10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2015; 11(7):813-824. doi:10.7150/ijbs.11797 This issue Cite
Research Paper
Functional Analysis of a c-di-AMP-specific Phosphodiesterase MsPDE from Mycobacterium smegmatis
State Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, PR China
Received 2015-2-5; Accepted 2015-5-4; Published 2015-5-30
Abstract
Cyclic di‑AMP (c-di-AMP) is a second signaling molecule involved in the regulation of bacterial physiological processes and interaction between pathogen and host. However, the regulatory network mediated by c-di-AMP in Mycobacterium remains obscure. In M. smegmatis, a diadenylate cyclase (DAC) was reported recently, but there is still no investigation on c-di-AMP phosphodiesterase (PDE). Here, we provide a systematic study on signaling mechanism of c-di-AMP PDE in M. smegmatis. Based on our enzymatic analysis, MsPDE (MSMEG_2630), which contained a DHH-DHHA1 domain, displayed a 200-fold higher hydrolytic efficiency (kcat/Km) to c-di-AMP than to c-di-GMP. MsPDE was capable of converting c-di-AMP to pApA and AMP, and hydrolyzing pApA to AMP. Site-directed mutations in DHH and DHHA1 revealed that DHH domain was critical for the phosphodiesterase activity. To explore the regulatory role of c-di-AMP in vivo, we constructed the mspde mutant (Δmspde) and found that deficiency of MsPDE significantly enhanced intracellular C12-C20 fatty acid accumulation. Deficiency of DAC in many bacteria results in cell death. However, we acquired the M. smegmatis strain with DAC gene disrupted (ΔmsdisA) by homologous recombination approach. Deletion of msdisA reduced bacterial C12-C20 fatty acids production but scarcely affected bacterial survival. We also provided evidences that superfluous c-di-AMP in M. smegmatis could lead to abnormal colonial morphology. Collectively, our results indicate that MsPDE is a functional c-di-AMP-specific phosphodiesterase both in vitro and in vivo. Our study also expands the regulatory network mediated by c-di-AMP in M. smegmatis.
Keywords: cyclic di‑AMP, Mycobacterium smegmatis, phosphodiesterase, diadenylate cyclase, fatty acids
INTRODUCTION
Bacterial cyclic nucleotide second messenger molecules regulate various cellular processes including carbon/nitrogen metabolism, cell cycle, motility, quorum sensing, biofilm formation, and even virulence [1, 2]. Cyclic di‑AMP (c-di-AMP) is a recently discovered second messenger molecule in bacteria [3]. An upsurge in c-di-AMP research has appeared, with many important cellular pathways mediated by c-di-AMP being revealed. However, these findings are only a tip of the iceberg on account of the broad distribution of c-di-AMP among bacteria and archaea. Bacterial intracellular c-di-AMP levels are maintained by two functionally opposite enzymes: diadenylate cyclases (DACs) and phosphodiesterases (PDEs). However, endogenous or environmental stimuli can disturb this mutual balance of enzymes. Further, the fluctuation in c-di-AMP level is a direct trigger for the signal transduction pathways which regulate different bacterial physiological processes.
The first DAC was indentified during the structural study on Thermotoga maritima DNA integrity scanning protein (DisA), which condensed two ATP molecules to form one c-di-AMP molecule [4]. Genes coding for functional DAC were identified in a wide range of bacteria, including Listeria monocytogenes [5], Staphylococcus aureus [6], and Streptococcus pyogenes [7]. These bacteria only possess one functional DAC, and disruption of any of these cyclases results in cell death [7]. The bacteria from Bacillus species, such as B. subtilis [8] and B. thuringiensis [9], have more than one gene coding for DAC. These genes are also essential for bacteria survival: bacteria with all the DACs deleted cannot survive [10].
