10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(1):15-24. doi:10.7150/ijbs.7490 This issue Cite
Research Paper
C1q-like Factor, a Target of miR-430, Regulates Primordial Germ Cell Development in Early Embryos of Carassius auratus
1. College of Fisheries, Key Laboratory of Freshwater Animal Breeding, Ministry of Agriculture, Freshwater Aquaculture Collaborative Innovation Center of Hubei Province, Huazhong Agricultural University, Wuhan, 430070, China.
2. State Key Laboratory of Freshwater Ecology and Biotechnology, Institute of Hydrobiology, Chinese Academy of Science, Wuhan 430072, China.
Received 2013-8-25; Accepted 2013-11-13; Published 2013-12-1
Abstract
C1q-like is a significant maternal factor of TNF/C1q super-family, and the abundant protein has been observed in both mature eggs of Carassius auratus and Carassius auratus gibelio, but its biological function in early embryo development has remained unclear. In this study, we firstly revealed a high level of maternal C1q-like transcript existence only in mature eggs of Carassius auratus, whereas no any maternal C1q-like transcript was observed in that of Carassius auratus gibelio. During embryonic development, the C1q-like zygotic expression begins around cardiopalmus stage in embryos of both Carassius auratus and Carassius auratus gibelio. Then, we examined the biological role of C1q-like by morpholino-mediated knockdown in early embryo development. Knockdown of CaOC1q resulted in a significant reduction of primordial germ cells (PGCs) in Carassius auratus, as shown by whole mount in situ hybridization with vasa-specific RNA probe, fluorescence immunostaining of vasa protein, and GFP imaging of the GFP-nanos1-3'UTR mRNA reporter. In vitro and in vivo evidence indicated that a microRNA, miR-430 could repress the C1q-like expression and PGC development. These data suggest that C1q-like should be a direct target of miR-430 and play an essential role in PGC development of Carassius auratus.
Keywords: C1q-like, microRNA, miR-430, knockdown, primordial germ cell, early embryogenesis
Introduction
Primordial germ cells (PGCs), the founder cells of both female and male gametes, initially appear outside the gonadal region, and then are specified and migrated to the genital ridges [1-3]. The first discovered molecular marker for PGCs in fish is vasa [4, 5], while several other types of factors, such as Kit ligand, FGFs, leukemia inhibitory factor (LIF) and interleukin-4 (IL4) [6-9], have been shown to stimulate PGCs proliferation and/or survival in vitro. However, it is not clear what roles these factors have in regulating PGC development in vivo. Previously, Kawase et al. observed that addition of TNF-α to culture medium stimulated proliferation of PGCs without transforming them into embryonic stem cells, and that its effect was specific for the PGCs at younger stages before and during their migration to gonads [10]. C1q-domain-containing proteins have been shown to be members of TNF/C1q super-family, and to possess diverse functions in immune response, cell activity and organ development [11-14], but their roles in PGC development have not been observed so far.
Gibel carp (Carassius auratus gibelio Bloch), one of the most important freshwater aquaculture species in China, has been extensively studied for its dual modes of unisexual and sexual reproduction [15-17]. Many key maternal factors, such as Vasa, Dazl, H2Af1o and Spindlin, have been found to play important roles in early embryonic development [18-23]. Significantly, two homologous ovary-specific C1q-like genes, CaOC1q (Carassius auratus ovary-specific C1q-like) and CagOC1q (Carassius auratus gibelio ovary-specific C1q-like), have been identified from gonochoristic Carassius auratus and gynogenetic Carassius auratus gibelio [11], respectively. They are highly conserved, and there are abundant maternal proteins to be associated with cortical granules in mature eggs [13]. As an essential maternal factor, however, its physiological role in early embryonic development is still unknown.
MicroRNAs (miRNAs) are a class of 22~24-nucleotide RNAs which usually bind to 3'UTR of target genes, and lead to target mRNA degradation or translation inhibition [24,25]. MiR-430, a well-characterized miRNA expressed at maternal-zygotic transition (MZT) stage, has been shown to target a large number of maternally expressed mRNAs and to promote their deadenylation and degradation [26, 27]. During zebrafish embryonic development, miR-430 regulates PGC development through controlling sdf1a, cxcr7, TDRD7 and nanos1 expression [28-30]. In this study, we found a significant difference of C1q-like transcriptional level in mature eggs between Carassius auratus and Carassius auratus gibelio, in which there was abundant maternal C1q-like protein in both mature eggs, but a high level of maternal C1q transcript existed only in mature eggs of Carassius auratus, whereas almost no any maternal transcript was observed in mature eggs of Carassius auratus gibelio. The finding provided a good chance to use morpholino-mediated knockdown in Carassius auratus to study physiological role of the C1q-like in early embryonic development. Thereby, we show that the C1q-like factor, which is a target of miR-430, regulates PGC development in early embryos of Carassius auratus.
