10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2013; 9(6):578-586. doi:10.7150/ijbs.6357 This issue Cite
Research Paper
Loop Nucleotide Polymorphism in a Putative miRNA Precursor Associated with Seed Length in Rice (Oryza sativa L.)
1. State Key Laboratory of Crop Genetics and Germplasm Enhancement, Nanjing Agricultural University, 210095, Nanjing, China;
2. Laboratory of Host-Pathogen Interaction, Temasek Life Sciences Laboratory, 1 Research Link, National University of Singapore, 117604, Singapore;
3. Department of Agronomy, Iowa State University, Ames, IA 50011, USA.
Received 2013-3-29; Accepted 2013-6-5; Published 2013-6-16
Abstract
The terminal loop region of primary miRNA transcripts is an important determinant controlling miRNA function in human, animals and plants. However, the effects of polymorphisms in loop region of miRNA precusors on phenotypic variation have not been reported, especially on agronomic traits in rice. From rice japonica variety Koshihikari and indica Guichao2, we isolated a precursor of putative miRNA osa-MIR2923a. We detected a GG/AA polymorphism in the loop structure from japonica and indica rice varieties, which was corresponding to japonica/indica rice differentiation. By using high-resolution melting (HRM) analysis, we measured the polymorphisms in a RIL (recombinant inbred lines) population derived from japonica variety Koshihikari and indica Guichao2. We found that the GG/AA polymorphism in the osa-MIR2923a loop was correlated to grain length and length-width ratio. We further found the significant association between seed length and GG/AA polymorphism in a population consisting of 72 rice landraces. Three targets were predicted, whose expressions showed significant differences between the two varieties. Our results suggested that the putative miRNA precursor and the three target genes could play functional roles for indica/japonica seed differentiation.
Keywords: rice, miRNA, polymorphism, seed length.
Introduction
MicroRNAs (miRNAs) are small, noncoding RNAs that can play crucial regulatory roles in eukaryotes [1]. The plant miRNAs represent an important class of endogenous small RNAs that control development and adaptation to stresses [2-4]. The terminal loop region of primary microRNA transcripts is an important determinant of microRNA biogenesis [5]. Loop nucleotides control the production of miRNAs, modulate the activity of the miRNA gene, and control miRNA function in human and animal [5, 6]. In Arabidopsis thaliana and rice, Dicer homologue Dicer-like 1 (AtDCL1 and OsDCL1) are involved in processing of terminal loop region to generate pri- and pre-miRNAs [7, 8].
However, the effects of polymorphisms in loop region of miRNA precusors on phenotypic variation have not been reported, especially on agronomic traits in rice. It is well known that miRNAs can control development and adaptation to stresses in plant, we therefore initiated to investigate the potential contributions of miRNAs to agronomic traits including yield, quality and pest/disease resistance in rice for further breeding use through marker assisted breeding. We report herein that an interesting miRNA precursor with GG/AA polymorphism in the loop structure associated with seed length, which could be useful for further breeding purpose. A flexible terminal loop region is investigated through high resolution melting (HRM) technique, which is gaining prominence as a method for discriminating DNA sequence variants. HRM analysis is performed with DNA amplicons post-PCR without additional processing and data analysis can be achieved automatically in a few minutes [9]. It has been used in BRCA1 and BRCA2 mutation detection for those who have a positive family history for breast or ovarian cancer [10]. However, it has not been commonly used in gene mutation scanning, especially miRNA gene genotyping in plant breeding.
We detected GG/AA polymorphism in the loop of osa-MIR2923a precursor through HRM analysis, and concluded that the GG/AA mutation affecting grain length of milled rice and length-width ratio. As the seed length differences between GG/AA genotypes are generally corresponding to indica/japonica differentiation, our results suggested that miRNAs could play functional roles in general japonica/indica seed type differentiation.
Results
Osa-MIR2923a identification
We isolated a putative miRNA precursor, consisting of 326nt with ΔG = -126.70 (Figure 1A). Next to the loop structure, a miRNA was found and underlined (Figure 1B), which was similar to osa-miR2923 and osa-miR2920 [11] (Figure 1C). We nominated the miRNA as osa-MIR2923a due to higher score 63 when comparing with osa-MIR2923 than osa-MIR2920.
Identification of osa-MIR2923a. A: putative miRNA precursor, consisting of 326nt with ΔG = -126.70; B: a miRNA was found and underlined next to the loop structure; C: similarity to osa-miR2923 and osa-miR2920. The miRNA was nominated as osa-MIR2923a due to higher score 63 when comparing with osa-MIR2923 than osa-MIR2920.

