10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2012; 8(6):882-890. doi:10.7150/ijbs.4421 This issue Cite
Research Paper
Transient and Stable GFP Expression in Germ Cells by the vasa Regulatory Sequences from the Red Seabream (Pagrus major)
1. Center of Biotechnology R&D, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, P.R. China;
2. Graduate school of Chinese Academy of Sciences, Beijing 100049, P.R. China;
3. Department of Biological Sciences, National University of Singapore, 10 Kent Ridge Crescent, Singapore 119260.
#Theses authors contributed equally to the work.
Received 2012-3-30; Accepted 2012-6-4; Published 2012-6-16
Abstract
Primordial germ cells (PGCs) are the precursors of gametes responsible for genetic transmission to the next generation. They provide an ideal system for cryopreservation and restoration of biodiversity. Recently, considerable attention has been raised to visualize, isolate and transplant PGCs within and between species. In fish, stable PGC visualization in live embryo and individual has been limited to laboratory fish models such as medaka and zebrafish. One exception is the rainbow trout, which represents the only species with aquaculture importance and has GFP-labeled germ cells throughout development. PGCs can be transiently labeled by embryonic injection of mRNA containing green fluorescence protein gene (GFP) and 3'-untranslated region (3'-UTR) of a maternal germ gene such as vasa, nos1, etc. Stable PGC labeling can be achieved through production of transgenic animals by some transcriptional regulatory sequences from germ genes, such as the vasa promoter and 3'-UTR. In this study, we reported the functional analyses of the red seabream vasa (Pmvas) regulatory sequences, using medaka as a model system. It was showed that injection of GFP-Pmvas3'UTR mRNA was able to label medaka PGCs during embryogenesis. Besides, we have constructed pPmvasGFP transgenic vector, and established a stable transgenic medaka line exhibiting GFP expression in germ cells including PGCs, mitotic and meiotic germ cells of both sexes, under control of the Pmvas transcriptional regulatory sequences. It is concluded that the Pmvas regulatory sequences examined in this study are sufficient for germ cell expression and labeling.
Keywords: Pagrus major, PGCs, vasa, transgene, GFP
Introduction
Primordial germ cells (PGC) are the precursors of the germ cell lineage, responsible for genetic transmission through generations. The inherent nature endows PGCs with numerous advantages of applications in fish bioengineering, such as cryopreservation and restoration of biodiversity [1]. PGCs can be specifically labeled and isolated for cell culture and transplantation, providing tools for reproduction of endangered species in close relatives [2]. The techniques open a new approach for genetic resource preservation and improve our understanding in germ line development [3]. However, in teleosts, successful surrogate breeding by germline cell transplantation has been limited to the rainbow trout [4, 5] and zebrafish [6, 7].
PGCs visualization and labeling is the first step to forward the application. They can be transiently labeled by embryonic injection of mRNA containing green fluorescence protein gene (GFP) and 3'-untranslated region (3'UTR) of a maternal germ gene such as vasa, nos1, etc. The 3'UTR sequences play a critical role in eliminating the mRNA from the somatic cell line via microRNA activity, while stabilize the mRNA in PGCs through the DND (dead end) protein [8]. Besides, the function of the 3'UTR is widely conserved among fish species. For example, the mRNA combining GFP and zebrafish nos1 3'UTR has been proved to be able to identify PGCs in a wide range of fish species [9]. Meanwhile, other chimeric mRNA consisting of GFP and vasa 3'UTR from Nibe croaker or zebrafish has been used to visualize PGCs in rainbow trout successfully [10], in spite that GFP-Olvas 3'UTR mRNA from the medaka is not able to visualize PGCs in either zebrafish or loach [9]. Recently, in cyprinid fish, nontransgenic labeled PGCs (zebrafish nos1 3'UTR) have been successfully applied to fluorescence-activated cell sorting (FACS) [11] and produce inter-species germ-line chimeras [7]. Although injection of chimeric mRNA labels PGCs transiently, this nontransgenic technique is especially useful for cryopreservation of PGCs and seed production by surrogate breeding.