Materials and methods
Materials
Carassius auratus gibelio and Carassius auratus were obtained from Guanqiao Experimental Station, Institute of Hydrobiology, Chinese Academy of Sciences. During reproduction season, the mature fish was selected and artificially induced into spawning by two intraperitoneal injections with a mixture of acetone-dried carp pituitary, HCG and LRH-A as described previously [13,22]. Experimental protocols used here were approved by the animal care and use committee of Institute of Hydrobiology, Chinese Academy of Sciences.
Virtual Northern Blot and RT-PCR analysis
Differential transcription of oocyte-specific C1q-like was analyzed by virtual Northern blotting as described previously [31]. Briefly, 1μg PCR product amplified by forward primer C1q-like-F (5'-GAGCTTGAGAAACTGAATGAC-3') and downstream primer C1q-like-R (5'-TGACTGCCATTGTTGTTCCAC-3') was labeled as the probe using DIG-High Primer system (Boehringer Mannheim). The α-tubulin control amplified by forward primer (5'-GTGCACTGGTCTTCAGGGGTT-3') and reverse primer (5'-GGGAAGTGGATGCGTGGGTAT-3') was done as the loading control. Semi-quantitative RT-PCR was carried out on 20 eggs and embryos at different stages of oogenesis and embryonic development as previously described [32]. The expression change of C1q-like regulated by miR-430 and miR-430 expression during embryo development was measured by quantitative RT-PCR as previously described [33]. α-tubulin was used as the internal control for RT-PCR. All the experiments were repeated at least three times.
Whole mount RNA in situ hybridization and antibody in situ staining
The antisense DIG-labeled RNA probes used in our experiments were generated from CaOC1q, CagOC1q and vasa mRNA sequences, and whole-mount RNA in situ hybridization was performed as described [34,35]. All the procedures were RNase-free until the hybridization finished. Finally, embryos were stained with BCIP/NBT and visualized with a microscope. The pictures were taken with a Leica camera.
Antibody immunofluorescence labeling was done as previously described using vasa specific antibody [18]. Finally, embryos were incubated with secondary FITC-labeled goat-anti-rabbit antibody in dark for 2h at room temperature and visualized with a Leica fluorescence microscope.
Morpholino microinjection
C1q-like morpholino (C1q-like MO) targeting the translation start codon of CaOC1q and control morpholino (Cont MO) were synthesized by Gene Tools, and subsequently injected into animal pole of fertilized eggs at the one-cell stage (10 ng per embryo, an optimal concentration without causing non-specific defects) as described previously [36]. The morpholino sequences were as follows: C1q-like MO, 5'-GCAGTACCATTTGTAAGACCACCAT-3'; Control MO (Cont MO), 5'-CCTCTTACCTCAGTTACAATTTATA-3'. MiR-430 mimic and control were purchased from Genepharma (Shanghai, China), and about 0.6 ng was injected in each embryo. After injection, embryos were incubated at 24.5°C in Holtfreter's medium, which was replaced by oxygen-enriched water one day later.
The full coding sequence of C1q-like was amplified and subcloned into pEGFP-N3 (provided by Dr. Qiya Zhang, Institute of Hydrobiology, Chinese Academy of Science) and pCS2+ vector (supplied by Dr. Shuo Lin, University of California, Los Angeles) to generate mammalian expression constructs. For rescue experiment, C1q-like ORF with 5-mismatch in the morpholino binding site (no change to the coding protein) was subcloned into the pCS2+ vector. The constructed vector was used for in vitro mRNA transcription to generate 5-mis rescue mRNA that could not bind to the morpholino. GFP-nanos1-3′UTR reporter was provided by Professor Erez Raz (Institute of Cell Biology, ZMBE, Germany). Capped RNAs were synthesized by SP6 RNA polymerase (Roche) with linearized plasmids, following the manufacturer's instructions. Then the RNAs were re-suspended in water and injected at a concentration of 100 ng/µL. The embryos of different development stages were detected by inverted fluorescence microscope (Leica) and photographed by CCD as described previously [37].
Luciferase reporter assay
The 3′ untranslated region (3′UTR) of CaOC1q, which contains one putative miR-430 binding site, was amplified by PCR and inserted into the SacI and XhoI sites of the pmir-GLO vector (Promega). The binding site (GTACTT) for miR-430 within the 3′UTR of the CaOC1q gene was replaced with GCTAGC to generate a mutant control for the luciferase reporter assays. Grass carp (Ctenopharyngodon idellus) ovary cells (abbreviated as CO cells) were transfected with plasmids and microRNA mimics using Lipofectamine 2000 (Invitrogen) in 48-well plates. 48h post transfection, the cells were lysed and assayed for dual reporter (Firefly and Renilla) activity using a Dual-Luciferase reporter assay system (Promega) as described [38,39]. Relative reporter activities were determined by normalizing Firefly activity to Renilla activity.
Statistical analysis
Data was shown as mean ± SD. For statistical analysis, non-parametric Mann-Whitney U tests and ANOVA tests were performed with SPSS software (SPSS Inc.). A probability (P) of <0.05 was considered statistically significant.