Sequencing analysis between indica and japonica varieties
Pre-miRNAs were isolated and sequenced from fifteen rice varieties including five japonica including Koshihikari, Nipponbare, Azucena, TP309 and Kitaki; and ten indica Guichao2, IR24, IR64, IRBB5, IRBB27, IRBB7 IR24, N22, Dular and YufengB. The polymorphism between indica/japonica was interrogated, and two-nt mutations were found, GG/AA. GG appeared in the five japonica rice, while AA in ten indica (Figure 2). The result revealed that the polymorphism in the loop structure of pri-miRNA could correlate with indica and japonica differentiation.
GG/AA genotyping through HRM
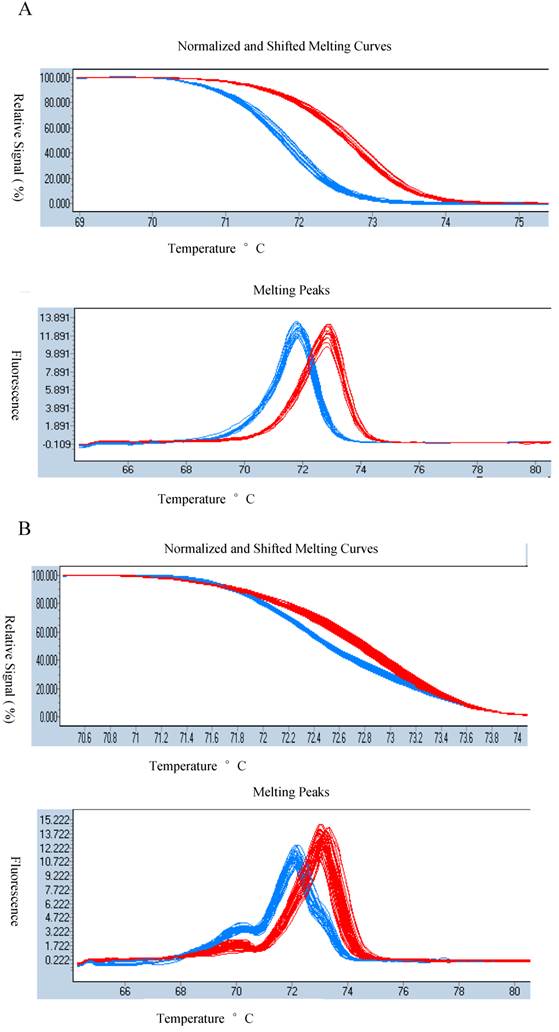
As GG/AA polymorphism was detected in the loop structure, we carried out nucleated high-resolution melting (HRM) assay for genotyping. The GG/AA polymorphism in loop structure was detected in the RIL population clearly through HRM (Figure 2A). Meanwhile we applied HRM in 72 rice landraces which could be useful for genotyping in rice germplasm identification (Figure 2B). The primer pairs were designed in loop structures (Table 1).
AA(blue)/GG(red) in loop structure of osa-MIR2923a precursors of indica/japonica landraces.

Primers for osa-MIR2923a and targets
| Primer | Sequence | Amplicon (bp) | Application |
|---|---|---|---|
| osa-MIR2923a Forward | CTTAATTTATCTATCCCCTTATTT | 310 | precursor cloning |
| osa-MIR2923a Reverse | ATGCCGTTTAAAAAGTTATCA | ||
| osa-MIR2923a Forward | GCAAATTCAAATTCAACTTATACAAGTAGA | 88 | HRM |
| osa-MIR2923a Reverse | TAACAAATTTAACCGCGAATTAAACG | ||
| Rim2 M53 Forward | GGTAATCTGCGCGGGACAA | 236 | real time PCR |
| Rim2 M53 Reverse | GGAGAGGCGTTGAAAAGTTATCAT | ||
| PPO Forward | AAGCTCCCCGCCGACGAC | 143 | real time PCR |
| PPO Reverse | CTGTGCCATGGGAAGAAGAGC | ||
| Os11g0153200 Forward | CCCCCGAGGGACAAAAATCCACT | 226 | real time PCR |
| Os11g0153200 Reverse | GAGCTCCAGCGGGTGCTTCTCC | ||
| Actin Forward | AGATGTGGATTGCCAAGGCTGAGT | 183 | endogenous reference |
| Actin Reverse | TAGGCCGGTTGAAAACTTTGTCCA |
Osa-MIR2923a affecting grain length
Through HRM analysis, we genotyped 93 individuals from a RIL population derived from indica(Guichao2)/japonica(Koshihikari) varieties.