Stable PGC labeling has been achieved through the production of transgenic animals by promoter and some regulatory sequences of germ genes, such as the vasa promoter and 3'-UTR. At present, transgenic fish lines driven by vasa promoter, which express GFP specifically in germ cell lineage, have been reported only in the model fish, such as zebrafish [12] and medaka [13]. The only exception is rainbow trout [14], a species with high aquaculture importance. It has immense benefits for basic research, making FACS, long-term tracing of donor PGCs in recipient individuals, PGCs cryopreservation and screening mutants affecting PGCs feasible. FACS has been successfully applied to isolate labeled PGCs from transgenic rainbow trout [15] and zebrafish [16]. However, it is not easy to apply this approach in commercial fish species for various reasons: intensive labor, time and space consumption of producing transgenic fish; risk of biological contamination caused by releasing of the transgenic fish; consumers' attitudes toward genetically modified fish.
Red seabream (Pagrus major) is an economically important marine species in China. The methods for sperm cryopreservation of the fish have been developed successfully. However, little progress has been achieved in the embryo cryopreservation. The PGCs cryopreservation, which can eventually be used to generate viable individuals by surrogate breeding, provides us an attractive alternative way. However, the technique for labeling the PGCs of red seabream has not been reported. In this study, we used medaka as a model system to investigate the function of the red seabream vasa (Pmvas) regulatory sequences, which will facilitate further research on labeling, isolation and cryopreservation of red seabream PGCs.
Materials and Methods
Fish breeding and embryo preparation
Fish breeding followed the guidelines on care and use of laboratory animals for scientific purpose, approved by National Advisory Committee in Singapore. The medaka strain: af (strain without self-fluorescence) was maintained under an artificial photoperiod of 14 h light to 10 h darkness at 26 ℃. Embryos were collected in the morning after mixing male and female fish which were separated one day before. Embryogenesis and oogenesis were staged according to Iwamatsu [17, 18].
Prepare of GFP-Pmvas 3'UTR and RFP-Olnos3 3'UTR mRNA
Chimeric mRNA containing GFP fused to 3'UTR of red seabream vasa gene (Fig. 1A) and RFP combined with 3'UTR of medaka nos3 gene (Fig. 1B) were synthesized by in vitro transcription. The template plasmid for RFP-Olnos3 3'UTR mRNA (which contained RFP and the 3' UTR of medaka nanos3 in PCS2 vector) was kindly supplied by Doctor Li Mingyou; The template plasmid for GFP-Pmvas 3'UTR was constructed as following procedures: 3'UTR of the red seabream vasa mRNA (Pmvasa, AB378581) was cloned into pGEM-T easy vector by primers and designed as vasa3U-T vector:
vasa3U-F: 5′-GCTGATGATGACGACTGGGATT-3′
vasa3U-R: 5′-AACAAATATTTATTTATTGGTGATC-3′
AatII and SacII sites were introduced at the 5' end and 3' end of GFP ORF, respectively, by a PCR reaction with primers:
EGFP-F: 5′-TATATTGACGTCCGCCACCATGGTGAGC-3′
EGFP-R: 5′-TATATTCCGCGGTTACTTGTACAGCTCGTC-3′
After double digestion, the fragment was cloned into the corresponding sites of vasa3U-T. The resultant plasmid was linearized by SalI digestion for in vitro transcription by Message machine T7 kit (Ambion Inc., Austin, TX).
Structure of chimeric mRNA and transgenic vector for PGCs labeling
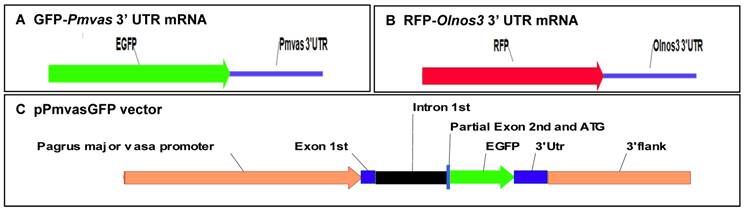
Construct of pPmvasGFP vector
The 5' and 3' flanking region sequences of the red seabream vasa gene were previously isolated by genome walking, basing on the vasa mRNA sequence (Pmvasa, AB378581) [19]. The procedures for construction of pPmvasGFP vector (Fig. 1C) were as follows:
The 3' flanking region downstream the stop codon (TAA) of the red seabream vasa gene was amplified from red seabream genome DNA by primers:
vasa3-F: 5′-TCTAGCGGCCGCGATGATGACGACTGGGATTA-3′
vasa3-R: 5′-TCTAGCGGCCGCAGGTCATGATTGCTGATATT-3′
The resultant 1.9 kb fragment was digested with NotI and cloned into the corresponding site of pEGFP-4.1 vector and designed as pEGFP-3V.
