10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2014; 10(2):225-235. doi:10.7150/ijbs.6797 This issue Cite
Research Paper
Vasa Identifies Germ Cells and Critical Stages of Oogenesis in the Asian Seabass
Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543.
Received 2013-5-30; Accepted 2013-10-15; Published 2014-2-6
Abstract
Germ cells produce sperm and eggs for reproduction and fertility. The Asian seabass (Lates calcarifer), a protandrous marine fish, undergoes male-female sex reversal and thus offers an excellent model to study the role of germ cells in sex differentiation and sex reversal. Here we report the cloning and expression of vasa as a first germ cell marker in this organism. A 2241-bp cDNA was cloned by PCR using degenerate primers of conserved sequences and gene-specific primers. This cDNA contains a polyadenylation signal and a full open reading frame for 645 amino acid residues, which was designated as Lcvasa for the seabass vasa, as its predicted protein is homologous to Vasa proteins. The Lcvasa RNA is maternally supplied and specific to gonads in adulthood. By chromogenic and fluorescent in situ hybridization we revealed germ cell-specific Lcvasa expression in both the testis and ovary. Importantly, Lcvasa shows dynamic patterns of temporospatial expression and subcellular distribution during gametogenesis. At different stages of oogenesis, for example, Lcvasa undergoes nuclear-cytoplasmic redistribution and becomes concentrated preferentially in the Balbiani body of stage-II~III oocytes. Thus, the vasa RNA identifies both female and male germ cells in the Asian seabass, and its expression and distribution delineate critical stages of gametogenesis.
Keywords: Asian seabass, gametogenesis, gene expression, germ cell, oogenesis, sex reversal, vasa.
Introduction
Germ cell development is the basis of sexual reproduction and fertility. In diverse animal species, primordial germ cells (PGCs) segregate from the soma early in embryogenesis. PGCs migrate into the developing gonad and become gonadal germ stem cells, oogonia in the ovary and spermatogonia in the testis [1]. At sexual maturation, germ stem cells undergo self-renewal and differentiation, meiosis entry and progression, culminating in the production of eggs in female and sperm in male [2]. The mechanism underlying how a germ cell makes the decision to undergo either egg or sperm production has long been a mystery.
Sex development involves sex determination and differentiation. Genetically, maleness in mammals is determined by gene Sry [3], and in the fish medaka by gene dmy [4] also called dmrt1y [5]. Although the final decision for sperm or egg production is ultimately made in and by germ cells, most studies on sex differentiation have so far focused on the gonadal somatic sex [6], Formation of testis-triggering Sertoli cells or ovary-triggering granulose or follicular cells is considered as determining the gonadal sex, which in turn determines the organism sex and germ cell sexuality [6]. It is generally accepted that a sexually bipotential germ cell pool makes the decision absolutely depending upon the gonadal sex. In fish, the sexual bipotentiality or plasticity of germ stem cells has been demonstrated by transplantation experiments [7]. Accumulating data argue for or against the involvement of germ cells in sex differentiation and reversal. A conserved interplay between germ cells and gonadal sex has indeed been reported in cattle, zebrafish and medaka [8], where animals with germ cells usually develop into two opposite sexes, while those without germ cells produce only one sex, male. We have recently revealed differential expression of germ genes boule and dazl in male and female gonial cells of the rainbow trout, pointing to germ cell sex prior to meiosis and possibly a role for germ cells in sex differentiation [9, 10].
In higher vertebrates including mammals and birds, stable sexual dimorphism prevails, because maleness or femaleness, once established, is usually retained throughout lifespan. In many invertebrates and lower vertebrates such as fish, hermaphroditism is commonplace, with one organism developing into both sexes. Simultaneous hermaphrodites have both the ovary and testis and thus produce eggs and sperm at the same time, such as Rivulus marmoratus [11]. Sequential hermaphrodites produce sperm and eggs or vice versa in different times. Many marine teleosts, in particular those of the order Perciformes, are either protandrous hermaphrodites (male first) or protogynous hermaphrodites (female first). In these hermaphroditic organisms, sex development also involves sex reversal. One of such protandrous perches is the Asian seabass (Lates calcarifer), which undergoes male-to-female sex reversal under natural conditions: It matures as male and changes to female [12]. In this organism, sex reversal accompanies major morphological changes of the gonad but lacks an intermediate gonadal status of ovotestis, and testicular and ovarian tissues cannot be found in one and the same gonad [13], Therefore, the seabass represents an excellent model to study the role and behavior of germ cells not only in sex determination and differentiation but also in sex reversal.
To study germ cell development, a germ cell marker is required. One of the best candidate genes is vasa, which was first identified in Drosophila by genetic screens for maternal-effect mutations. vasa is essential for the anterior-posterior polarity and germ cell formation [14]. Vasa is an RNA helicase of the DEAD box family and resides in the cytoplasm for involvement in translational control. The Drosophila vasa homolog has been identified in the hydra [15], silkworm [16], grasshopper [17], oyster [18], Xenopus [19], chicken [20], mouse [19], human [21] and several teleost species, such as zebrafish [22, 23], trout [24], tilapia [25], medaka [26], gibel carp [27] and grass carp [28]. vasa is a germ cell marker in many species examined so far, such as zebrafish [22, 23, 29], trout [24], tilapia [25], medaka [26], Gymnogobius [30], gibel carp [27] and rare minnow [31]. In the fish species examined in detail by in situ hybridization (ISH), vasa expression occurs in both mitotic and meiotic germ cells of both sexes [27, 32]. A similar observation has been described also for other germ genes such as boule and dazl in medaka [32] and dazl in trout [9]. One exception is boule, whose expression takes place in male gonia but not in female gonia [9]. In medaka, vasa has been reported essential for PGC migration [33].
