consistent with detection based on a species-level chronogram employing cytochrome b (cytb) nucleotide sequences using MEDUSA [77], however, the disaccordance may be derived from differences of phylogenetic relationship within Odontoceti and different sampling scheme. As to bats, the vespertilionids seem to have experienced rapid radiation throughout their evolutionary progress (r=0.12 sp/My, rate shift 9 in Fig. 2). Actually, such scenario consists with by a recent comparative analysis on evolutionary substitution rates, which indicated that vespertilionid lineages had significantly faster rates in contrast with New World phyllostomids, an other successful radiation group within Chiroptera, which diverged at approximately the same time [78]. Unlike the largely tropical and subtropic distribution of other bats, the vespertilionids seem to be able overcome the energetic constraints and inhabit beyond tropic and subtropic areas [12, 79]. We suggest that ecological differences between tropical and temperate environment as well as some intrinsic adaptive traits (e.g., torpor, hibernation, migration, delayed fertilization, and multiple offspring) may be triggering factors for the rapid diversification through opening the unsaturated ecospace in temperate zone [78-80]. However, due to the impossibility of identifying rate shifts in unresolved branches, the shift point may be tipward along the branch with increasing phylogenetic resolution [4, 56]. As to Primates, although the exceptional rate shift of Simiiformes in the Late Oligocene seems to be odd since its net diversification rate was declined in contrast with boreoeutherian background rate (from 0.062 sp/My to 0.021 sp/My), the high relative extinction rate (ε=0.99) actually implies high speciation and high turnover (b=2.10 sp/My, d=2.08 sp/My) have synchronously dominated macroevolution of this group, and differential extinctions among lineages have played important role in shaping current uneven biodiversity pattern. In fact, climatic shifts have been regarded as one of the major factors causing extinction of ancestral primates [81, 82], if so, their present uneven biodiversity pattern may be attributed to the climatic and fauna changes in the Neogene and Quaternary [58, 83, 84].
Caveats of MEDUSA
How to explain the uneven biodiversity pattern within mammal has long intrigued biologists [1-5]. MEDUSA integrating likelihood from phylogenetic tree shape and species richness provides avenue and framework to identify general diversification patterns and exceptional rate shifts in light of phylogenetic relationship, divergence times and current biodiversity at a broad and comprehensive scale [2, 4]. It is a basilic step to evaluate potential causal links to changes in the magnitude and timing of diversification throughout the evolutionary history.
Although there were evidences of rapid radiation within the three most species-rich mammalian families (e.g., Muridae, Cricetidae, and Vespertilionidae; Fig. 2), not all species-rich groups exhibited exceptional diversification rates. In fact, no diversification bursts were detected from the crown of Rodentia (2278 species), Chiroptera (1116 species), Primate (376 species), Carnivora (286 species), Sciuridae (278 species), or Soricidae (376 species) (Figs. 1 and 2), all of which represent recognized mammalian radiations with high biodiversity [11, 14]. Furthermore, there was also no signal of early rapid radiation in the Chiroptera or Cetacea stems (Figs. 1 and 2), which have been hypothetically regarded as successful mammalian groups entering into novel adaptive zones [9, 10]. Although these results conflict with the notion about triggering role of transition into novel habitats in diversification [7-10, 18], they actually mean that their biodiversity patterns observed either in the family- or order-level trees were not much higher than the model's expectation, and that background or clade diversification rate is sufficient to explain the heterogeneous patterns of diversity observed nowadays [2, 4]. In other words, it is premature to conclude that those lineages with average diversification rates have not experienced rapid cladogenesis merely based on inferences from MEDUSA. In fact, the distinct and disparity ecomorphological forms of crown whales have provided the evidences of early adaptive radiation in the evolutionary history of cetaceans [77], even though this pattern has not been identified by MEDUSA in the present study. Another example showing this shortcoming is the observed unusual diversification pattern of the ruminant clade (rate shift 3 in Fig. 2). The separation of giraffids and antilocaprids into lineages with unusual shifts was mostly attributed to the low species richness of these two lineages compared to their species-rich sister clade. When the paleontological diversity of Giraffidae (16 extinct species) and Antilocapridae (63 extinct species) derived from Paleobiology Database on 21 November, 2011 is considered in MEDUSA, these two families are included in those clades with exceptional diversification rates. Moreover, the short length of the branches of these five ruminant lineages could be another sign of rapid radiation (Fig. 2). These results emphasize the key role of extinction in erasing the signal of historical radiation and unexceptional diversification rates [77]. We suggest that the examination of the tempo of diversification across particular clades (e.g., using lineage-through-time plots) or of the disparity in ecomorphological patterns (e.g., morphological disparity index) among clades may be therefore more efficient to detect early rapid radiation signals in those cases where MEDUSA did not reveal exceptional diversification patterns.