The 5' flanking region upstream the start codon (ATG) of the vasa gene was amplified by primers:
Vasa5-F: 5′-TAGCAAGCTTCCAACTGTCGCCGCTGAT-3′
Vasa5-R: 5′-GTCAGGATCCTCTTCTTCCCACTCGTCCAT-3′
The resultant 3.8 kb fragment was digested with BamHI and HindIII and cloned into the corresponding site of pEGFP-3V to produce the pPmvasGFP transgenic vector.
Microinjection
The purified vector pPmvasGFP (25 ng/µl in 10mM Tris-Cl PH7.5, 0.02% Phenol Red) was injected into fertilized eggs of af strain at 1 cell stage. GFP-Pmvas 3'UTR and RFP-Olnos3 3'UTR mRNA (100 ng/µl each, DEPC water, 0.02% Phenol Red) were co-injected into fertilized eggs of af strain at 1 cell stage, to confirm that the chimeric mRNA containing Pmvas 3'UTR label PGCs specifically. The injected embryos were incubated in 1*ERM at 28 ℃ for GFP or RFP observation and photography under fluorescent microscope (Leica MeFIII) at different stages.
Selection of the transgenic line
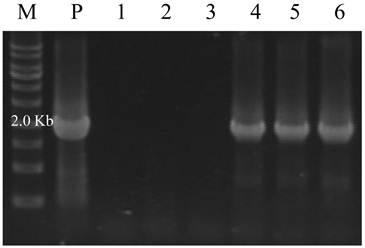
Sexually mature medaka from injected embryos were mated with nontransgenic fish to obtain F1 fish. Embryos were screened to indentify germline transmitting founder (F0) by both fluorescence microscopy and genomic PCR, using primers for 3' flanking region (1.9 kb) of the red seabream vasa gene. The transgenic offspring (F1) with GFP were raised to maturity and crossed with sibling transgenic F1 to confirm the F2 had a GFP segregation ratio following Mendelian genetics.
Cryosection
In order to confirm that GFP expression cells in gonads of F1 transgenic fish were germ cells, cryosections of transgenic ovaries and tests were performed. The method was described previously [20].
Results
Visualization of medaka PGCs by GFP-Pmvas 3'UTR mRNA
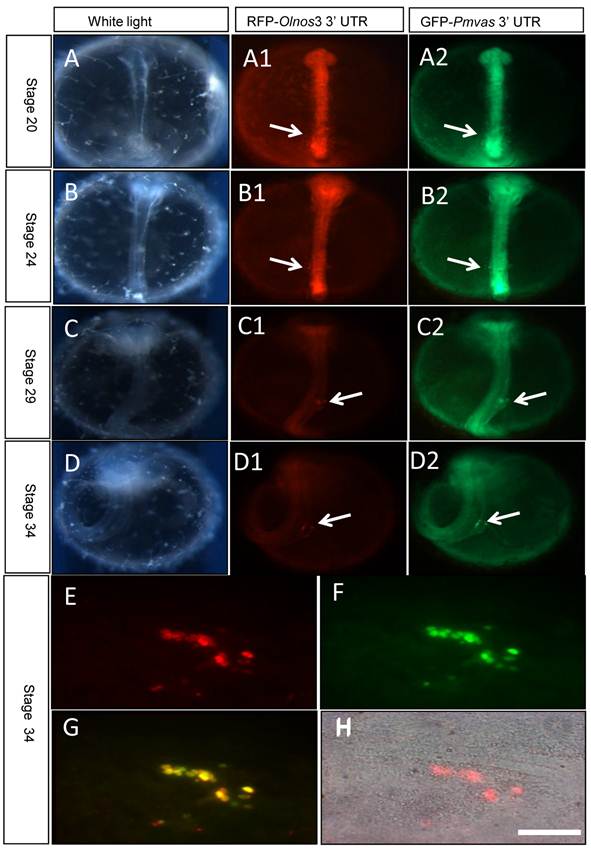
In order to confirm that 3'UTR of Pagrus major vasa shares similar function with that of medaka nanos3 gene, so that can be used to visualize PGCs, GFP-Pmvas 3'UTR mRNA was co-injected into fertilized embryos with RFP-Olnos3 3'UTR mRNA, which has been used to trace migration of PGCs in medaka [21]. Results showed that the GFP fluorescence didn't differentiate between somatic cells and PGCs until early segmentation period (Fig. 2A). As embryogenesis proceeded, GFP in PGCs became more obvious since the fluorescence of somatic cells reduced gradually, resembling the situation of RFP-Olnos3 3'UTR mRNA injection (Fig. 2B-D). Cells acquiring fluorescence were rounder and had larger size, which were typical characteristics of PGCs (Fig. 2E-H). However, it's interesting that GFP signal distributed uniformly throughout entire cell and the nucleus couldn't be identified, while most RFP signal distributed in cytoplasm and the nucleus could be easily identified. In spite of that, both chimeric mRNA can visualize and reveal the migration route of PGCs in medaka during embryogenesis.