vasa has been cloned in two marine teleosts . In the gilthead seabream (Sparus aurata), vasa was examined by Northern blot analysis and chromogenic ISH, where vasa was present in the ovary, where vasa expression was prominent in vitellogenic oocytes but barely detectable in oogonia and previtellogenic oocytes [34]. In the European seabass (Dicentrarchus labrax), RT-PCR analyses revealed a high level of vasa expression in the ovary and testis as well as a remarkably reduced level of expression in 10 somatic organs [35]. A marker capable of detecting germ cells at different stages of development in both sexes of marine teleosts has remained to be established by ISH and/or immunohistochemistry
In this study, we cloned Lcvasa, the gene encoding LcVasa, the seabass homolog of highly conserved Vasa proteins, as a first germ cell marker in the seabass. By both chromogenic and fluorescent ISH, Lcvasa expression delineates germ cells throughout gametogenesis of both sexes. We reveal for the first time that Lcvasa exhibits highly dynamic expression and subcellular distribution, which allows for identification of critical stages of gametogenesis.
Materials and Methods
Fish
Experiments with fish were conducted strictly following the Guide for the Care and Use of Laboratory Animals of the National Advisory Committee for Laboratory Animal Research in Singapore and approved by this committee (Permit Number:067/12). The seabass and its embryos were collected from a local fish farmer. The embryos were maintained in aerated seawater at ambient temperature. Seabass embryogenesis proceeds rapidly and hatching takes place before 24 h post fertilization (hpf). Embryos were collected at an interval of 2 h upon first cleavage till the late somitogenesis for RT-PCR analysis, and staged as previously described [36]. The maturity of seabass gonads was classified as described [13]. In this study, the testis was at stage M3 (Male stage 2 according to [13] Table 1) and contains numerous cysts of sperm and spermatids, fewer spermatocytes and sparely dispersed spermatogonia. The ovary was at stage F2 (Female stage 2 according to [13] Table 1) and composed mainly of oogonia, previtellogenic and early vitellogenic oocytes.
Total RNA extraction
Total RNA from adult organs (100 mg) and embryos (20~30) at different stages of development (2~30 h post fertilization) were isolated with the Trizol Reagent (Invitrogen). RNA samples were treated with the RNase-free DNase (Promega) to eliminate genomic DNA contamination.
Isolation of cDNA sequence
A cDNA library was synthesized from 3 μg of testicular RNA by using the RACE cDNA Amplification Kit (BD BioSciences). A cDNA fragment of ~750 bp was amplified from the cDNA libraries with degenerate primers corresponding to the conserved amino acid sequences MDDWEEE and MACAQTG. The sequence of this fragment was used to design a gene-specific primer covering the initiation codon (ATGGACGACTGGGAGGAAGAGACTGC, broken underline in Figure 1) for 3'-RACE in combination with the SMART library 3' adapter primer (5'-AAGCAGTGGTAACAACGCAGAGTAC(T)30 as described [27]. The 3'-RACE product was cloned into pGEM-T. Recombinant plasmids in three colonies were found to contain one and the same insert of 2241 bp as analyzed by sequencing, and thus named pLvasa. Similarly, an 854-bp fragment was amplified from the cDNA library by using a pair of primers (defined by solid underline in Figure 1) and cloned, resulting in pLvasa854. Correct cloning was analyzed by test digestion and/or sequencing (see below).
Nucleotide sequence and its deduced protein of Lcvasa cDNA. Shown in bold are the translation start codon, stop codon and putative polyadenylation signal. Underlined are RGG repeats. Primers for cloning and 3'-RACE (dash), RT-PCR analysis and riboprobe (solid) are underlined with arrows depicting their extension directions.

Sequence analysis
Sequences were determined by using the BigDye Terminator v3.1 Cycle Sequencing Kit on ABI PrismR 3100 Genetic Analyzer (Applied Biosystems). Sequence alignment was run on the Vector NTI Suite 11 (Invitrogen). Phylogenetic trees were constructed by using the neighbor-joining algorithm on the DNAMAN package (Lynmon Biosoft).
RT-PCR analysis
cDNA synthesis was primed by oligo(dT)12-18 anchored as primer by using the Powerscript TM Reverse Transcriptase kit (Clontech, USA). Lcvasa was amplified by using cDNA-specific primers RT1 (GACCAGCAACCAGACCTCACCTTCAC) and RT2 (GAGTATGGCTGATTGTAGGGGCTGC) spanning a 431-bp cDNA fragment. As a control, an 800-bp fragment of the β-actin cDNA was amplified by using primers MA1 and MA2 (TGGAGCCTCCAATCCAGACAGAGTATT). PCR was run for 35 cycles for Lcvasa and 25 cycles for β-actin at 94°C for 10 sec, 60°C for 20 sec and 72°C for 60 sec. PCR products were analyzed by agarose gel electrophoresis as described [32].