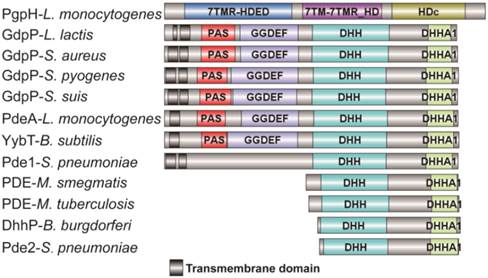
Most of the reported PDEs belong to GdpP protein family (GGDEF domain protein-containing phosphodiesterase) which consists of an N-terminal signal regulatory module linked to a degenerate GGDEF domain and a C-terminal DHH-DHHA1 domain module [11, 12]. The DHH-DHHA1 domain is essential for PDE activity. Yybt in B. subtilis, GdpP in S. aureus, and PdeA in L. monocytogenes etc., all belong to GdpP protein family and perform a PDE function to maintain the intracellular c-di-AMP level [5, 6, 9, 11]. DhhP in Borrelia burgdorferi and MtPDE (also referred as CnpB) in M. tuberculosis are also functional c-di-AMP-specific PDE which only contain a core DHH-DHHA1 domain, and both are soluble proteins and essential for virulence [13, 14] (Fig. 1). Recently, Huynh and co-workers identified L. monocytogenes protein PgpH as a novel PDE which specifically hydrolyzed c-di-AMP to 5'-pApA via a catalytic His-Asp (HD) domain [15]. PgpH homologs are conserved in many c-di-AMP synthesizing microorganisms [15]. PDEs from different families might exhibit cooperative activities in regulating c-di-AMP level.
c-di-AMP regulates various cellular pathways by binding with specific receptor proteins or RNAs [3]. This binding allosterically alters the activity or conformation of receptors and thus triggers signal cascades related to various cellular processes. Unlike c-di-GMP receptors, c‑di‑AMP receptors are more diverse.
Even though the potassium transport component KtrA, cation proton antiporter A (CpaA), as well as the histidine kinase protein KdpD of S. aureus, are all c‑di‑AMP receptor proteins involved in potassium homeostasis regulation, these proteins have different binding domains for c-di-AMP [16]. M. smegmatis TetR family transcription factor DarR and S. aureus cytoplasmic protein PstA are also reported to be c‑di‑AMP receptor proteins but these are different from each other in terms of structure and function [3, 16, 17]. A riboswitch class called ydaO was recently discovered to sense c-di-AMP [18-20]. ydaO selectively responds to c-di-AMP with sub-nanomolar affinity and controls numerous genes in a wide variety of bacteria [18, 20]. c-di-AMP also controls important signaling cascades of host during infection. For example, c-di-AMP secreted by L. monocytogenes through multidrug efflux pumps (MDRs) was able to induce cytosolic host response [21], and this response was likely to be mediated by STING and DDX41 [22, 23]. Moreover, c-di-AMP secreted by M. tuberculosis directly targeted macrophages and led to host inflammation response [24]. The elevated c-di-AMP levels in macrophage induced host to produce much higher level of IFN-β and resulted in attenuated virulence of M. tuberculosis [24].
Domain architecture of c-di-AMP PDEs. The reported PDEs belong to three different families, one is GdpP protein family (GGDEF domain protein-containing phosphodiesterase) which consists of an N-terminal signal regulatory module linked to a degenerate GGDEF domain and a C-terminal DHH-DHHA1 domain module, an other family only contains a DHH-DHHA1 domain, and the third is 7TMR family which contains a HD domian [7, 11, 15, 33, 38-40]. Accession numbers: PgpH-L. monocytogenes: 4S1B_A; GdpP-L. lactis: CAL98387; GdpP-S. aureus: YP_498622; GdpP-S. pyogenes: NP_270093; GdpP-S. suis: ABP93318; PdeA-L. monocytogenes: CAC98267; YybT-B. subtilis: NP_391931; Pde1-S. pneumonia: ABJ55207; PDE-M. smegmatis: YP_886967; PDE-M. tuberculosis: P71615; Dhhp-B. burgdorferi: NP_212753; Pde2-S. pneumonia: SPD_1153.
In M. tuberculosis, which causes human tuberculosis, both DAC and c-di-AMP-specific PDE were identified. MtdisA (also referred as DacA) is able to utilize both ATP and ADP to synthesize c-di-AMP [25]. Moreover, the DAC activity of MtdisA is allosterically regulated by high concentration of ATP [13]. These findings provide new insights into the substrates and the catalytic mechanism of DACs. On the other hand, MtPDE was demonstrated to possess c-di-AMP PDE activity. It was capable of hydrolyzing c-di-AMP exclusively to AMP, and deletion of mtpde led to reduced virulence in mouse [24].