Transient PGCs labeling by co-injection of RFP-Olnos3 3'UTR (Red) and GFP-Pmvas 3'UTR (Green) mRNA. PGCs were indicated by white arrows. A-D) embryos of stage 20, stage 24, stage 29 and stage34, respectively, E-H) the gonad region of stage 34 was squashed into cell lever and observed under fluorescence microscopy, Scale bar, 50 µm.
Establishment of transgenic medaka line with germ cells expressing GFP
GFP fluorescence was observed in 78% of 257 injected embryos. Among those, 54 embryos (21%) carrying GFP expression in PGCs (Table 1). The injected embryos with GFP expression were raised to maturity and crossed with nontransgenic strain to screen the germ-line transmitting founder (F0) using fluorescence microscopy combined with genomic PCR of the F1 progeny (Fig. 3). 3 out of 26 fish from injected embryos were confirmed to be germ-line transmitting founder (F0), with appearance ratio of GFP expression in F1 embryos ranging from 9.8% to 13.4% (Table 2). The F2 offspring represented approximately 75% of GFP expression segregation ratio, from an F1 transgenic/ F1 transgenic cross (Table 3). The results demonstrated that pPmvasGFP was stably transmitted to offspring following the Mendelian genetics. Therefore, pPmvasGFP transgenic medaka line was successfully established.
Result of microinjection of pPmvasGFP vector in medaka
| Strain | No. of injected embryos | GFP expression | GFP positive in PGCs | No. of hatching embryos | Sexual maturity founder |
|---|---|---|---|---|---|
| af | 257 | 200 (78%) | 54 (21%) | 110 (43%) | 83 |
Result of transgenic founder screening
| No. of Screened fish | No. of germline transmitting founder | Ration of GFP expression in F1 |
|---|---|---|
| 26 | 3 (12%) | |
| #1 | 37/296(12.5%) | |
| #2 | 46/467(9.8%) | |
| #3 | 31/231(13.4%) | |
| Mean | 114/994(11.5%) |
GFP expression ratio in offspring (F2) from F1 transgenic cross
| No. of embryos | GFP positive embryos | GFP negative embryos |
|---|---|---|
| 129 | 96 (74.4%) | 33 (25.6%) |
Identification of germline transmitting progeny by genomic PCR. M) DNA marker; P) positive control from pPmvasGFP vector; 1-3) F1 embryos without GFP expression; 4-6) F1 embryos with GFP expression.