In situ hybridization
In situ hybridization on whole mount sample and sections were performed as described [27, 32] with some minor modifications. Adult gonads were dissected and cut into small pieces, fixed in 4% paraformaldehyde in 0.1 M phosphate buffer (PBS, pH 7.4) (4% PFA-PBS) at 4°C overnight, followed by buffer changes with 0.5 M sucrose in PBS at 4°C overnight. The samples were embedded in Optimal Cutting Temperature and cryosectioned at 8 μm for ovaries and 5 μm for testes on the Leica RM2135 Microtomes (Leica, Germany). The cryosections were mounted on frost glass slides (Fishery, USA) and stored at -80°C before use. pLcvasa854 containing the 854-bp Lcvasa cDNA fragment was linearized with Apa I and Sac II for the synthesis of sense and anti-sense riboprobes from Sp6 or T7 promoter by using the digoxigenin (DIG) or FITC RNA Labeling Kit (Roche). The probes were treated with RNase-free TURBO DNase (Ambion) and purified. Chromogenic and fluorescent in situ hybridization (FISH) was carried as described [32]. After extensive washes in PBS in darkness, the slides were stained for nuclei with DAPI (1 µg/ml) for 10 min at room temperature, washed three times in PBS and mounted for microscopy.
Microscopy and Photography
Microscopic analyses were performed as previously described [27, 32]. Briefly, whole mount samples were observed in a large field using a Stereo- Fluorescence Systems (Leica MZFIII, Switzerland) and photographed. Cryosections were visualized under a Zeiss Axiovert upright microscope and photographed using a Zeiss AxioCam MRc digital camera.
Results
Isolation of seabass vasa cDNA
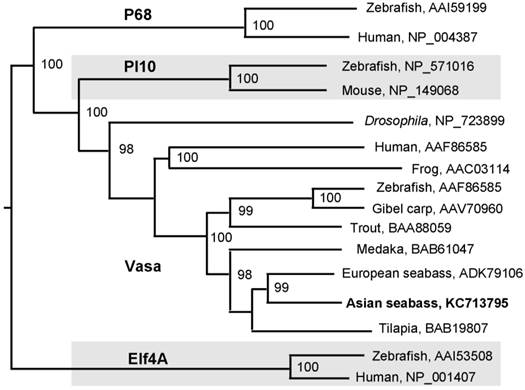
A 776-bp cDNA fragment was first obtained by using degenerate primers, which were designed on the basis of two conserved regions of vertebrate Vasa proteins. The sequence information from this fragment was used for 3'-RACE, leading to the cloning of a 2241 bp cDNA, designated as Lcvasa for the seabass gene encoding the Vasa homolog LcVasa. Lcvasa contains an open reading frame (ORF) of 1938 nt for 645 amino acid (aa) residues, a polyA signal and a 3'-untranslated region (UTR) of 302 bp (FIG.1). A blast search against databases revealed a maximal identity of LcVasa to known Vasa proteins. It is 65-83% and 52% identical to Vasa proteins from teleost fish and human respectively (FIG.2). Like Vasa homologs, LcVasa is characterized by an RGG-rich N-terminus and eight diagnostic motifs, namely AQTGSGKT as the ATPase motif A, PTRELA, GG, TPGR and DEAD as the ATPase motif B, SAT and ARGXD (F for any residue) for RNA unwinding, and HRIGRTGR for RNA binding. The RGG repeats at the N-terminus possibly functions in RNA binding [37] and in subcellular protein localization [38]. These eight motifs are highly conserved between LcVasa and representative Vasa proteins from invertebrates and vertebrates on a sequence alignment (FIG. 2). On a phylogenetic tree (FIG. 3), LcVasa is clustered together with Vasa but not its related family members pl10, Elf4 and p68. Therefore, LcVasa appears to be a Vasa homolog of the seabass.
Gonad- and embryo-specific expression of vasa RNA
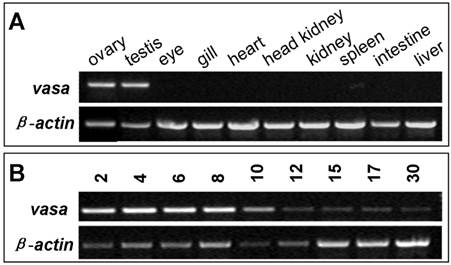
In many organisms including zebrafish, medaka and gibel carp, vasa RNA is supplied maternally and zygotic expression is restricted to germ cells. In the seabass, adult expression of vasa RNA was examined by RT-PCR to be high in the ovary and testis, but absent in any somatic organs examined, such as the eye, gill, heart, kidney, spleen, intestine and liver (FIG. 4A). Therefore, vasa RNA expression is restricted to the gonads. During embryogenesis, the vasa transcript is high in early stages, while persists at a reduced and detectable level in late stages till 30 hpf (FIG. 4B). Since the onset of zygotic transcription in fish occurs after midblastula transition, the vasa transcript is maternally inherited in seabass.
Sequence alignment of Vasa proteins. All the sequences of Vasa homologues were retrieved from NCBI (http://www-ncbi-nlm-nih-gov.libproxy1.nus.edu.sg/protein/?term=Vasa ). Positions of identical residues are highlighted yellow, those of shared residues blue and those of different residues green. Protein numbering is shown to the left. RGG repeats at the N-terminus are underlined. The eight conserved domains are framed. Degenerate primers are positioned by two arrows. The species' names and percentage identity values of the LcVasa to its homologs are given at the end of alignment. For accession numbers of sequences see FIG. 3.