We investigated the effects of GG/AA polymorphism in osa-MIR2923a on GL (grain length), GW (grain width), LWR (length-width ratio) and TSW (thousand-seed weight). We found that the GG/AA polymorphism in the osa-MIR2923a loop probably affected grain length and length-width ratio (Table 2), although not significant on seed width or thousand seed weight (data not shown). It was suggested that the seed length GL and LWR were highly correlated with GG/AA in loop structure of osa-MIR2923a.
Effect of osa-MIR2923a on grain traits in RIL population
| Traits | osa-MIR2923a | Mean | SE | Bonferroni test ( P<0.05) |
|---|---|---|---|---|
| GL | AA | 7.53 | 0.07 | a |
| GG | 7.30 | 0.07 | b | |
| LWR | AA | 2.26 | 0.05 | a |
| GG | 2.10 | 0.05 | b |
Significant at P<0.01 of Bonferroni test. GW and GH were not significant (data not shown).
Association between GG/AA polymorphism and seed type differentiation
We further compared the GG/AA polymorphism in 72 rice germplasm including 33 with AA genotype and 39 GG. Out of 33 with AA type, 27 were grown with indica type long seeds. In contrast, out of 39 GG, 34 were japonica short seeds. Chi-square test showed that the GG/AA polymorphism was significantly associated with seed length at P<0.001 (Table 3).
Chi-square test for association between GG/AA and seed types of japonica/indica
| AA | GG | P | |
|---|---|---|---|
| Long seed of indica type | 27 | 5 | < 0.0001 |
| Short seed of japonica type | 6 | 34 |
We then compared total nucleotide diversity (π) of the precursor between and within indica/japonica types. π was 0.01678 in the mixed population, much higher than 0.00907 and 0.00872 in the populations of indica and japonica types, thereby indicating that the two types differed at the given nucleotide site, dependent upon the identity and differentiation of the precursor. Tajima's D was positive high (2.75205), significant in the mixed population, while not significant in the two subpopulations of indica/japonica types (0.22217 and 0.24515) (Table 4). This indicated that the hypothesis of selective neutrality in the mixed population could be rejected, while selective neutrality could not be rejected in the two sub-populations. There is a high possibility that the AA/GG mutation played an important role in indica/japonica seed type differentiation.
In the 72 landraces, the grain length, width and length width ratio of the seed with indica AA type were significantly different with japonica GG. Grain height was almost the same (Table 5). This was consistent with the general fact that indica rice grain was longer than japonica. The grains with AA genotype generally showed the indica rice grain, i.e. longer and thinner with high LWR, in contrast with GG japonica, shorter and wider with low LWR.
Tajima's D statistics measure the departure from neutrality for the mutations.
| All the landraces | indica types | japonica types |
|---|---|---|
| 2.75205 | 0.22217 | 0.24515 |
| P<0.01 | P>0.10 | P>0.10 |
Association between AA/GG and seed types of the 72 landraces
| Traits | osa-MIR2923a | Mean | SE | Bonferroni test ( P<0.05) |
|---|---|---|---|---|
| GL | AA | 8.43 | 0.17 | a |
| GG | 7.49 | 0.16 | b | |
| GW | AA | 2.70 | 0.04 | a |
| GG | 3.11 | 0.03 | b | |
| GH | AA | 2.02 | 0.02 | a |
| GG | 2.09 | 0.02 | a | |
| LWR | AA | 3.16 | 0.082 | a |
| GG | 2.43 | 0.078 | b |
Significant at P<0.01 of Bonferroni test.
Expression profiles of targets
Three putative targets were predicted as following (Figure 4), TPA exp Oryza sativa transposon Rim2 M53 (BK000333.1), Polyphenol oxidase (PPO) gene (DQ532432.1) and Oryza sativa Os11g0153200 mRNA (NM_001072308.1). PPOs may also have protective roles in plants against pathogens and environmental stress in addition to a role in black/brown pigment biosynthesis [12]. The cDNA of Os11g0153200 encodes a putative carbonic anhydrase (CA; EC 4.2.1.1) catalyzes the interconversion of CO2 and HCO3-. CA is a major protein constituent of the C3 higher plant chloroplast where it is presumed to play a role in photosynthetic carbon assimilation [13].