The other caveat of MEDUSA is its dependence on phylogenetic topology and resolution. MEDUSA estimates per-lineage diversification rates from combined phylogenetic and taxonomic data through maximizing likelihood of the functions, which could avoid the problem of generating null model from traditional sister-group tests [86, 87]. However, it is still a little similar to the sister-group tests of key innovation hypotheses in explaining the taxonomic disparity of different groups. Hence, the fact that both log likelihood scores of the functions and identified shift points from MEDUSA may be influenced when the biodiversity of sister group changes [2] indicates that to confirm the shift points within the diversity tree, diversification analyses using MEDUSA should be performed either upon a 'correct' phylogeny or upon all possible proposed topologies. Moreover, the incapability of identifying rate shifts in unresolved branches implies that caution should be taken when interpreting the observation of terminal clades with exceptional diversification rates (e.g., rate shifts 6, 8 and 9 in Fig. 2), as this pattern may change and the shift point may be tipward with increasing phylogenetic resolution, even though the significant rates shift within the subclades is indeed confirmed.
In summary, this study provided a general picture of mammalian differential diversification through MEDUSA. However, to scrabble up the puzzle of mammalian macroevolution, various novel diversification approaches that integrate paleontological and neontological information, involving binary and quantitative key traits, biogeographic history and phylogenetic analysis, are solely needed in the future.
Acknowledgements
We thank Zuo Chen and Tong Shen for helpful discussion. Two anonymous reviewers provided helpful comments on earlier versions of this manuscript. This study is supported by the National Natural Science Foundation of China (30830016, 30670277, 31172045, 31110103910), the Program for New Century Excellent Talents in University, the Ministry of Education of China (NCET-07-0445), the major project of the Natural Science Foundation of the Jiangsu Higher Education Institutions of Jiangsu Province, China (07KJA18016), the Priority Academic Program Development of Jiangsu Higher Education Institutions to G.Y, and the Research Innovation Program for College Graduates of Jiangsu Province (CXZZ11_0883).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Rabosky DL. Primary controls on species richness in higher taxa. Syst Biol. 2010;59:634-645
2. Rabosky DL, Donnellan SC, Talaba AL. et al. Exceptional among-lineage variation in diversification rates during the radiation of Australia's most diverse vertebrate clade. Proc R Soc Lond B Biol Sci. 2007;274:2915-2923
3. Ricklefs RE. Speciation, extinction and diversity. In: (ed.) Butlin R, Bridle J, Schluter D. Speciation and patterns of diversification. Cambridge: Cambridge University Press. 2009:257-277
4. Alfaro ME, Santini F, Brock C. et al. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc Natl Acad Sci USA. 2009;106:13410-13414
5. Mooers AO, Heard SB. Inferring evolutionary process from the phylogenetic tree shape. Q Rev Biol. 1997;72:31-54
6. Slowinski JB, Guyer C. Testing whether certain traits have caused amplified diversification: an improved method based on a model of random speciation and extinction. Am Nat. 1993;142:1019-1024
7. Yoder JB, Clancey E, Des Roches S. et al. Ecological opportunity and the origin of adaptive radiations. J Evolution Biol. 2010;23:1581-1596
8. Schluter D. The ecology of adaptive radiation. Oxford: Oxford University Press. 2000
9. Simpson GG. Tempo and mode in evolution. New York: Columbia University Press. 1949
10. Simpson GG. The major features of evolution. New York: Columbia University Press. 1953
11. Macdonald DW. The Encyclopedia of Mammals. New York: Oxford University Press. 2010