Phylogenetic tree of Vasa proteins. All the sequences of Vasa homologues were retrieved from NCBI (http://www-ncbi-nlm-nih-gov.libproxy1.nus.edu.sg/protein/?term=Vasa). The neighbor-joining algorithm was used for tree construction. Numerals at nodes are bootstrap values. Species are followed by accession numbers of sequences.
RT-PCR analysis of vasa expression in seabass. (A) Adult tissues. (B) Embryos. Stages given above lanes are hours post fertilization (hpf). β-actin serves as a normalization factor.
Germ cell-specific expression of the vasa RNA in the testis
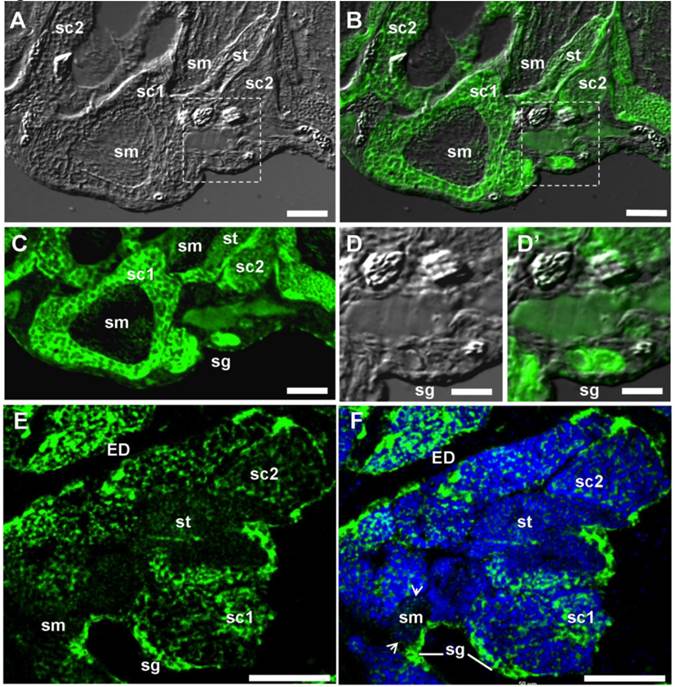
Restriction of the vasa transcript to the gonads suggests its germ cell-specific expression. To confirm this, spatial expression was analyzed by ISH in adult gonads. The adult seabass testis is mainly composed of spermatogonia and spermatogenic cells at different stages. Spermatogonia are at the most peripheral region of seminiferous cysts. Spermatogenesis proceeds synchronously within each cyst and cysts containing germ cells at progressively advanced stages of development are closer to the efferent duct. Chromogenic ISH revealed that vasa RNA expression in the testis was exclusively in germ cells, with the signal being strong in spermatogonia and spermatocytes, reduced in spermatids and barely detectable in sperm (Supplementary Material: FIG. S1). By FISH, the hybridization signal was found also strong in spermatogonia and spermatocytes, and weak in spermatids and sperm (FIG. 5A, B, C, E and F). Interestingly, the vasa RNA are distributed not only in the cytoplasm but also in the nucleus (FIG. 5D and F). At higher magnification, Lcvasa was detected in the chromatoid body (CB) of maturing sperm (FIG. 5F). Therefore, Lcvasa expression in the adult testis is limited to germ cells.
Germ cell-specific expression of the vasa RNA in the ovary
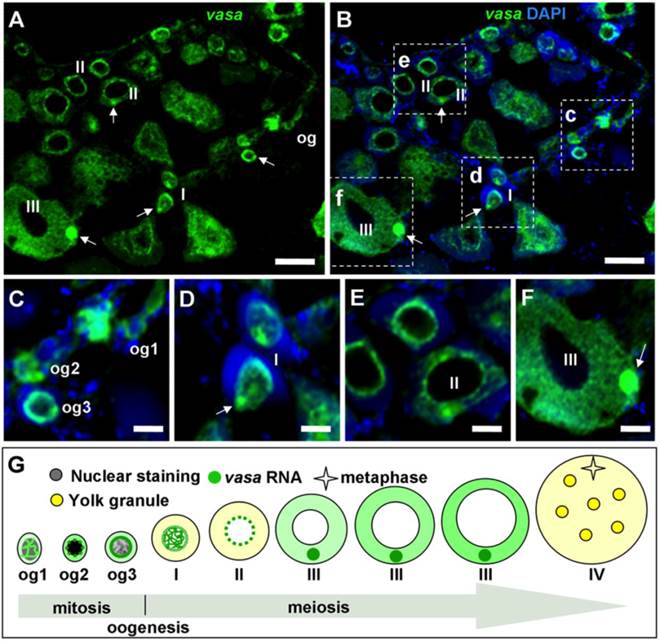
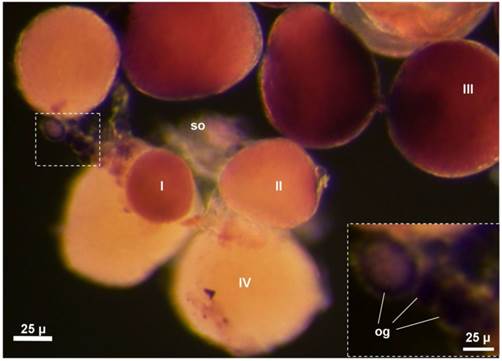
In the seabass, the adult ovary is composed of somatic follicle cells, a small number of oogonia and numerous oocytes at various stages of development. Chromogenic ISH with an antisense vasa riboprobe revealed that vasa expression in the adult ovary was limited to female germ cells but absent in somatic cells on whole mount samples, where stage-III oocytes and certain oogonia displayed the strongest signal, whereas stage-IV oocytes exhibited weak signal (FIG. 7). On ovarian sections, the strongest signal was present in stage-II~III oocytes and certain oogonia (Supplementary Material: FIG. S2A). A closer inspection revealed three types of oogonia exhibiting a strong, moderate and weak signal, which are called type I (og1), II (og2) and III oogonia (og3), respectively (FIG. S2A). Distinction of three types of oogonia became more evident at higher magnification (FIG. S2B). These results suggest that Lcvasa expression in the ovary is specific to germ cells.