Results
Significant difference of maternal C1q-like transcript in mature eggs between Carassius auratus and Carassius auratus gibelio
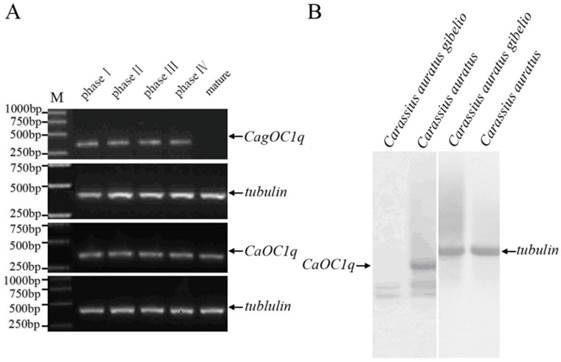
Previous studies have observed abundant maternal C1q-like protein existence during oocyte maturation and early embryo development of both Carassius auratus and Carassius auratus gibelio [11,13]. To further elucidate their transcription pattern and corresponding difference, we performed RT-PCR and virtue Northern blot analyses. As shown in Fig. 1A, high level transcription of CagOC1q and CaOC1q occurs during oogenesis and oocyte maturation from phase I to phase IV, but a significant expression difference is observed from mature eggs between Carassius auratus and Carassius auratus gibelios, in which a high level of maternal C1q transcript exists only in mature eggs of Carassius auratus, whereas almost no any maternal C1q transcript is detected in mature eggs of Carassius auratus gibelio. Virtue Northern Blot further showed a 1.2kb band in Carassius auratus mature eggs which was consistent with the molecular weight of the full length sequence of CaOC1q cDNA, while no any corresponding signal was appeared in Carassius auratus gibelio mature eggs except some non-specific signals (Fig.1B). These data indicate that a high level of maternal C1q-like transcript exists in mature eggs of Carassius auratus, whereas there is no any maternal C1q-like transcript in that of Carassius auratus gibelio.
Significant difference of maternal C1q-like transcript in mature eggs between Carassius auratus and Carassius auratus gibelio. (A) RT-PCR detection of C1q-like during oogenesis and oocyte maturation from phase I to phase IV oocytes and in mature eggs. (B) Virtue Northern Blot detection of CagOC1q and CaOC1q in mature eggs.
Zygotic expression pattern of C1q-like during embryonic development in Carassius auratus and Carassius auratus gibelio
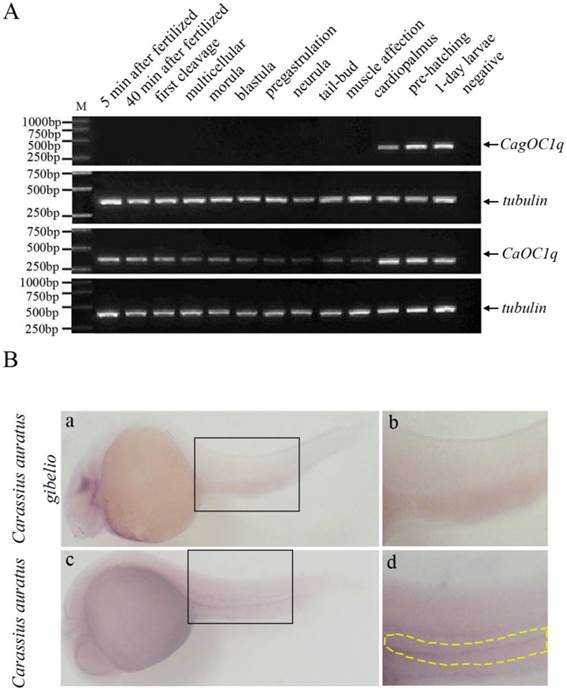
We performed RT-PCR analysis to determine the expression pattern of C1q-like during embryo development. As shown in Fig. 2A, the zygotic expression of C1q-like occurs around cardiopalmus stage during embryonic development in both Carassius auratus and Carassius auratus gibelio, but the maternal C1q-like transcript exists only in early embryos of Carassius auratus, and holds to cardiopalmus stage until the zygotic expression has begun. However, no any maternal C1q-like transcript is observed in early embryos of Carassius auratus gibelio before the zygotic expression. Whole mount in situ hybridization was also used to investigate the spatial and temporal pattern difference of ovary-specific C1q-like in early embryos between Carassius auratus and Carassius auratus gibelio. As shown in Fig. 2B, no any positive signal is observed at the corresponding region of Carassius auratus gibelio embryos at that stage (Fig.2B-a, b), whereas positive CaOC1q transcript signal is detected in the corresponding ventral genital ridge region of the embryo trunk of Carassius auratus at muscle affection stage (Fig.2B-c, d). The data implicate that the maternal ovary-specific C1q-like transcript might be localized into genital ridge region before the zygote CaOC1q expression.
(A) RT-PCR detection of C1q-like expression during embryonic development in Carassius auratus gibelio and Carassius auratus. (B) Whole mount RNA in situ hybridization of Carassius auratus gibelio (a) and Carassius auratus (c) embryos at muscle affection stage. Panels b and d show the corresponding amplification signals of genital ridge in the selected region of panels a and c. Embryos are shown laterally with anterior to the left.