GFP expression pattern in transgenic line
Examination of transgenic embryos by fluorescence microscopy showed that the expression pattern of the pPmvasGFP transgenic fish was similar to the previous reported transgenic line by Tanaka [13]. The female transmitting progeny exhibited GFP fluorescence in every cell before gastrulation (Fig. 4A). After later gastrulation stage (stage 16), GFP fluorescence became intensive in germ ring and embryonic shield (Fig. 4B). Zygotic expression of GFP in male transmitting progeny was not visible until early segmentation. In segmentation period, the GFP fluorescence of embryonic body became stronger. However, obvious PGCs with GFP expression couldn't be detected (Fig. 4C) in the stage. By stage 29, two PGCs clusters formed aligning on both sides of the trunk around the 10th to 12th somites (Fig. 4D). The GFP fluorescent cells migrated dorsally from the surface of yolk mass and formed a single clump at the dorsal region of the intestine by stage 34 (Fig. 4E). Then, the clump moved bilaterally and formed two rows of cells dorsolateral to the digestive tract until stage 38 (Fig. 4F). After hatching, GFP positive cells could still be obviously detected on the dorsolateral side of the gut (Fig. 5A-B). Two months after hatching, GFP continued to be present in gonad region of transgenic medaka (Fig. 5C-F). In particular, GFP showed strong expression in gonad, little or none expression in gill, heart, spleen and kidney (Fig. 6A2-B2). However, weak expression of GFP was also detected in brain, gut and liver (Fig. 6A1-B1). In ovary, GFP was found abundant in oogonia and early stage oocytes, and reduced or diluted in large oocytes (Fig. 7A-B). However, in testis, GFP seemed to be expressed abundantly in spermatogonia, while little in later stages of spermatogenesis (Fig. 7C-D). It suggested that the adult pPmvasGFP expression might be germ cell specific in both sexes of medaka, and it might be more specific in testes.
Germline Expression of GFP in transgenic medaka before hatching. GFP was monitored by fluorescence microscopy. A-F showed the expression pattern of the transgenic offspring following female transmission. The PGCs were indicated by white arrows. White square box represented the presumptive gonad region under white light. A) stage 10; B) stage 16; C) Stage 24; D) stage 29; E ) stage 34; F) stage 38.
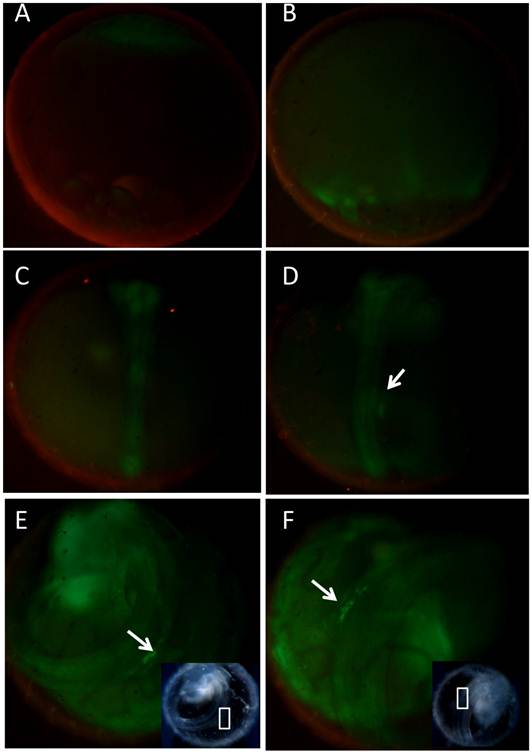
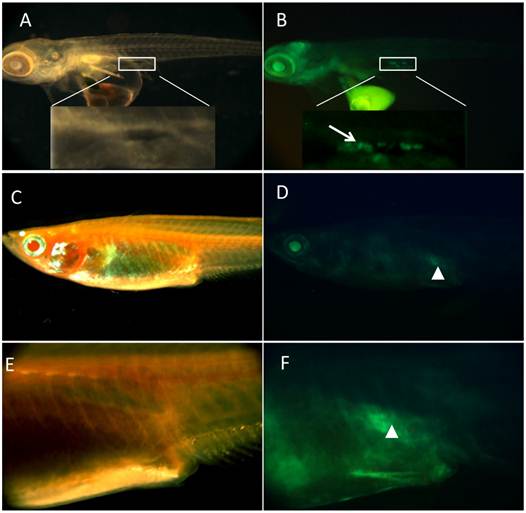
Germline Expression of GFP in transgenic medaka after hatching. A-B) medaka of two days after hatching, with PGCs expression GFP, located at dorsal side of intestine; C-D) medaka of two months after hatching, with gonad expressing GFP; E-F) magnification of corresponding area of C and D, respectively. Tissue with germ cells was encompassed by white square box. PGCs were indicated by white arrow. Gonad with GFP was indicated by white arrowhead.