We furthered our analysis of vasa RNA expression by fluorescent ISH (FISH), which provides improved sensitivity and simultaneous detection of two or more different signals [9, 32, 39]. After FISH, the hybridization signal was seen again throughout oogenesis (FIG. 6A and B). Its intensity varies dramatically at different stages of oogenesis. The signal in oogonia is moderate for nuclear staining and strongest for vasa staining, og2 displays the most intense nuclear staining and reduced vasa signal, and og3 shows a remarkable reduction in both nuclear staining and vasa signal.
The vasa RNA is cytoplasmic and nuclear in og1 and becomes mainly cytoplasmic in og2 and og3 (FIG. 7; FIG. 6C), suggesting increase of its expression in stage-I oocytes and richest stage-II~III oocytes (FIG. 6B). Observations at higher magnification revealed that the vasa RNA exhibited highly dynamic redistribution during different stages of oogenesis (FIG. 6C-G). Three types of oogonia (og1~og3) were easily identifiable (FIG. 6C). The vasa staining in Og1 showed that vasa RNA could undergo nuclear-to-cytoplasmic redistribution when og1 develops into og2. Interestingly, the signal was seen predominantly in the nucleus of stage-I oocytes (FIG. 6D), implying that the vasa RNA could undergo cytoplasmic-to-nuclear redistribution when og3 enters into meiosis to become oocytes. Moreover, the signal concentrates in the perinuclear cytoplasm of stage-II oocytes (FIG. 6E), indicating that the export of bulky vasa RNA from the nucleus to cytoplasm when oogenesis transits from stage I to stage II of oocytes. Later at stage II, the perinuclear signal begins to radiate in the cytoplasm in a nucleus-periphery direction (FIG. 6E). In stage-III oocytes, the signal was found evenly in the cytoplasm except a spherical structure called the Balbiani body (BB) [32], In BB, the vasa RNA was most abundant (FIG. 6F). Therefore, the vasa RNA undergo a series of sequential events during oogenesis (FIG. 6G). These include nucleocytoplasmic export, cytoplasmic concentration and perinuclear localization during oogonial development, nuclear concentration at the meiotic entry, and nucleocytoplasmic export, cytoplasmic redistribution and localization into the BB during oocyte development. Therefore, the level and subcellular distribution of the vasa RNA delineate with critical major stages of oogonial and oocyte development.
In summary, by sequence and pattern of RNA expression, we conclude that Lcvasa encodes a Vasa ortholog whose RNA expression is a reliable germ cell marker in the Asian seabass. In combination with position, morphology and nuclear staining, lcvasa expression and subcellular distribution allow for identification of the critical stages of female germ cell, such as self-renewal, differentiation, meiotic entry and progress.
vasa expression in the adult testis by fluorescent ISH. Testicular cryosections after fluorescent ISH (green) were analyzed by microscopy. Nuclei were stained blue with DAPI. (A) Bright field micrograph showing the testicular structure. (B) Merge micrograph between bright field and fluorescent optics. (C-F) Fluorescent micrograph. Large magnification of the area boxed in (A and B), along with (E and F) highlighting spermatogonia at the periphery. Arrow, CB; sg, spermatogonium; sc1 and sc2, primary and secondary spermatocyte; sm, sperm, st, spermatid; ED, efferent ductile; Scale bars, 50 µm in (A-C, E, F) and 25 µm in (D and D').
vasa RNA expression in the adult ovary by fluorescent ISH. (A and B) Micrographs showing the vasa RNA expression and distribution at the major stages. Nuclei were stained blue with DAPI. (C-F) Large magnification of the areas framed in (B), highlighting intracellular distribution of the vasa RNA at critical stages of oogenesis. (C-F) Large magnification of the areas framed c-f in (B), highlighting differential staining and subcellular distribution at representative stages of oogonia (C) and oocytes (D-F). Arrow, BB; og1~3, stages of oogonia; I~IV, stages of oocytes, so, somatic cells. Scale bars, 40 µm in (A and B) and 10 µm in (C-F). (G) Oogenic expression and subcellular distribution of Lcvasa. The signal intensity correlates with the level of abundance.
vasa RNA expression in the whole mount adult ovary by chromogenic ISH. An antisense vasa riboprobe (purple) was used for ISH. Different stages of oocytes (I~IV) and oogonia (og) are seen. The hybridization signal is strong in oogonia (inset) and stage-I~III oocyte.