Being a "fast grower" and non-pathogenic, M. smegmatis is used as a research model for M. tuberculosis. In M. smegmatis, c-di-AMP is synthesized by a sole DAC MsDisA [26]. Overexpression of msdisA induces cell expansion and aggregation, as well as inhibits bacterial motility [26]. A c-di-AMP receptor DarR has been recently identified, and it acts as a repressor of fatty acid synthesis genes [17]. However, there is no report on c-di-AMP-specific PDE in M. smegmatis. In addition, the way M. smegmatis maintains its c-di-AMP homeostasis is also obscure.
MsPDE (MSMEG_2630) is a DHH-DHHA1 domain protein which lacks any other regulatory domains conserved in GdpP protein family. MsPDE has affinity for wide substrates in vitro because of its unique subunit packing and large domain interface [27]. In this research, we explored the enzyme kinetic parameters of MsPDE and confirmed that MsPDE is a c-di-AMP-specific PDE and the DHH domain is critical for PDE activity in vitro. To understand the physiological function of c-di-AMP in M. smegmatis, we constructed the deletion mutants of msdisA and mspde and found that disruption of these genes affected the intracellular concentration of C12-C20 fatty acids. Our results also provided new evidences that M. smegmatis tended to maintain the intracellular c-di-AMP concentration at low level, and superfluous c-di-AMP would lead to abnormal cellular processes.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions
E. coli DH5α and E. coli BL21(DE3) were used for gene cloning and protein expression respectively (Table 1). Both strains were grown in lysogeny broth (LB) at 37 °C. M. smegmatis MC2 155 (NC_008596) was grown at 37 °C in 7H9 Middlebrook broth or on 7H10 agar plates. Where required, antibiotics were added as follows: kanamycin, 50 μg/ml; ampicillin, 100 μg/ml and hygromycin, 100 μg/ml.
DNA manipulations
Overlap PCR was performed for site-directed mutation of mspde gene and the primers are shown in Supplementary Table S1. The corresponding genes (mspde, mtpde, msdisA, mspdeDHH-AAA, mspdeGGGH-AAAA, btyybT320-657, btytqI, btrecJ) were cloned into pET28b and transformed into E. coli BL21(DE3) (Table 1). All the constructs were verified by DNA sequencing.
Protein expression and purification
E. coli BL21(DE3) cells harboring recombinant plasmids were grown up to OD600 0.8 at 37 °C and then induced by adding 0.5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) at 28 °C for 8 h. Harvested cells were re-suspended in binding buffer (25 mM Tris, 500 mM NaCl, pH 8.0) before sonication. The cell extracts were further clarified by centrifugation and hexahistidine-tagged protein was purified from the supernatant using Ni-NTA affinity column at room temperature. Fractions were dialyzed overnight to remove the residual imidazole. The purified protein was analyzed by 12% SDS-PAGE and quantified by Coomassie brilliant blue assay.
Construction of mspde and msdisA mutant strains
The mutant Δmspde with mspde (msmeg_2630) deletion and mutant △mspde with msdisA (msmeg_6080) deletion were constructed using the method of homologous recombination [28]. The pMind-derived suicide plasmid, which carried hygromycin resistance gene (hyg) and a sacB-lacZ cassette as selection markers, was constructed and electroporated into M. smegmatis. The allelic-exchange mutant strain was selected on 7H10 agar containing 50 μg/ml hygromycin, 2% sucrose, and 200 μg/ml X-gal. Selected mutants were verified by PCR and sequencing.
Construction of mspde and msdisA overexpression strains
The mspde and msdisA genes were amplified by PCR (Supplementary Table S1) and cloned into pMV261 plasmid at the NcoI and XhoI sites, respectively [29]. The expression plasmids pMV261-mspde and pMV261-msdisA were then electroporated into M. smegmatis to create the overexpression strains OEmspde and OEdisA, respectively (Table 1).