C1q-like knockdown impairs PGC development in Carassius auratus
The existence of maternal C1q-like transcript in early embryos of Carassius auratus gave us a good chance to use morpholino-mediated knockdown to study functional role of the C1q-like in early embryonic development of fish. Firstly, we checked the efficacy of the designed morpholino by co-injecting morpholino with a CaOC1q expression construct fused with green fluorescent protein (GFP) into the fertilized eggs of Carassius auratus. C1q-like MO specifically reduced expression of GFP at cardiopalmus stage in 100% of embryos (n=100) (Fig. 3A). In comparison with strong GFP signal in the control group (Fig. 3A-a), there was no detectable signal when co-injected with the C1q-like morpholino (n=100) (Fig.3A-b). Finally, the inhibition efficacy of C1q-like MO was the same in Carassius auratus gibelio (n=100, data not shown). This revealed the ability of the C1q-like MO to inhibit protein production from its target sequence. Further, no obvious developmental abnormalities or mortalities were observed in either the C1q-like MO or Cont MO injected embryos (n=100, data not shown).
Knockdown of C1q-like results in defects of PGCs development in Carassius auratus. (A) Validation of knockdown efficiency of C1q-like morpholino. Embryos were injected with Cont MO (a) or C1q-like MO (b) with C1q-like-GFP fusion protein expression vector. Vasa RNA in situ hybridization (B) and antibody immunostaining (C) to identify PGCs in cardiopalmus stage embryos. (a, d) Cont MO injected embryos; (b, e) C1q-like MO injected embryos; (c, f) C1q-like MO + 5-mis mRNA injected embryos. (D) Quantification of vasa-positive PGC number in the Cont MO, C1q-like MO and C1q-like MO + 5-mis mRNA injected embryos. (E) Identification of PGCs by GFP-nanos1-3′UTR reporter in Carassius auratus.
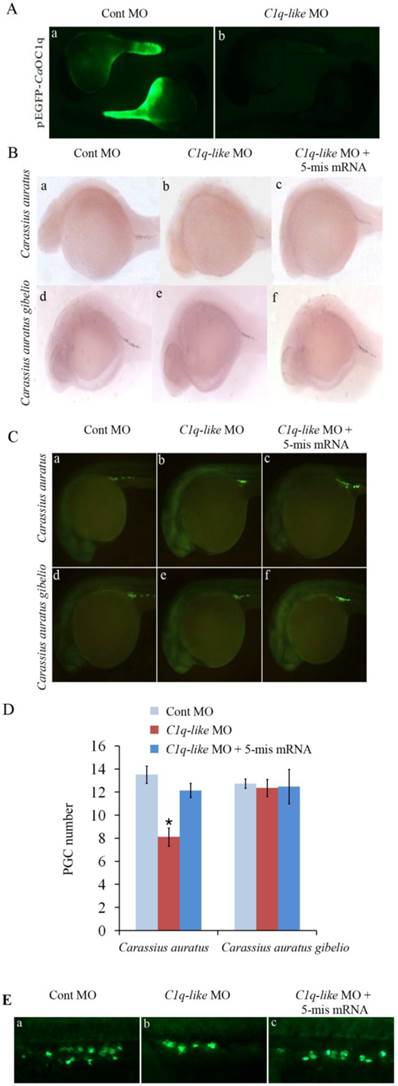
In order to analyze the function of C1q-like on PGC development during early embryogenesis, we injected C1q-like MO into the fertilized eggs of Carassius auratus and Carassius auratus gibelio, and PGCs were recognized through whole mount in situ hybridization with vasa-specific RNA probe (Fig.3B). Consistent with the absence of maternal C1q-like transcript in Carassius auratus gibelio, knockdown of CagOC1q didn't obviously change the number of PGCs (Fig.3B-d, e, f). In contrast, CaOC1q knockdown significantly reduced vasa-labeled PGCs in Carassius auratus (Fig.3B-b). Moreover, the reduction of PGC signals could be recovered by 5-mis CaOC1q mRNA (Fig.3B-c). In order to further determine the number of PGCs, they were identified and quantified by fluorescence immunostaining of vasa protein (Fig.3C). Similar to the result in Fig.3B, knockdown of C1q-like didn't obviously change the number of PGCs in Carassius auratus gibelio (Fig.3C-d, e, f and D). However, the average number of PGCs in the C1q-like MO injected embryos in Carassius auratus (Fig.3C-b and D) was 39.8% lower than that of the Cont MO injected embryos (Fig.3C-a and D). The specific effect of C1q-like MO on PGCs was confirmed through analyzing rescue of PGCs by overexpression of 5-mis CaOC1q mRNA (Fig.3C-c and D). To further address this, we injected GFP-nanos1-3′UTR mRNA with morpholino into fertilized embryo, and then imaged the fluorescence of GFP. The GFP-nanos1-3′UTR reporter further revealed that C1q-like plays an important role in PGC development in Carassius auratus (Fig.3E).