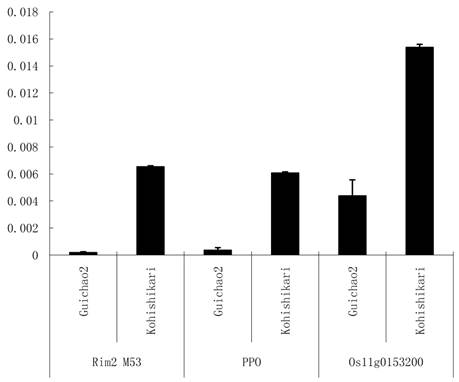
The significant differences of the three targets were detected between the two varieties (Figure 5). The primer pairs were designed for the three targets (Table 1). It was suggested that the seed traits could be affected by the three targets of osa-MIR2923a.
Difference graphs for AA(red)/GG(blue) in RIL population: melting curves (A above) and melting peaks (A down); and in indica/japonica type landraces: melting curves (B above) and melting peaks (B down).
Predicted targets of osa-MIR2923a. Target1: Oryza sativa transposon Rim2 M53 (BK000333.1); Target2: Polyphenol oxidase (PPO) gene (DQ532432.1) and Target3: Oryza sativa Os11g0153200 mRNA (NM_001072308.1). The nucleotides different among the genes are highlighted in red.

Targets expression profile in seeds of Koshihikari and Guichao2. Rim2 M53: Oryza sativa transposon Rim2 M53 (BK000333.1); PPO: Polyphenol oxidase gene (DQ532432.1) and Os11g0153200: Oryza sativa Os11g0153200 mRNA (NM_001072308.1). The Y-axis indicates the ratio of gene expression comparing to Actin gene.
Discussion
Although the terminal loop region of primary microRNA transcripts has been proved to be an important determinant of microRNA biogenesis and control miRNA function in human, animals and plants [5-8], the effects of polymorphisms in loop region of miRNA precusors on phenotypic variation have not been reported, especially on agronomic traits in rice. The grain length and width of the seed with indica AA type of osa-MIR2923a were significantly different with japonica GG, which was consistent with the general fact that indica rice grain was longer than japonica. It was suggested that miRNAs could play important roles in indica/japonica seed type differentiation through regulating the three targets. Seed size is a major determinant of crop yield and is a target trait for plant breeding. The GG/AA polymorphism could be expected to be applied in seed size improvement.
The terminal loop region of primary microRNA transcripts is an important determinant of microRNA biogenesis[5]. In rice, OsDCL1 is reported to be involved in processing of terminal loop region to generate pri- and pre-miRNAs [8]. So far no rice HYPONASTIC LEAVES1 and SERRATE homologs was identified. It will be interesting to identify the detail mechanism of osa-MIR2923a maturation and steps affected by this GG/AA mutation.
Since one of the osa-MIR2923a's targets is the mRNA of one CA which plays a role in photosynthetic carbon assimilation, it is possible that the miRNA might be involved in the source activity regulation and then the possible source-stock effect on seed size. On the other hand, Polyphenol oxidases (PPOs) are implicated in a range of biological functions in diverse systems. In addition to a role in black/brown pigment biosynthesis, PPOs may also have protective roles in plants against pathogens and environmental stress[14]. This miRNA could regulate its putative target PPO which was proved to be responsible for differentiating two subspecies[12]. Transcriptome studies have shown that large portions of the eukaryotic genomic sequences are transcribed to long noncoding RNAs (lncRNAs)[15], and miRNAs can regulate expression of the lncRNA [16]. How osa-MIR2923a affects rice growth, development or other traits through repressing the putative target genes or lncRNAs remains to be further investigated to illustrate the regulation mechanism.
For GG/AA genotyping for the pri-miRNA, dCAPs marker was developed at the GG/AA mutation site; however the dCAPs marker could not be recognized in electrophoresis due to the stem-loop structure. Meanwhile dCAPs used for the detection of sequence variants is laborious as they require post-PCR manipulation. Here we carried out genotyping the polymorphism in the loop structure through HRM approach, which has been commonly used in clinical chemistry, human pathology and patient genotyping since it was invented in 2003 [1]. It has been introduced into genotyping of crop plants including rice [17]. Its advantage is that it is performed in a real-time PCR device, and the PCR amplification and HRM analysis are performed in a closed tube, and effectively single step[18]. The simplicity, robustness and high throughput of HRM analysis is very time and cost effective to genotype thousands of plants in marker assisted breeding and germplasm screen.