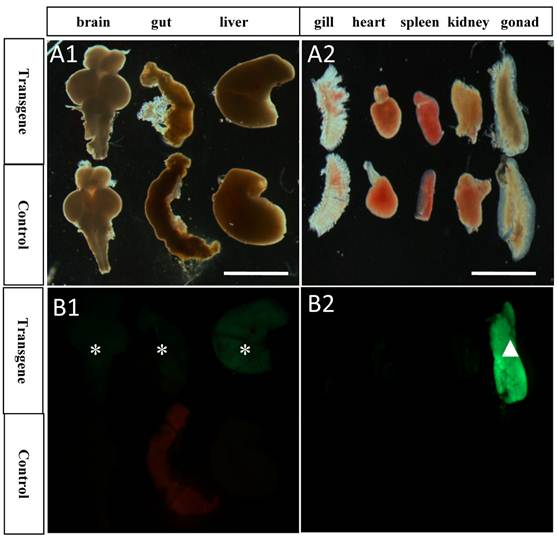
GFP expression pattern in different tissues of transgenic and control medaka. A1-B1) GFP showed some weak expression in brain, gut and liver of transgenic tissues, while no expression in the control; A2-B2) GFP showed strong expression in gonad, little or no expression in gill, heart, spleen and kidney, while no expression in the control. Asterisks represent tissues with weak GFP expression; White arrowhead represents tissue with strong GFP expression; Scale bar, 2 mm.
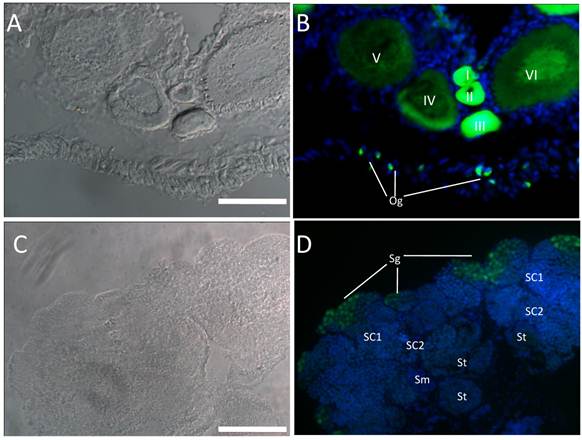
GFP expression pattern in germ cells of gonads. Cryosections of ovary (A-B) and testis (C-D) were used to detect endogenous GFP. Nuclei was stained by DAPI (Blue). In ovary, GFP expression was abundant in early stages of oogenesis (oogonia, oocytes of I-III stage) and reduced in late stages oocytes (IV-VI); In testis, Abundant GFP expression was detected in spermatogonia, while little expression was detected in other stages of spermatogenesis. Og, oogonia; I-VI, different stages of oocytes; Sg, spermatogonia; Sc1 and Sc2, primary and secondary spermatocytes; St, spermatids; Sm, sperm; Scale bar, 100 µm.
Discussion
In this study, we demonstrated that medaka PGCs could be visualized by microinjection of GFP-Pmvas 3' UTR chimeric mRNA. Besides, we also successfully established a stable transgenic medaka line expressing GFP in germ cells including PGCs, mitotic and meiotic germ cells of both sexes, under control of the transcriptional regulatory sequences from a highly diverged species, the red seabream (Pagrus major).
It is known that the 3' UTR of vasa plays critical role for stabilization of chimeric RNA in PGCs but not in somatic cells [22]. In rainbow trout, a construct containing the SV40 polyadenylation signal, instead of the 3'UTR of the vasa gene, failed to label PGCs with GFP, suggesting that the region was essential for visualizing PGCs in the species [10]. Our results also indicated that the 3'UTR of the red seabream vasa gene could stabilize the GFP chimeric mRNA in PGCs of medaka embryos, similar to the 3'UTR of medaka nanos3 gene. Therefore, we inferred that the red seabream shared a similar mechanism with medaka that specific stabilization of RNA in PGCs by vasa 3'UTR, although we couldn't rule out the other mechanism of vasa 3'UTR's RNA localization function [23]. To distinguish the possibility, whole mount in situ hybridization of the vasa gene in oocytes and embryos of the red seabream will be performed in the further studies.