Discussion
In this study, we have identified Lcvasa, the Asian seabass vasa, as a germ cell marker in both sexes. Three lines of evidence support that Lcvasa is a true Vasa homolog. First, its predicted LcVasa protein shows the highest sequence homology to Vasa proteins among the DEAD family members described so far, and contains the eight diagnostic motifs of Vasa proteins. In addition LcVasa shares with known Vasa proteins an important feature, namely the presence of multiple N-terminal RGG-repeats that are thought to be a putative RNA-binding motif [40]. The RGG-rich N-terminus of the zebrafish Vasa protein has been implicated in subcellular localization of the fused green fluorescent protein in PGCs [38]. Second, ovarian Lcvasa RNA expression is restricted to female germ cells and barely detectable in somatic cells. Finally, testicular Lcvasa RNA expression is similarly limited to male germ cells and hardly detectable in somatic cells.
Also, the embryonic expression of Lcvasa RNA is coincident with that of Vasa homolog. It is maternally inherited since it is already abundant in early stages of embryos before zygotic transcription commences. A high level of RNA persists in subsequent stage before it declines to a low level afterwards. This temporal expression pattern is reminiscent of situations in other fish species. In zebrafish, vasa RNA is also high until the early gastrula stage at 6 h post fertilization and decreases sharply to a low level [38], and is a germ plasm component and segregates with PGCs [29]. A similar expression pattern has also been reported for vasa RNA in medaka [26] and gibel carp [27]. Embryonic expression patterns similar to that of Lcvasa have been described for RNAs of other germ cell genes such as boule and dazl [32]. Future work will determine whether Lcvasa identifies PGCs in the Asian seabass embryo.
The gilthead seabream represents so far the only marine teleost analyzed for germ cell-specific expression of vasa RNA by chromogenic ISH [34]. In this fish, vasa expression was examined in the ovary and found to be restricted to vitellogenic oocytes and absent in oogonia and previtellogenic oocytes, and whether vasa is expressed also in male germ cells is unclear. In this study, germ cell-specific vasa expression is evident throughout spermatogenesis and oogenesis by both chromogenic and fluorescent ISH. Specifically, the signal in oogonia is easily detectable after chromogenic ISH on whole mount samples and ovarian cryosections, and becomes even more evident after fluorescent ISH on sections. The vasa expression pattern in the Asian seabass observed in this study is similar to that reported in tilapia, in which the hybridization signal is strong in both spermatogonia and oogonia but barely detectable in spermatids [25].
Gametogenesis is a complicated process and involves a series of events, including germ stem cell self-renewal and differentiation, meiosis entry and progression in both sexes, and post meiotic morphogenesis called spermiogenesis in male [41]. Understanding of the mechanisms underlying these events requires unambiguous staging of these events, which is a challenge in most animal species due to a lack of stage-specific germ cell markers. In teleosts such as zebrafish [42], oogenesis proceeds from oogonia to oocytes of different stages. Distinction between oogonia and oocytes is a precondition to study female germ stem cells' self-renewal and meiotic differentiation. This distinction is usually made on the basis of difference of DAPI staining between mitotic oogonia and meiotic oocytes. This dye does not stain the oocyte nucleus in many teleost species including medaka [32] and trout [9, 39]. Another dye is propidium iodide, which produces a similarly differential staining in zebrafish [29], gibel carp [27]. In the Asian seabass, DAPI also does not stain the oocyte nuclei. This dye, however, exhibits also cytoplasmic staining in growing oocytes of the Asian seabass. Future work will determine whether such staining is unique to this organism or common to marine perches.
One striking observation described in this study is the dynamic of vasa mRNA expression and distribution. vasa mRNA expression and distributions are dynamic during animal gametogenesis. Similar dynamic expression has been reported in medaka fish [26], tilapia [25], gibel carp [27] and zebrafish [43], also dynamic expression has been studied in zebrafish [43]. Moreover, this kind of dynamics expression might be associated with its functions in germ cells as discussed in a review [44].
However, vasa-RNA's dynamic distribution, such as nucleocytoplasmic redistribution, was first reported here. The dynamic nucleocytoplasmic distribution of lcvasa is reminiscent to that of CTN-RNA. CTN-RNA through different poly(A) site usage is diffusely distributed in nuclei as well as in paraspeckles, while under physiologic stress CTN-RNA is posttranscriptionally cleaved to produce protein-coding mouse cationic amino acid transporter 2 (mCAT2) mRNA and becomes distributed into cell cytoplasm [45]. Recent studies showed that paraspeckles contributes to transcriptional regulation of genes' expression [46] and functions as a reservoir for A-to-I edited mRNAs, being released into the cytoplasm under certain stress conditions [47]. At the same time, nuclear transcriptomes are reported to be longer and contain many more editing events (A-to-G or A-to-I) than cytosolic transcriptomes, and most of the sites exhibiting nucleus-specific editing are in introns or novel intergenic transcripts, also RNA editing is globally associated with the modification of microRNA regulation in 3′ untranslated regions [48, 49]. Evidences showed intron retention (IR) is a consequence of mis-splicing that leads to failed excision of intronic sequences from pre-messenger RNAs, leading to reduced mRNA and protein levels [50, 51]. Physiological IR may serve a precautionary role in dynamic gene expression through nuclear retention of mRNA during stressful situations [52, 53]. Similarly, in our study, lcvasa RNA undergoes dynamic nuclear-cytoplasm distributions during oogenesis including nucleocytoplasmic export, cytoplasmic concentration and perinuclear localization during oogonial development, nuclear concentration at the meiotic entry, and nucleocytoplasmic export, cytoplasmic localization. Maybe this kind of dynamic distribution is also associated with physiological IR or other RNA editing machinery, to address this issue, intensive investigations need to be conducted and this will provide the possibility for us to obtain new insights on understanding vasa gene functions in animal gametogenesis.