In conclusion, we identified GG/AA mutation in loop structure of a putative miRNA osa-MIR2923a which could affect the expressions of its targets. The GG/AA polymorphism could be associated with seed length and indica/japonica seed type differentiation.
Experimental procedures
Plant materials
Rice landraces for sequencing included five japonica varieties, Koshihikari, Nipponbare, Azucena, TP309 and Kitaki; and ten indica, Guichao2, IR24, IR64, IRBB5, IRBB27, IRBB7 IR24, N22, Dular and YufengB. RIL population (F10) was derived from Koshihikari and Guichao2, consisting of 93 individuals. Seventy two rice landraces were used for HRM genotyping including 32 with long seed of indica type and 40 with short seed of japonica type.
DNA sequencing
Total DNA from leaves was extracted and purified using the DNeasy plant mini kit (QIAGEN, Germany). The PCR program precursor amplifications on PTC-100 PCR machines (MJ Research, CA, USA) consisted of the following steps: 94°C for 2 min followed by 37 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 2min, then a final step of 72°C for 10 min. Each PCR reaction consisted of 16 PCR buffer (Finnzymes, Espoo, Finland) with 1.5 mM MgCl2, 200 nM of each PCR primer, 50 mM of each dNTP, 10 ng genomic DNA and one unit of DNA-polymerase (Finnzymes, Espoo, Finland). PCR products were sequenced in both directions using the same primers for amplifying the genomic DNA on the ABI3730xl DNA sequencher (ABI, CA, USA) using the BigDye V3.0 kit. Sequences of three varieties generated by each primer pair were aligned using Sequencher and SNPs were identified by visual inspection.
Prediction for stem-loop structures of precursor and targets of miRNA
Secondary structures were predicted by using the mfold program (http://mfold.rit.albany.edu/?q=mfold/RNA-Folding-Form) with the default parameters[19].
The miRNA sequence was subjected to BLAST analysis against rice genome. Target predictions were performed by searching for miRNA complementary sequences (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html) with the default parameters.
PCR and HRM conditions
Samples were run in duplicate using 10 ng of genomic DNA, 250 nM of forward and reverse primers, mastermix and PCR grade water in a total reaction volume of 25 uL in LightCycler® 480 Real-Time PCR System. Amplification conditions were 95°C for 5 min followed by 40 cycles of 95°C for 15 s and 60°C for 60s. The primer sequences used are listed in Table 1. The primers were used at 500 nM final concentration.
Trait and osa-MIR2923a association analysis
An RIL population derived from japonica variety Koshihikari and indica Guichao2 was used for trait and osa-MIR2923a association analysis. Traits investigated were as following GL: grain length (mm); GW: grain width (mm); LWR: length-width ratio; TSW: thousand-seed weight (g).
Chi-square test was used for association analysis in the population of 72 landraces. DnaSP version 4.0 [20] was used to estimate total nucleotide diversity per nucleotide site (π) [21]. Tajima's D test compares nucleotide diversity with the number of segregating sites, which are expected to be equal if mutations are selectively neutral [22].
To investigate associations between phenotypic traits and genotypes (GG/AA) of osa-MIR2923a, mean phenotypic values of traits were calculated for those progeny with the alternate alleles, inherited from the Koshihikari (GG), alleles inherited from the Guichao2 (AA). A two-way ANOVA was performed on the progeny using two genotypes. This was conducted by using the general linear model (GLM) procedure of SAS (SAS Institute) and the Bonferroni method of multiple comparisons with α < 0.01.
Synthesis of cDNA and Real time PCR
To investigate expressions of the precursor and targets of osa-MIR2923a, total RNA was isolated from seed using plant RNA purification reagent (Invitrogen). Seed are maturing ones ready for harvest. Poly(A) tails were added to the 3' end of the RNAs by poly(A) polymerase (Ambion), and the polyadenylated RNAs were reverse transcribed by Super-Script II reverse transcriptase (Invitrogen) with the oligo (dT) 3'-RACE adaptor (Ambion). Real-time PCR was done in Real-Time PCR machine (I-Cycle, BioRad), with each reaction containing 200 ng of first-strand cDNAs, 0.5 μL of 10 mmol L-1 gene-specific primers, and 12.5 μL of real-time PCR SYBR MIX (iQ™ SYBR® Green Supermix, Bio-Rad). Amplification conditions were 95°C for 5 min followed by 40 cycles of 95°C for 15 s and 60°C for 60s. The rice Actin was selected as the endogenous reference. The PCR specificity is examined by 3% agarose gel using 5µl from each reaction to check the right product length and make sure no primer dimer or non-specific amplicons. The primers for real time PCR together with genomic DNA amplification were listed in Table 2.