miR-430 regulates PGCs development in Carassius auratus
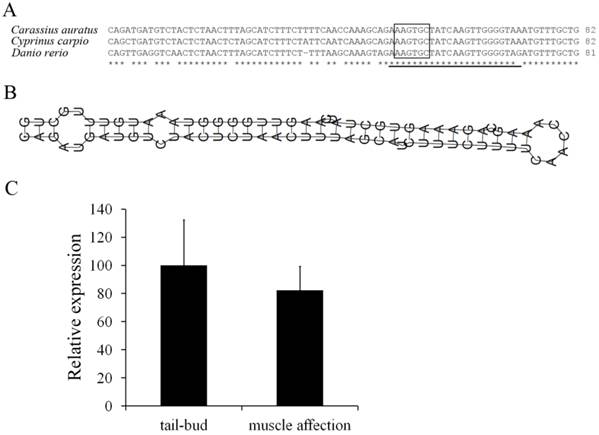
One miR-430 precursor was found from ovary transcriptome of Carassius auratus gibelio sequenced by 454 GS-FLX platform, and its sequence was found to be identical to that in Carassius auratus (data not shown). Compared with Cyprinus carpio and Danio rerio, the miR-430 precursor in Carassius auratus has very high identity to miR-430b, and the mature microRNA sequence was consistent with that in Cyprinus carpio (Fig. 4A). Moreover, the miR-430 precursor in Carassius auratus showed typical stem-loop structure (Fig. 4B) and RT-PCR analysis detected its expression in embryos at tail-bud stage and muscle affection stage (Fig. 4C).
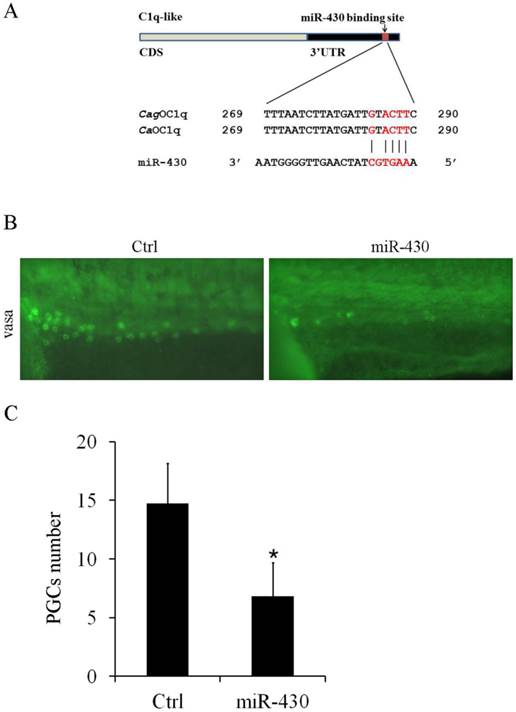
Furthermore, we observed one binding site for miR-430b in C1q-like 3'UTR, which contains only one mismatch within the seed-matched site (Fig. 5A). In teleost fish, miR-430 is essential for primordial germ cell development by targeting sdf1a, cxcr7, TDRD7 and nanos1 mRNA [28-30]. Although miR-430 was widely studied in zebrafish development, its function was unknown in Carassius auratus. Therefore, we evaluated the effect of miR-430 over-expression on PGCs development in Carassius auratus. Ectopic expression of miR-430 apparently reduced vasa-labeled PGCs in Carassius auratus compared with the control miRNA mimic, as shown by vasa antibody in situ staining (Fig.5B). The reduced level was above 50% indicated by the quantified data (Fig.5C).
Molecular characterization of miR-430 and its embryonic expression in Carassius auratus. (A) Multiple alignments of Carassius auratus, Cyprinus carpio and Danio rerio miR-430 precursors. Boxed letters represent seed sequence of miR-430 and the lined letters represent miR-430 mature sequence. (B) Stem-loop structure of miR-430 precursor in Carassius auratus. (C) Real-time PCR detection of miR-430 expression in embryos at tail-bud stage and muscle affection stage.
miR-430 regulates PGCs development in Carassius auratus. (A) Sequence alignment of C1q-like 3'UTR and miR-430. Red color letters represent seed sequence of miR-430 and its binding position. (B) Representative images of PGCs development in cardiopalmus stage embryos, as shown by vasa antibody immuostaining. (C) Quantification of PGCs numbers in the embryos injected with miR-430 mimic and its control.