12. Wilson DE, Reeder DM. Mammal Species of the World. A Taxonomic and Geographic Reference (3rd ed): Johns Hopkins University Press. 2005
13. Kemp TS. The Origin and Evolution of Mammals. New York: Oxford University Press. 2005
14. Vaughan TA, Ryan JM, Czaplewski NJ. Mammalogy. Sudbury, Massachusetts: Jones and Bartlett Publishers. 2010
15. Woodburne MO, Rich TH, Springer MS. The evolution of tribospheny and the antiquity of mammalian clades. Mol Phylogenet Evol. 2003;28:360-385
16. Ricklefs RE, Losos JB, Townsend TM. Evolutionary diversification of clades of squamate reptiles. J Evolution Biol. 2007;20:1751-1762
17. Roelants K, Gower DJ, Wilkinson M. et al. Global patterns of diversification in the history of modern amphibians. Proc Natl Acad Sci USA. 2007;104:887-892
18. de Queiroz A. Interpreting sister-group tests of key innovation hypotheses. Syst Biol. 1998;47:710-718
19. Pagel M. Inferring evolutionary processes from phylogenies. Zool Scr. 1997;26:331-348
20. Barraclough TG, Nee S, Harvey PH. Sister-group analysis in identifying correlates of diversification - Comment. Evol Ecol. 1998;12:751-754
21. Murphy WJ, Eizirik E, O'Brien SJ. et al. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348-2351
22. Murphy WJ, Pringle TH, Crider TA. et al. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413-421
23. Springer MS, Burk-Herrick A, Meredith R. et al. The adequacy of morphology for reconstructing the early history of placental mammals. Syst Biol. 2007;56:673-684
24. Springer MS, Murphy WJ, Eizirik E. et al. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA. 2003;100:1056-1061
25. Springer MS, Stanhope MJ, Madsen O. et al. Molecules consolidate the placental mammal tree. Trends Ecol Evol. 2004;19:430-438
26. Meredith RW, Janecka JE, Gatesy J. et al. Impacts of the Cretaceous Terrestrial Revolution and KPg Extinction on mammal diversification. Science. 2011;334:521-524
27. Amrine-Madsen H, Scally M, Westerman M. et al. Nuclear gene sequences provide evidence for the monophyly of australidelphian marsupials. Mol Phylogenet Evol. 2003;28:186-196
28. Janecka JE, Miller W, Pringle TH. et al. Molecular and genomic data identify the closest living relative of primates. Science. 2007;318:792-794
29. Madsen O, Scally M, Douady CJ. et al. Parallel adaptive radiations in two major clades of placental mammals. Nature. 2001;409:610-614
30. Meredith RW, Westerman M, Springer MS. A phylogeny of Diprotodontia (Marsupialia) based on sequences for five nuclear genes. Mol Phylogenet Evol. 2009;51:554-571
31. Teeling EC, Springer MS, Madsen O. et al. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science. 2005;307:580-584
32. Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web Servers. Syst Biol. 2008;57:758-771
33. Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). 2010:1-8
34. Nilsson MA, Churakov G, Sommer M. et al. Tracking marsupial evolution using archaic genomic retroposon insertions. Plos Biol. 2010;8(7):e1000436
35. Beck RMD. A dated phylogeny of marsupials using a molecular supermatrix and multiple fossil constraints. J Mammal. 2008;89:175-189
36. Meredith RW, Westerman M, Case JA. et al. A phylogeny and timescale for marsupial evolution based on sequences for five nuclear genes. J Mamm Evol. 2008;15:1-36
37. Nilsson MA, Arnason U, Spencer PBS. et al. Marsupial relationships and a timeline for marsupial radiation in South Gondwana. Gene. 2004;340:189-196
38. Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 2007;24:1586-1591
39. Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301-302
40. Burnham KP, Anderson DR. Model selection and multimodel inference: a practical information-theoretic approach. New York: Springer. 2002
41. Nishihara H, Okada N, Hasegawa M. Rooting the eutherian tree: the power and pitfalls of phylogenomics. Genome Biol. 2007;8:R199