Phosphodiesterase assays
The purified protein was incubated with 10 nM nucleotide substrate (cAMP, cGMP, c-di-AMP, c-di-GMP, pApA or pGpG) at 25 °C. The assay was performed in initial reaction mixture, which contained 100 mM Tris (pH 7.8), 100 mM NaCl, and 10 mM MgCl2. The reaction was terminated by boiling for 10 min and then centrifuged at 16,000 g for 15 min to remove the denatured protein. Subsequently, the supernatant was loaded onto an Agilent 1200 series HPLC system (Agilent Technologies, USA) to monitor the degradation products. The samples were separated by an Elite Hypersil BDS C18 column (4.6 ×200 mm, 5 μm particle sizes) with the mobile phase containing 90% phosphate buffer (30 mM K2HPO4 and 20 mM KH2PO4, pH 6.0) and 10% methanol. The column temperature was controlled at 25 °C and the flow rate was set at 1 mL/min.
The optimum reaction temperature was determined by testing PDE activity of MsPDE at various temperatures (4 to 65 °C) in the initial reaction mixture. 100 mM MES (pH 5.5-6.5) or Tris (pH 7.0-9.5) buffer was added to explore the optimum reaction pH of MsPDE. The metal dependence of MsPDE was assayed by replacing Mn2+ with other divalent metal cations (Ca2+, Zn2+, Co2+, Mg2+, Fe2+, Ni2+, and Cu2+) in the initial condition. For optimization of Mn2+ and Mg2+ concentration, experiments were conducted by adding MnCl2 or MgCl2 to the reaction mixture at various final concentrations (0-100 mM). The effect of salt concentration on the PDE activity was evaluated by changing the NaCl concentrations from 0 to 500 mM.
Identification of the reaction product by LC/Q-TOF
The products of MsPDE were identified by LC/Q-TOF analysis, which was performed on Agilent 1260 LC system (Agilent Technologies, USA) coupled to an ultra high definition quadrupole time-of-flight mass spectrometer Model 6540 (Agilent Technologies, USA) equipped with a dual source electrospray ionization ion source.
The reaction products were separated on Agilent C18 reverse-phase column, with the binary mobile phase composed of 2% methanol and 98% water (containing 0.2% ammonium acetate and 0.1% acetic acid) being set at a constant flow rate of 300 μL/min and column temperature of 30 °C.
Q-TOF parameters were as follows: ionization mode, positive mode; capillary voltage: 4,000 V; drying gas: 9 L/min; nebulizer pressure: 40 psig; gas temperature: 350 °C; skimmer voltage: 65 V; octopole RF Peak voltage: 750 V, fragmentor voltage: 150 V. LC/MS accurate mass spectra were recorded across a range of 100-1000 m/z at the MS scan rate 1.5 spectra/s. Accurate mass measurements of each peak from the total ion chromatograms were obtained by an automated calibrant delivery system using a low flow of a calibrating solution (calibrant solution A, Agilent Technologies), which contains the internal reference masses purine (C5H4N4) at m/z 121.0509 and HP-921 [hexakis-(1H, 1H, 3H-tetrafluoro-pentoxy) phosphazene] (C18H18O6N3P3F24) at m/z 922.0098.