Here, we identify three types of oogonia, on the basis of positioning in the germinal epithelium, nuclear staining and more importantly, vasa staining and its subcellular distribution. In addition, differential vasa staining and distribution also enable distinction between stage I and II. Future work is needed to determine whether type I, II and III of oogonia are self-renewing oogonial stem cells, proliferating oogonia and differentiating oogonia, respectively. The identification of these 5 earliest stages of oogenesis provides a basis to study female germ stem cell self-renewal, proliferation, differentiation and entry into meiosis in the Asian seabass as a model of marine teleosts.
Supplementary Material
Fig.S1-S2.
Abbreviations
aa, amino acid(s); bp, base pair(s); ISH, in situ hybridization; hpf, hour(s) post fertilization; nt, nucleotide(s); M3, male stage 3; F3, female stage 3.
Acknowledgements
We thank J. Deng for fish samples and CM Foong for laboratory management. This work was supported by the National Research Foundation of Singapore (NRF-CRP7-2010-03).
Authors' contributions
H.X., M.L., and M.D. performed the research; H.X. analyzed the data; H.X. and Y.H. designed the research and wrote the paper.
Competing Interests
No competing financial interests exist.
References
1. Wylie C. Germ cells. Cell. 1999;96:165-74
2. Raz E. Primordial germ-cell development: the zebrafish perspective. Nat Rev Genet. 2003;4:690-700
3. Fechner PY. The role of SRY in mammalian sex determination. Acta Paediatr Jpn. 1996;38(4):380-9
4. Matsuda M, Nagahama Y, Shinomiya A. et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559-63
5. Nanda I, Kondo M, Hornung U. et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci U S A. 2002;99:11778-83
6. Kocer A, Reichmann J, Best D, Adams IR. Germ cell sex determination in mammals. Mol Hum Reprod. 2009;15:205-13
7. Okutsu T, Suzuki K, Takeuchi Y. et al. Testicular germ cells can colonize sexually undifferentiated embryonic gonad and produce functional eggs in fish. Proc Natl Acad Sci U S A. 2006;103:2725-9
8. Kurokawa H, Saito D, Nakamura S. et al. Germ cells are essential for sexual dimorphism in the medaka gonad. Proc Natl Acad Sci U S A. 2007;104:16958-63
9. Li M, Shen Q, Wong FM. et al. Germ cell sex prior to meiosis in the rainbow trout. Protein Cell. 2011;2:48-54
10. Bejar J, Hong Y, Schartl M. Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development. 2003;130:6545-53
11. Sola L, Marzovillo M, Rossi AR. et al. Cytogenetic analysis of a self-fertilizing fish, Rivulus marmoratus: remarkable chromosomal constancy over a vast geographic range. Genome. 1997;40:945-9
12. Yue GH, Xia JH, Liu F, Lin G. Evidence for Female-Biased Dispersal in the Protandrous Hermaphroditic Asian Seabass, Lates calcarifer. PLoS One. 2012;7:e37976
13. Guiguen Y, Jalabert B, Thouard E, Fostier A. Changes in plasma and gonadal steroid hormones in relation to the reproductive cycle and the sex inversion process in the protandrous seabass, Lates calcarifer. Gen Comp Endocrinol. 1993;92:327-38
14. Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611-7
15. Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev Genes Evol. 2001;211:299-308
16. Nakao H. Isolation and characterization of a Bombyx vasa-like gene. Dev Genes Evol. 1999;209:312-6
17. Chang CC, Dearden P, Akam M. Germ line development in the grasshopper Schistocerca gregaria: vasa as a marker. Dev Biol. 2002;252:100-18
18. Fabioux C, Huvet A, Lelong C. et al. Oyster vasa-like gene as a marker of the germline cell development in Crassostrea gigas. Biochem Biophys Res Commun. 2004;320:592-8
19. Fujiwara Y, Komiya T, Kawabata H. et al. Isolation of a DEAD-family protein gene that encodes a murine homolog of Drosophila vasa and its specific expression in germ cell lineage. Proc Natl Acad Sci U S A. 1994;91:12258-62
20. Tsunekawa N, Naito M, Sakai Y. et al. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127:2741-50
21. Castrillon DH, Quade BJ, Wang TY. et al. The human VASA gene is specifically expressed in the germ cell lineage. Proc Natl Acad Sci U S A. 2000;97:9585-90
22. Olsen LC, Aasland R, Fjose A. A vasa-like gene in zebrafish identifies putative primordial germ cells. Mech Dev. 1997;66:95-105
23. Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157-65
24. Yoshizaki G, Sakatani S, Tominaga H, Takeuchi T. Cloning and characterization of a vasa-like gene in rainbow trout and its expression in the germ cell lineage. Mol Reprod Dev. 2000;55:364-71
25. Kobayashi T, Kajiura-Kobayashi H, Nagahama Y. Differential expression of vasa homologue gene in the germ cells during oogenesis and spermatogenesis in a teleost fish, tilapia, Oreochromis niloticus. Mech Dev. 2000;99:139-42