42. Kriegs JO, Churakov G, Kiefmann M. et al. Retroposed elements as archives for the evolutionary history of placental mammals. Plos Biol. 2006Apr;4(4):e91
43. Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289-290
44. Harmon LJ, Weir JT, Brock CD. et al. GEIGER: investigating evolutionary radiations. Bioinformatics. 2008;24:129-131
45. Rabosky DL. LASER: A maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol Bioinform. 2006;2:247-250
46. Vos R, Caravas J, Hartmann K. et al. BIO::Phylo-phyloinformatic analysis using perl. BMC Bioinformatics. 2011;12:63
47. Huerta-Cepas J, Dopazo J, Gabaldon T. ETE: a python Environment for Tree Exploration. BMC Bioinformatics. 2010;11:24
48. Rutschmann F. Molecular dating of phylogenetic trees: A brief review of current methods that estimate divergence times. Diversity Distrib. 2006;12:35-48
49. Hug LA, Roger AJ. The impact of fossils and taxon sampling on ancient molecular dating analyses. Mol Biol Evol. 2007;24:1889-1897
50. Hallström BM, Janke A. Mammalian evolution may not be strictly bifurcating. Mol Biol Evol. 2010;27:2804-2816
51. Nilsson MA, Harlid A, Kullberg M. et al. The impact of fossil calibrations, codon positions and relaxed clocks on the divergence time estimates of the native Australian rodents (Conilurini). Gene. 2010;455:22-31
52. Roos J, Aggarwal RK, Janke A. Extended mitogenomic phylogenetic analyses yield new insight into crocodylian evolution and their survival of the Cretaceous-Tertiary boundary. Mol Phylogenet Evol. 2007;45:663-673
53. Luo ZX, Yuan CX, Meng QJ. et al. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011;476:442-445
54. Nilsson MA, Gullberg A, Spotorno AE. et al. Radiation of extant marsupials after the K/T boundary: Evidence from complete mitochondrial genomes. J Mol Evol. 2003;57:S3-S12
55. Martin HA. Cenozoic climatic change and the development of the arid vegetation in Australia. J Arid Environ. 2006;66:533-563
56. Burk A, Springer MS. Intergeneric relationships among Macropodoidea (Metatheria: Diprotodontia) and the chronicle of kangaroo evolution. J Evolution Biol. 2000;7:213-237
57. Springer MS, Kirsch JAW, Case JA. The chronicle of marsupial evolution. In: (ed.) Givinish T, Sytsma K. Molecular Evolution and Adaptive Radiation. New York: Cambridge University Press. 1997:129-161
58. Zachos J, Pagani M, Sloan L. et al. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686-693
59. Archibald JD, Deutschman DH. Quantitative analysis of the timing of the origin and diversification of extant placental orders. J Mamm Evol. 2001;8:107-124
60. Bininda-Emonds ORP, Cardillo M, Jones KE. et al. The delayed rise of present-day mammals. Nature. 2007;446:507-512
61. Steeman ME, Hebsgaard MB, Fordyce RE. et al. Radiation of extant cetaceans driven by restructuring of the oceans. Syst Biol. 2009;58:573-585
62. Stanley SM. Macroevolution: Pattern and Process. Baltimore: Johns Hopkins University Press. 1998
63. Alroy J. Speciation and extinction in the fossil record of North American mammals. In: (ed.) Butlin R, Bridle J, Schluter D. Speciation and Patterns of Diversity. Cambridge: Cambridge University Press. 2009:301-323
64. Purvis A, Fritz SA, Rodriguez J. et al. The shape of mammalian phylogeny: patterns, processes and scales. Proc R Soc Lond B Biol Sci. 2011;366:2462-2477
65. Pybus OG, Harvey PH. Testing macro-evolutionary models using incomplete molecular phylogenies. Proc R Soc Lond B Biol Sci. 2000;267:2267-2272
66. Rabosky DL, Lovette IJ. Density-dependent diversification in North American wood warblers. Proc R Soc Lond B Biol Sci. 2008;275:2363-2371
67. Rabosky DL, Lovette IJ. Explosive evolutionary radiations: Decreasing speciation or increasing extinction through time? Evolution. 2008;62:1866-1875
68. Cardillo M. Phylogenetic structure of mammal assemblages at large geographical scales: linking phylogenetic community ecology with macroecology. Philos T R Soc B. 2011;366:2545-2553