It has been reported that PGCs of fishes could be visualized by chimeric RNAs fused to vasa 3' UTR from highly diverged taxonomic groups. For example, PGCs of rainbow trout (belonging to Salmoniformes) could be visualized by mRNA containing vasa 3'UTR from both Nibe croaker (a marine fish belonging to Perciformes) and zebrafish (belonging to Cypriniformes) [10]. However, GFP chimeric mRNA containing medaka (belonging to Beloniformes) vasa 3'UTR did not identify PGCs in either zebrafish or loach embryos, although it did enable visualization of the PGCs in medaka embryos. Similarly, in the study, medaka PGCs can be visualized by chimeric mRNA containing vasa 3'UTR sequences from the red seabream (belonging to Perciformes). It has been demonstrated that euteleosts ( including medaka, trout, .et al) but not ostariophysans (including zebrafish, carp, .et al) vasa 3'UTRs have lost the basal function of RNA localization [23]. Considering all of those, we speculated that vasa 3'UTRs possessing the basal function of RNA localization could label a larger range of fish species than those losing the function.
In transgenic F1 fish, GFP labeling PGCs couldn't be detected until they clustered bilaterally at stage 29. However, GFP showed nonspecific expression in brain, gut and liver regions, different from the exclusive expression pattern in germ cells of the previous report by Tanaka [12]. This phenomenon may ascribe to phylogenetic distance between medaka and red seabream, as well as lack of some transcription regulatory sequences in the pPmvasGFP construct. In gonads, GFP seemed to be present in early stages of gametogenesis. GFP was abundant in oogonia and early stage oocytes, and reduced or diluted in large oocytes, mimicking the expression pattern of endogenous vasa in situ hybridization results [24-26]. Interestingly, in testes, GFP seemed to be present abundantly in spermatogonia, while little in other stages of spermatogenesis, different from the expression pattern of endogenous vasa mRNA: abundant in spermatogonia, gradually reduced in spermatocytes and absent in sperm [24-26]. By this aspect, the transgenic line showed more specific expression pattern in gonads and might facilitate the cell culture of spermatogonia.
In conclusion, we have proved that vasa transcriptional regulatory sequences from the red seabream examined in this study are sufficient for germ cells expression and labeling. This work in the model fish indicated that medaka and red seabream shared similar mechanism that specific stabilization of RNA in PGCs by vasa 3'UTR. Both pPmvasGFP construct and GFP-Pmvas 3'UTR mRNA can be applied to visualize PGCs of the red seabream, which will facilitate further research on PGCs isolation, cryopreservation, and surrogate breeding of the species.
Acknowledgements
We thank J. Deng for fish breeding and embryos collections. This work was supported by the National Natural Science Foundation of China (No.31072212 & No.41076100), Fundamental Research Project of Technology Program of Qingdao, (12-1-4-8-(7)-jch), Biomedical Research Council of Singapore (R-08-1-21-19-585) and the National Research Foundation of Singapore (R-154-000-529-281).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Yoshizaki G, Kobayashi T, Takeuchi T. Primordial germ cell: a novel tool for fish bioengineering. Fish physiol Biochem. 2003;28:453-457
2. Xu HY, Li MY, Gui JF, Hong YH. Fish germ cells. Sci China Life Sci. 2010;53:435-446
3. Okutsu T, Yano A, Nagasawa K. et al. Manipulation of Fish Germ Cell: Visualization, Cryopreservation and Transplantation. J Reprod Dev. 2006;52:685-693
4. Takeuchi Y. Generation of Live Fry from Intraperitoneally Transplanted Primordial Germ Cells in Rainbow Trout. Biol Reprod. 2003;69:1142-1149
5. Okutsu T. Testicular germ cells can colonize sexually undifferentiated embryonic gonad and produce functional eggs in fish. Proc Natl Acad Sci. 2006;103:2725-2729