26. Shinomiya A, Tanaka M, Kobayashi T. et al. The vasa-like gene, olvas, identifies the migration path of primordial germ cells during embryonic body formation stage in the medaka, Oryzias latipes. Dev Growth Differ. 2000;42:317-26
27. Xu H, Gui J, Hong Y. Differential expression of vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp (Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate. Dev Dyn. 2005;233:872-82
28. Li CJ, Liu L, Chen XH. et al. Identification of a vasa homologue gene in grass carp and its expression pattern in tissues and during embryogenesis. Comp Biochem Physiol B Biochem Mol Biol. 2010;157:159-66
29. Knaut H, Pelegri F, Bohmann K. et al. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149:875-88
30. Saito T, Otani S, Fujimoto T. et al. The germ line lineage in ukigori, Gymnogobius species (Teleostei: Gobiidae) during embryonic development. Int J Dev Biol. 2004;48:1079-85
31. Cao M, Yang Y, Xu H. et al. Germ cell specific expression of Vasa in rare minnow, Gobiocypris rarus. Comp Biochem Physiol A Mol Integr Physiol. 2012;162:163-70
32. Xu H, Li Z, Li M. et al. Boule is present in fish and bisexually expressed in adult and embryonic germ cells of medaka. PLoS One. 2009;4:e6097
33. Li M, Hong N, Xu H. et al. Medaka vasa is required for migration but not survival of primordial germ cells. Mech Dev. 2009;126:366-81
34. Cardinali M, Gioacchini G, Candiani S. et al. Hormonal regulation of vasa-like messenger RNA expression in the ovary of the marine teleost Sparus aurata. Biol Reprod. 2004;70:737-43
35. Blazquez M, Gonzalez A, Mylonas CC, Piferrer F. Cloning and sequence analysis of a vasa homolog in the European sea bass (Dicentrarchus labrax): tissue distribution and mRNA expression levels during early development and sex differentiation. Gen Comp Endocrinol. 2011;170:322-33
36. Lim M. Isolation and expression analyses of dazl, vasa and sox3 genes in the seabass (Lates calcarifer). Master thesis, National University of Singapore. 2005
37. Hay B, Jan LY, Jan YN. Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development. 1990;109:425-33
38. Wolke U, Weidinger G, Koprunner M, Raz E. Multiple levels of posttranscriptional control lead to germ line-specific gene expression in the zebrafish. Curr Biol. 2002;12:289-94
39. Li M, Shen Q, Xu H. et al. Differential conservation and divergence of fertility genes boule and dazl in the rainbow trout. PLoS One. 2011;6:e15910
40. Liang L, Diehl-Jones W, Lasko P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 1994;120:1201-11
41. Goto R, Saito T, Takeda T. et al. Germ cells are not the primary factor for sexual fate determination in goldfish. Dev Biol. 2012
42. Bally-Cuif L, Schatz WJ, Ho RK. Characterization of the zebrafish Orb/CPEB-related RNA binding protein and localization of maternal components in the zebrafish oocyte. Mech Dev. 1998;77:31-47
43. Kosaka K, Kawakami K, Sakamoto H, Inoue K. Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev. 2007;124:279-89
44. Boulanger L, Kocer A, Daniel N. et al. Attempt to rescue sex-reversal by transgenic expression of the PISRT1 gene in XX PIS-/- goats. Sexual development: genetics, molecular biology, evolution, endocrinology, embryology, and pathology of sex determination and differentiation. 2008;2:142-51
45. Prasanth KV, Prasanth SG, Xuan Z. et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249-63
46. Schuldt A. Proteomics of the nucleolus. Nat Cell Biol. 2002;4:E35
47. Nakagawa S, Hirose T. Paraspeckle nuclear bodies--useful uselessness? Cell Mol Life Sci. 2012;69:3027-36
48. Solnestam BW, Stranneheim H, Hallman J. et al. Comparison of total and cytoplasmic mRNA reveals global regulation by nuclear retention and miRNAs. BMC genomics. 2012;13:574
49. Chen L. Characterization and comparison of human nuclear and cytosolic editomes. Proc Natl Acad Sci U S A. 2013;110:E2741-7
50. Takemura R, Takeiwa T, Taniguchi I. et al. Multiple factors in the early splicing complex are involved in the nuclear retention of pre-mRNAs in mammalian cells. Genes to cells: devoted to molecular & cellular mechanisms. 2011;16:1035-49
51. Yap K, Lim ZQ, Khandelia P. et al. Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention. Genes Dev. 2012;26:1209-23
52. Kallehauge TB, Robert MC, Bertrand E, Jensen TH. Nuclear retention prevents premature cytoplasmic appearance of mRNA. Molecular cell. 2012;48:145-52
53. Wong JJ, Ritchie W, Ebner OA. et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583-95
Author contact
![]() Corresponding author: Prof. Yunhan Hong, Email: dbshyhedu.sg. Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543. Fax: +65 6779 2486; Tel: +65 6516 2915
Corresponding author: Prof. Yunhan Hong, Email: dbshyhedu.sg. Department of Biological Sciences, National University of Singapore, 14 Science Drive 4, Singapore 117543. Fax: +65 6779 2486; Tel: +65 6516 2915

 Global reach, higher impact
Global reach, higher impact