69. Morley RJ. Creaceous and Tertiary climate changes and the past distribution of megathermal rainforests. In: (ed.) Bush MB, Flenley J. Tropical rainforest response to climatic changes. Chichester: Praxis Publishing. 2007
70. Hassanin A, Douzery EJP. Molecular and morphological phylogenies of Ruminantia and the alternative position of the Moschidae. Syst Biol. 2003;52:206-228
71. Prothero DR, Heaton TH. Faunal stability during the early Oligocene climatic crash. Palaeogeogr Palaeoclimatol Palaecol. 1996;127:257-283
72. Meng J, McKenna MC. Faunal turnovers of Palaeogene mammals from the Mongolian Plateau. Nature. 1998;394:364-367
73. Novello A, Blondel C, Brunet M. Feeding behavior and ecology of the Late Oligocene Moschidae (Mammalia, Ruminantia) from La Milloque (France): Evidence from dental microwear analysis. Comptes Rendus Palevol. 2010;9:471-478
74. Nikaido M, Matsuno F, Hamilton H. et al. Retroposon analysis of major cetacean lineages: the monophyly of toothed whales and the paraphyly of river dolphins. Proc Natl Acad Sci USA. 2001;98:7384-7389
75. Fordyce RE. Whale evolution and Oligocene southern ocean environments. Palaeogeogr Palaeoclimatol Palaeoecol. 1980;31:319-336
76. Fordyce RE, de Muizon C. Evolutionary history of cetaceans: a review. In: (ed.) Mazin JM, de Buffrénil V. Secondary adaptations of tetrapods to life in water. München, Germany: Verlag Dr. Friedrich Pfeil. 2001
77. Slater GJ, Price SA, Santini F. et al. Diversity versus disparity and the radiation of modern cetaceans. Proc R Soc Lond B Biol Sci. 2010;277:3097-3104
78. Lack JB, Van Den Bussche RA. Identifying the confounding factors in resolving phylogenetic relationships in Vespertilionidae. J Mammal. 2010;91:1435-1448
79. Stevens RD. Untangling latitudinal richness gradients at higher taxonomic levels: familial perspectives on the diversity of New World bat communities. J Biogeogr. 2004;31:665-674
80. Altringham J. Bats biology and behaviour. New York: Oxford University Press. 1996
81. Gingerich PD. Plesiadapis and the delineation of the order Primates. In: (ed.) Wood B, Martin L, Andrews P. Major topics in primate and human evolution. Cambridge: Cambridge University. 1986:32-46
82. Fleagle JG. Primate adaptation and evolution, 2nd edition. San Diego: Academic Press. 1999
83. Retallack GJ. Neogene expansion of the North American prairie. PALAIOS. 1997;12:380-390
84. Lourens L, Hilgen F, Shackleton NJ. et al. The neogene period. In: (ed.) Gradstein F, Ogg J, Smith AG. Geologic Time Scale. Cambridge: Cambridge University Press. 2004
85. Harmon LJ, Schulte JA, Larson A. et al. Tempo and mode of evolutionary radiation in Iguanian Lizards. Science. 2003;301:961-964
86. Mitter C, Farrell B, Wiegmann B. The phylogenetic study of adaptive zones: Has phytophagy promoted insect diversification? Am Nat. 1988;132:107-128
87. Slowinski JB, Guyer C. Testing whether certain traits have caused amplified diversification: an improved method based on a model of random speciation and extinction. Am Nat. 1993;142:1019-1024
Author contact
![]() Corresponding author: Dr. Guang Yang; Jiangsu Key Laboratory for Biodiversity and Biotechnology, College of Life Sciences, Nanjing Normal University, Nanjing 210046. China; Phone: 86-25-85891163; Fax: 86-25-85891163; E-mail: gyangedu.cn. Dr. Yi Wu; College of Life Sciences, Guangzhou University, Guangzhou 510006, China. Phone: 86-20-39366919; Email: wuyiedu.cn.
Corresponding author: Dr. Guang Yang; Jiangsu Key Laboratory for Biodiversity and Biotechnology, College of Life Sciences, Nanjing Normal University, Nanjing 210046. China; Phone: 86-25-85891163; Fax: 86-25-85891163; E-mail: gyangedu.cn. Dr. Yi Wu; College of Life Sciences, Guangzhou University, Guangzhou 510006, China. Phone: 86-20-39366919; Email: wuyiedu.cn.

 Global reach, higher impact
Global reach, higher impact