6. Saito T, Goto-Kazeto R, Arai K, Yamaha E. Xenogenesis in Teleost Fish Through Generation of Germ-Line Chimeras by Single Primordial Germ Cell Transplantation. Biol Reprod. 2007;78:159-166
7. Saito T, Goto-Kazeto R, Fujimoto T. et al. Inter-species transplantation and migration of primordial germ cells in cyprinid fish. Int J Dev Biol. 2010;54:1479-1484
8. Ketting RF. A Dead End for MicroRNAs. Cell. 2007;131:1226-1227
9. Saito T, Fujimoto T, Maegawa S. et al. Visualization of primordial germ cells in vivo using GFP-nos1 3'UTR mRNA. Int J Dev Biol. 2006;50:691-699
10. Yoshizaki G. Green Fluorescent Protein Labeling of Primordial Germ Cells Using a Nontransgenic Method and Its Application for Germ Cell Transplantation in Salmonidae. Biol Reprod. 2005;73:88-93
11. Goto-Kazeto R, Saito T, Takagi M. et al. Isolation of teleost primordial germ cells using flow cytometry. Int J Dev Biol. 2010;54:1485-1490
12. Krøvel AV, Olsen LC. Expression of a vasEGFP transgene in primordial germ of the zebra fish. Mech Dev. 2002;116:141-150
13. Tanaka M. Establishment of medaka (Oryzias latipes) transgenic lines with the expression of green fluorescent protein fluorescence exclusively in germ cells: A useful model to monitor germ cells in a live vertebrate. Proc Natl Acad Sci. 2001;98:2544-2549
14. Yoshizaki G, Takeuchi Y, Sakatani S, Takeuchi T. Germ cell-specific expression of green fluorescent protein in transgenic rainbow trout under control of the rainbow trout vasa-like gene promoter. Int J Dev Biol. 2000;44:323-326
15. Kobayashi T, Yoshizaki G, Takeuchi Y, Takeuchi T. Isolation of highly pure and viable primordial germ cells from rainbow trout by GFP-dependent flow cytometry. Mol Reprod Dev. 2004;67:91-100
16. Fan L, Moon J, Wong TT. et al. Zebrafish Primordial Germ Cell Cultures Derived fromvasa::RFP Transgenic Embryos. Stem Cells Dev. 2008;17:585-598
17. Iwamatsu T, Oshima E, Sakai N. Oogenesis in the Medaka, Oryzias latipes: Stages of Oocyte Development: Developmental Biology. Zool Sci. 1996;5:873-882
18. Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 2004;121:605-618
19. Lin F, Liu QH, Li J. et al. Molecular cloning and construction of the 5' flanking region expression vector of the vasa gene of Red seabream (Pagrus major). Oceanol Limnol (Chineses Edition). 2011;42:186-192
20. Xu HY, Gui JF, Hong YH. Differential expression of vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp (Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate. Dev Dynam. 2005;233:872-882
21. Kurokawa H, Aoki Y, Nakamura S. et al. Time-lapse analysis reveals different modes of primordial germ cell migration in the medaka Oryzias latipes. Dev Growth Differ. 2006;48:209-221
22. Wolke U, Koprunner M, Raz E. Multiple Levels of Posttranscriptional Control Lead to Germ Line-Specific Gene Expression in the Zebrafish. Curr Biol. 2002;12:289-294
23. Knaut H, Steinbeisser H, Schwarz H, Nusslein-Volhard C. An Evolutionary Conserved Region in the vasa 3'UTR Targets RNA Translation to the Germ Cells in the Zebrafish. Curr Biol. 2002;12:454-466
24. Rusche L, Xu HY, Li Z. et al. Boule Is Present in Fish and Bisexually Expressed in Adult and Embryonic Germ Cells of Medaka. PLoS ONE. 2009;4:e6097
25. Liu L, Hong N, Xu HY. et al. Medaka dead end encodes a cytoplasmic protein and identifies embryonic and adult germ cells. Gene Expr Patterns. 2009;9:541-548
26. Shinomiya A, Kobayashi T, Nagahama Y, Hamaguchi S. The vasa-like gene, olvas identifies the migration path of primordial germ cells during embryonic body formation stage in the medaka, Oryzias latipes. Dev Growth Differ. 2000;42:317-326
Author contact
![]() Corresponding author: 1. Prof. Jun Li, junliac.cn Tel: +86 532 82898718; Fax: +86 532 82898718; 2. Prof. Yunhan Hong, dbshyhedu.sg Tel: +65 6516 2915; Fax: +65 6779 2486.
Corresponding author: 1. Prof. Jun Li, junliac.cn Tel: +86 532 82898718; Fax: +86 532 82898718; 2. Prof. Yunhan Hong, dbshyhedu.sg Tel: +65 6516 2915; Fax: +65 6779 2486.

 Global reach, higher impact
Global reach, higher impact