Acknowledgements
This project is financially supported by supported by A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 010809001), and the Fundamental Research Funds for the Central Universities (KYZ201202-6).
Authors' contributions
CMW conceived of the experiments, performed the experiments, and wrote the manuscript; JMW supervised this project and revised the manuscript; JY, WJT, ZYL and MQW performed the experiments and analyzed the data; CSZ analyzed the data. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Reed GH, Kent JO, Wittwer CT. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics. 2007;8:597-608
2. Wang J W, Czech B, Weigel D. miR156-Regulated SPL Transcription Factors Define an Endogenous Flowering Pathway in Arabidopsis thaliana. Cell. 2009;138:738-49
3. Wang CM, Liu P, Sun F, Li L, Ye J, Yue GH. Isolation and Identification of miRNAs in Jatropha curcas. Int J Biol Sci. 2012;8:418-29
4. Fujii H, Chiou T-J, Lin S-I, Aung K, Zhu J-K. A miRNA Involved in Phosphate-Starvation Response in Arabidopsis. Current Biology. 2005;15:2038-43
5. Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic acids research. 2010;38:7689-97
6. Yue Sibiao, Robin Deis Trujillo Yujie Tang, William E. O'Gorman,Changzheng Chen. Loop nucleotides control primary and mature miRNA function in target recognition and repression. RNA Biology. 2011;8:1115-23
7. Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12753-8
8. Liu B, Li PC, Li X, Liu CY, Cao SY, Chu CC. et al. Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant physiology. 2005;139:296-305
9. Vossen RH, van Duijn M, Daha MR, den Dunnen JT, Roos A. High-throughput genotyping of mannose-binding lectin variants using high-resolution DNA melting analysis. Human Mutation. 2010;31:E1286-E93
10. Hondow HL, Fox SB, Mitchell G, Scott RJ, Beshay V, Wong SQ. A high-throughput protocol for mutation scanning of the BRCA1 and BRCA2 genes. BMC cancer. 2011;11:265
11. Sanan-Mishra N, Kumar V, Sopory SK, Mukherjee SK. Cloning and validation of novel miRNA from basmati rice indicates cross talk between abiotic and biotic stresses. Molecular Genetics and Genomics. 2009;282:463-74
12. Yu Y, Tang T, Qian Q, Wang Y, Yan M, Zeng D. et al. Independent losses of function in a polyphenol oxidase in rice: differentiation in grain discoloration between subspecies and the role of positive selection under domestication. The Plant Cell Online. 2008;20:2946-59
13. Ferreira FJ, Guo C, Coleman JR. Reduction of plastid-localized carbonic anhydrase activity results in reduced Arabidopsis seedling survivorship. Plant physiology. 2008;147:585-94
14. Winters A, Heywood S, Farrar K, Donnison I, Thomas A, Webb KJ. Identification of an extensive gene cluster among a family of PPOs in Trifolium pratense L.(red clover) using a large insert BAC library. BMC plant biology. 2009;9:94
15. Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J. et al. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proceedings of the National Academy of Sciences. 2012;109:2654-9
16. Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S. et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750-6
17. Tan Y-Y, Fu H-W, Zhao H-J, Lu S, Fu J-J, Li Y-F. et al. Functional molecular markers and high-resolution melting curve analysis of low phytic acid mutations for marker-assisted selection in rice. Molecular Breeding. 2012:1-12
18. Lilliebridge RA, Tong SYC, Giffard PM, Holt DC. The Utility of High-Resolution Melting Analysis of SNP Nucleated PCR Amplicons—An MLST Based Staphylococcus aureus Typing Scheme. PLoS One. 2011;6:e19749
19. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research. 2003;31:3406-15
20. Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496-7
21. Nei M. Molecular Evolutionary Genetics. Columbia: New York. 1987
22. Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585-95
Author contact
![]() Corresponding authors: Chunming Wang and Jianmin Wan.
Corresponding authors: Chunming Wang and Jianmin Wan.

 Global reach, higher impact
Global reach, higher impact