Strains and plasmids used in this study
| Strain and plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | A cloning host | Novagen |
| E. coli BL21(DE3) | Protein expression host | Novagen |
| BL21/pET28-mspde | BL21(DE3) with pET28-mspde, used for protein purification, | This work |
| BL21/pET28-mtpde | BL21(DE3) with pET28-mtpde, used for protein purification | This work |
| BL21/pET28-msdisA | BL21(DE3) with pET28-msdisA, used for protein purification | This work |
| BL21/pET28-mspdeDHH-AAA | BL21(DE3) with pET28-mspdeDHH-AAA, used for protein purification | This work |
| BL21/pET28-mspdeGGGH-AAAA | BL21(DE3) with pET28-mspdeGGGH-AAAA, used for protein purification | This work |
| pET28-btyybT320-657 | BL21(DE3) with pET28- yybT320-657, used for protein purification | This work |
| BL21/pET28-btytqI | BL21(DE3) with pET28- ytqI, used for protein purification | This work |
| BL21/pET28-btrecJ | BL21(DE3) with pET28- recJ, used for protein purification | This work |
| M. smegmatis | Wild type of M. smegmatis MC2155 strain (NC_008596) | [28] |
| Δmspde | mspde deleted strain of M. smegmatis MC2155 | This work |
| ΔmsdisA | msdisA deleted strain of M. smegmatis MC2155 | This work |
| Msm/pMV261 | M. smegmatis MC2 155 with pMV261 | This work |
| OEmspde | M. smegmatis MC2 155 with pMV261-mspde, mspde overexpression strain | This work |
| OEdisA | M. smegmatis MC2 155 with pMV261-disA, msdisA overexpression strain | This work |
| Plasmids | ||
| pET28b (+) | T7-driven expression vector, KmR | Novagen |
| pET28-mspde | mspde in NcoI and XhoI sites of pET28b | This work |
| pET28-mtpde | mtpde in NcoI and XhoI sites of pET28b | This work |
| pET28-msdisA | msdisA in NcoI and NotI sites of pET28b | This work |
| pET28-mspdeDHH-AAA | mspde-(D134A-H135A-H136A) in NcoI and XhoI sites of pET28b | This work |
| pET28-mspdeGGGH-AAAA | mspde-(G312A-G313A-G314A-H315A) in NcoI and XhoI sites of pET28b | This work |
| pET28-btyybT320-657 | yybT320-657 in NcoI and XhoI sites of pET28b | This work |
| pET28- btytqI | ytqI in NcoI and XhoI sites of pET28b | This work |
| pET28- btrecJ | recJ in NcoI and XhoI sites of pET28b | This work |
| pMV261 | KanR, pAL5000 replicon, colE1 replicon, hsp60 promoter, expression vector | [28] |
| pMV261-mspde | mspde in EcoRI and HindIII sites of pMV261, mspde overexpression vector | This work |
| pMV261-msdisA | msdisA in EcoRI and HindIII sites of pMV261, msdisA overexpression vector | This work |
Determination of intracellular c-di-AMP concentration of M. smegmatis by LC-MS/MS
M. smegmatis strains were grown to logarithmic phase (16 h) at 37 °C in 7H9 broth. The cells were immediately harvested at 4 °C and washed twice by distilled water. Nucleotide was extracted from the cell pellets using the method reported by Burhenne and Kaever [30]. Detection of c-di-AMP was performed on a Finnigan Surveyor Plus liquid chromatography system followed by a Thermo Scientic TSQ Quantum Ultra EMR tandem mass spectrum system (San Jose, CA, USA). Intracellular c-di-AMP level was normalized by the corresponding wet cell weight.
Measurements of intracellular fatty acids by gas chromatography (GC)
M. smegmatis strains were grown to logarithmic phase at 37 °C in 7H9 broth medium, and the cells were harvested and lyophilized immediately. Four basic steps (saponification, methylation, extraction, and base wash) were performed in the preparation of GC-ready extracts from the harvested cells according to the standard method [31]. The test samples, as well as calibration standard samples were loaded on Agilent 6890 gas chromatograph for fatty acids analysis. When the run was complete, the retention time and response of each peak were calculated by the ChemStation. Each peak from the chromatographic analysis was listed by retention time, response, Equivalent Chain Length (ECL) value and name. If response of fatty acids in test strain was more than 120% when compared with that of wild type strain, the fatty acid was thought to be up-regulated, and vise versa.