CaOC1q is an important target of miR-430
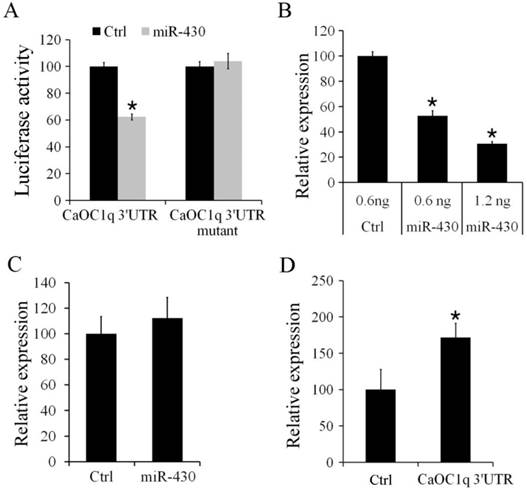
To determine the regulation of CaOC1q by miR-430, we firstly performed a luciferase reporter assay by linking the 3′UTR of CaOC1q to the C-terminus of Firefly luciferase present in pmirGLO vector. PmirGLO/3′-UTR and its mutant vectors (as described in methods) were co-transfected with miR-430 or control microRNA mimic into CO cells. Compared to control microRNA-mimics, luciferase activity was significantly reduced (up to 38%) when the PmirGLO/3′-UTR reporter was co-transfected with miR-430. In contrast, miR-430 did not change the activity of a luciferase reporter when the binding site for miR-430 in 3′UTR of CaOC1q was mutated (Fig. 6A). Furthermore, in vivo study showed that over-expression of miR-430 resulted in a significant decrease of CaOC1q expression compared with the control miRNA mimic in 30 hpf embryos (Fig. 6B). However, no difference in expression levels of CaOC1q were detected in 30 hpf embryos when CaOC1q express construct without its specific 3'UTR was co-injected with the control and miR-430 mimic (Fig. 6C). In addition, we injected pmirGLO/ CaOC1q 3′-UTR reporter at 1-cell stage, and found that CaOC1q mRNA expression was increased (Fig. 6D), for the reporter could bind to endogenous miR-430 and block CaOC1q degradation. Hence, the regulation of CaOC1q expression is specific to miR-430.
CaOC1q is a direct target of miR-430. (A) miR-430 suppressed the activity of pmirGLO reporter that is linked to the 3'UTR of CaOC1q, but not mutant 3'UTR of CaOC1q. The Firefly activity was normalized to Renilla expression, which was used as a control for transfection efficiency. (B) Over-expression of miR-430 reduced endogenous CaOC1q mRNA transcripts, as determined by qRT-PCR. (C) miR-430 had no effect on ectopic CaOC1q expression when embryos were injected with CaOC1q express construct without its specific 3'UTR. (D) CaOC1q mRNA expression was increased when embryos were injected with reporter which ectopically expressed CaOC1q 3′-UTR.
Discussion
In recent years, TNF family members have been reported to be involved in the process of oogenesis and embryogenesis, including primordial germ cell development [10, 40, 41]. As an important member of TNF/C1q super-family reported in fishes, C1q-like has been studied in oogenesis and oocyte maturation in Carassius auratus and Carassius auratus gibelio [11, 13]. In this study, we revealed the different expression pattern of C1q-like between Carassius auratus and Carassius auratus gibelio. Furthermore, we have identified and characterized C1q-like as a direct target of miR-430, and investigated its function on PGC development through the morpholino-mediated knockdown.
In this study, we have observed that C1q-like has basically consistent transcription pattern from stage I oocytes to stage IV oocytes in both species, but a high level of CaOC1q transcript exists only in mature eggs of Carassius auratus, whereas there is no any maternal CagOC1q mRNA in mature eggs of Carassius auratus gibelio (Fig.1). The observation implicates that maternal CagOC1q mRNA may be predominantly expressed by follicle cells and gone along with the follicular cell degeneration and deletion [13]. However, CaOC1q maternal mRNA is expressed by oocytes and localized in the cytoplasm, and kept its high transcription level during follicular cell apoptosis [11]. RT-PCR analysis indicated that the zygotic transcription started around cardiopalmus (Fig. 2). In addition, knockdown of CagOC1q has no effect on PGC development of early embryo (Fig. 3). These findings suggest that maternal CagOC1q protein should be sufficient for normal early embryo development in Carassius auratus gibelio. Previously, many maternal factors to control early embryo development had been identified by maternal-effect mutant screening in zebrafish [42]. And, some maternal factors had been demonstrated to be required for zebrafish PGCs formation [43, 44]. A unique finding of the current study was that the maternal CaOC1q mRNA might be involved in PGC development, because the knockdown of CaOC1q resulted in significant decrease of PGC number, implicating that the maternal CaOC1q mRNA translation might be necessary for maintaining sufficient level of CaOC1q protein to keep normal PGC development. Moreover, MiR-430, which was known to control normal PGC development [26-30], was also revealed to directly target CaOC1q and guide its expression at an optimal level in the early embryo development of Carassius auratus. Furthermore, several other maternal factors involved in oogenesis and early embryo development of Carassius auratus gibelio, such as Cyclin A2, Dazl and SNRPC [19, 45, 46], were also predicted to be targeted by miR-430 (data not shown). In future, it would be interesting to elucidate the relationship between miR-430 and maternal mRNA in Carassius auratus gibelio and Carassius auratus.
Recent studies have revealed several signaling pathways that regulate the proliferation, survival and migration of fish PGCs. For example, chemokine CXCL12 was found to regulate directional migration of PGCs through the G-protein-coupled receptor CXCR4 [47, 48]. IGF1Rb was shown to potentially regulate PGC development via cross-talk with chemokine signaling pathways [49]. In our previous work, zebrafish C1q-like was also demonstrated to be required for cell survival during embryonic development [14], and the sequence identity between zebrafish C1q-like and CagOC1q was relatively high (~70%). Our preliminary analysis indicated that in vitro over-expression of CagOC1q was able to protect FHM (fathead minnow) cells from ACTD induced apoptosis (data not shown). Further research will be undertaken to explore whether CaOC1q is essential for the cell survival of PGCs in vivo, and how C1q-like interacts with other molecular signals to control early embryo development.
Acknowledgements
This work was supported by grants to JF Gui from the National Key Basic Research Program (2010CB126301), the earmarked fund for Modern Agro-industry Technology Research System (NYCYTX-49), the Innovation Project of Chinese Academy of Sciences (KSCX3-EW-N-04), the Autonomous Project of the State Key Laboratory of Freshwater Ecology and Biotechnology (2011FBZ17), and by grant to J Mei from the Fundamental Research Funds for the Central Universities (52204-12018, 2013PY068), and the National Natural Science Foundation of China (31301931). Many thanks to Professor Erez Raz (Institute of Cell Biology, ZMBE, Germany) for kindly providing GFP-nanos1-3′UTR reporter.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Sawatari E, Shikina S, Takeuchi T, Yoshizaki G. A novel transforming growth factor-beta superfamily member expressed in gonadal somatic cells enhances primordial germ cell and spermatogonial proliferation in rainbow trout (Oncorhynchus mykiss). Dev Biol. 2007;301:266-75
2. Raz E. Guidance of primordial germ cell migration. Curr Opin Cell Biol. 2004;16:169-73
3. Xu H, Li M, Gui J, Hong Y. Fish germ cells. Sci China Life Sci. 2010;53:435-46
4. Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157-65
5. Yi M, Hong N, Li Z, Yan Y, Wang D, Zhao H. et al. Medaka fish stem cells and their applications. Sci China Life Sci. 2010;53:426-34
6. Cooke JE, Heasman J, Wylie CC. The role of interleukin-4 in the regulation of mouse primordial germ cell numbers. Dev Biol. 1996;174:14-21
7. Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841-7
8. Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550-1
9. Ross A, Munger S, Capel B. Bmp7 regulates germ cell proliferation in mouse fetal gonads. Sex Dev. 2007;1:127-37
10. Kawase E, Yamamoto H, Hashimoto K, Nakatsuji N. Tumor necrosis factor-alpha (TNF-alpha) stimulates proliferation of mouse primordial germ cells in culture. Dev Biol. 1994;161:91-5
11. Chen B, Gui J. Identification of a novel C1q family member in color crucian carp (Carassius auratus) ovary. Comp Biochem Physiol B Biochem Mol Biol. 2004;138:285-93
12. Mei J, Gui J. Bioinformatic identification of genes encoding C1q-domain-containing proteins in zebrafish. J Genet Genomics. 2008;35:17-24
13. Mei J, Chen B, Yue H, Gui JF. Identification of a C1q family member associated with cortical granules and follicular cell apoptosis in Carassius auratus gibelio. Mol Cell Endocrinol. 2008;289:67-76
14. Mei J, Zhang QY, Li Z, Lin S, Gui JF. C1q-like inhibits p53-mediated apoptosis and controls normal hematopoiesis during zebrafish embryogenesis. Dev Biol. 2008;319:273-84
15. Gui J, Zhou L. Genetic basis and breeding application of clonal diversity and dual reproduction modes in polyploid Carassius auratus gibelio. Sci China Life Sci. 2010;53:409-15
16. Jiang FF, Wang ZW, Zhou L, Jiang L, Zhang XJ, Apalikova OV. et al. High male incidence and evolutionary implications of triploid form in northeast Asia Carassius auratus complex. Mol Phylogenet Evol. 2013;66:350-9
17. Gui J, Zhu Z. Molecular basis and genetic improvement of economically important traits in aquaculture animals. Chin Sci Bull. 2012;57:1751-60
18. Xu H, Gui J, Hong Y. Differential expression of vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp (Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate. Dev Dyn. 2005;233:872-82
19. Peng JX, Xie JL, Zhou L, Hong YH, Gui JF. Evolutionary conservation of Dazl genomic organization and its continuous and dynamic distribution throughout germline development in gynogenetic gibel carp. J Exp Zool B Mol Dev Evol. 2009;312:855-71
20. Wu N, Yue HM, Chen B, Gui JF. Histone H2A has a novel variant in fish oocytes. Biol Reprod. 2009;81:275-83
21. Sun M, Li Z, Gui JF. Dynamic distribution of spindlin in nucleoli, nucleoplasm and spindle from primary oocytes to mature eggs and its critical function for oocyte-to-embryo transition in gibel carp. J Exp Zool A Ecol Genet Physiol. 2010;313:461-73
22. Xie J, Wen JJ, Chen B, Gui JF. Differential gene expression in fully-grown oocytes between gynogenetic and gonochoristic crucian carps. Gene. 2001;271:109-16
23. Yue HM, Li Z, Wu N, Liu Z, Wang Y, Gui JF. Oocyte-Specific H2A Variant H2af1o Is Required for Cell Synchrony Before Mid-Blastula Transition in Early Zebrafish Embryos. Biol Reprod. 2013;89(4):82
24. Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol. 2010;17:1169-74
25. Zhao S, Liu MF. Mechanisms of microRNA-mediated gene regulation. Sci China C Life Sci. 2009;52:1111-6
26. Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K. et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75-9
27. Svoboda P, Flemr M. The role of miRNAs and endogenous siRNAs in maternal-to-zygotic reprogramming and the establishment of pluripotency. EMBO Rep. 2010;11:590-7
28. Staton AA, Knaut H, Giraldez AJ. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat Genet. 2011;43:204-11
29. Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4:e7513
30. Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF. et al. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135-42
31. Zhang YB, Hu CY, Zhang J, Huang GP, Wei LH, Zhang QY. et al. Molecular cloning and characterization of crucian carp (Carassius auratus L.) interferon regulatory factor 7. Fish Shellfish Immunol. 2003;15:453-66
32. Xia W, Zhou L, Yao B, Li CJ, Gui JF. Differential and spermatogenic cell-specific expression of DMRT1 during sex reversal in protogynous hermaphroditic groupers. Mol Cell Endocrinol. 2007;263:156-72
33. Mei J, Bachoo R, Zhang CL. MicroRNA-146a inhibits glioma development by targeting Notch1. Mol Cell Biol. 2011;31:3584-92
34. Liu J, Hu B, Wang Y, Gui JF, Xiao W. Zebrafish eaf1 and eaf2/u19 mediate effective convergence and extension movements through the maintenance of wnt11 and wnt5 expression. J Biol Chem. 2009;284(24):16679-16692
35. Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature protocols. 2008;3:59-69
36. Wang Y, Zhou L, Li Z, Li WH, Gui JF. Apolipoprotein C1 regulates epiboly during gastrulation in zebrafish. Sci China Life Sci. 2013;56(11):975-84
37. Xu S, Xia W, Zohar Y, Gui JF. Zebrafish dmrta2 regulates the expression of cdkn2c in spermatogenesis in the adult testis. Biol Reprod. 2013;88:14
38. Huang W, Zhou L, Li Z, Gui JF. Expression pattern, cellular localization and promoter activity analysis of ovarian aromatase (Cyp19a1a) in protogynous hermaphrodite red-spotted grouper. Mol Cell Endocrinol. 2009;307:224-36
39. Zhu R, Zhang YB, Zhang QY, Gui JF. Functional domains and the antiviral effect of the double-stranded RNA-dependent protein kinase PKR from Paralichthys olivaceus. J Virol. 2008;82:6889-901
40. Kishore U, Gaboriaud C, Waters P, Shrive AK, Greenhough TJ, Reid KB. et al. C1q and tumor necrosis factor superfamily: modularity and versatility. Trends Immunol. 2004;25:551-61
41. Bornstein SR, Rutkowski H, Vrezas I. Cytokines and steroidogenesis. Mol Cell Endocrinol. 2004;215:135-41
42. Abrams EW, Mullins MC. Early zebrafish development: it's in the maternal genes. Curr Opin Genet Dev. 2009;19:396-403
43. Hashimoto Y, Maegawa S, Nagai T, Yamaha E, Suzuki H, Yasuda K. et al. Localized maternal factors are required for zebrafish germ cell formation. Dev Biol. 2004;268:152-61
44. Koprunner M, Thisse C, Thisse B, Raz E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001;15:2877-85
45. Xie J, Wen JJ, Yang ZA, Wang HY, Gui JF. Cyclin A2 is differentially expressed during oocyte maturation between gynogenetic silver crucian carp and gonochoristic color crucian carp. J Exp Zool A Comp Exp Biol. 2003;295:1-16
46. Wang HY, Zhou L, Gui JF. Identification of a putative oocyte-specific small nuclear ribonucleoprotein polypeptide C in gibel carp. Comp Biochem Physiol B Biochem Mol Biol. 2007;146:47-52
47. Knaut H, Werz C, Geisler R, Nusslein-Volhard C. A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature. 2003;421:279-82
48. Doitsidou M, Reichman-Fried M, Stebler J, Koprunner M, Dorries J, Meyer D. et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647-59
49. Schlueter PJ, Sang X, Duan C, Wood AW. Insulin-like growth factor receptor 1b is required for zebrafish primordial germ cell migration and survival. Dev Biol. 2007;305:377-87
Author contact
![]() Corresponding author: jfguiac.cn and jmeihzau.edu.cn
Corresponding author: jfguiac.cn and jmeihzau.edu.cn

 Global reach, higher impact
Global reach, higher impact