RESULTS
MsPDE is a functional PDE
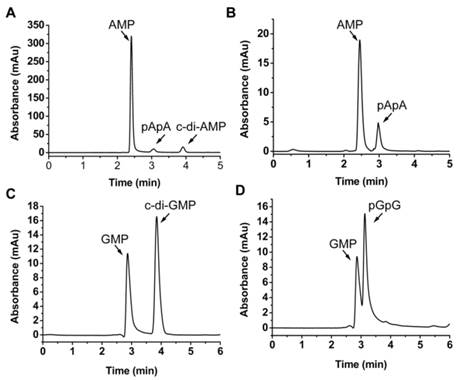
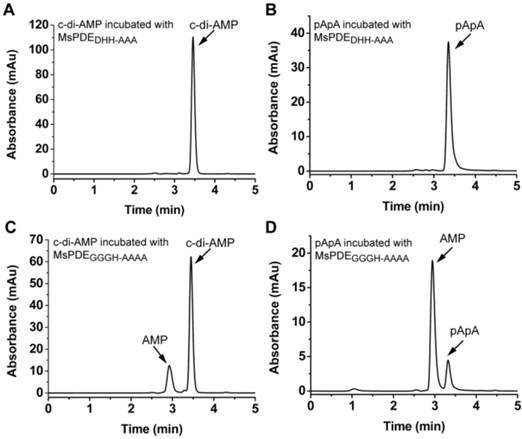
MsPDE shares 73% identity to MtPDE in M. tuberculosis and 25% identity to YybT333-659 in B. subtilis (Supplementary Fig. S1). To investigate whether MsPDE has the PDE activity, a C-terminal His-tagged recombinant protein was expressed and purified. MsPDE was then incubated with several naturally occurring nucleotide substrates (cAMP, c-di-AMP, cGMP, c-di-GMP, pApA and pGpG) in standard reaction mixture following by HPLC analysis. No degradation products were detected when MsPDE was incubated with cAMP and cGMP (data not shown). However, new product peaks appeared when MsPDE was incubated with c-di-AMP, c-di-GMP, pApA and pGpG (Fig. 2). MsPDE hydrolyzed c-di-AMP to pApA and AMP (Fig. 2A), and was also capable of converting pApA to AMP (Fig. 2B). LC/Q-TOF analysis further confirmed the hydrolysis products (Supplementary Fig. S2). These results indicated that pApA was the intermediate product when MsPDE hydrolyzed c-di-AMP to AMP, which was consistent with the observation on MtPDE [24]. Furthermore, MsPDE showed much higher PDE activity than MtPDE in vitro. When incubated with MsPDE, most of c-di-AMP was hydrolyzed into AMP, with trace amount of substrate and intermediate product remained, however, the PDE activity of MtPDE was much lower than that of MsPDE under the same condition (Supplementary Fig. S3). MsPDE also exhibited PDE activity toward c-di-GMP and pGpG, hydrolyzing them to GMP (Fig. 2C and 2D). MsPDE might hydrolyze c-di-GMP directly into GMP as we failed to identify pGpG when c-di-GMP was incubated with MsPDE.
PDE activity of MsPDE in vitro. MsPDE (0.7 μM) was incubated at 37 °C overnight in standard reaction mixture with 10 μM of c-di-AMP (A), pApA (B), c-di-GMP (C) and pGpG (D). Enzymatic reaction products were determined by HPLC. The standard reaction mixture contains 100 mM Tris (pH 7.5), 100 mM NaCl and 10 mM MnCl2.
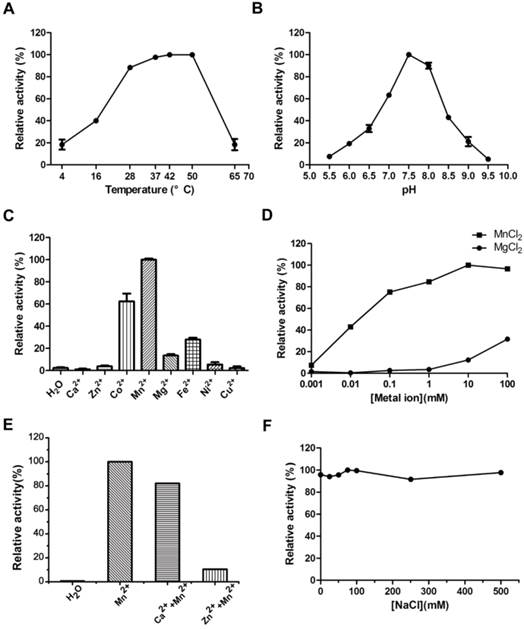
Optimization of MsPDE reaction conditions. (A) Temperature tolerance of MsPDE-catalyzed c-di-AMP hydrolysis. (B) pH dependence of MsPDE-catalyzed c-di-AMP hydrolysis. (C) Hydrolysis of c-di-AMP by MsPDE in the presence of indicated divalent metal cations. (D) Hydrolysis of c-di-AMP by MsPDE in the presence of Mn2+ or Mg2+. (E) PDE activity of MsPDE was inhibited by Ca2+ or Zn2+. (F) The effect of ionic strength on PDE activity of MsPDE. The error bars indicates the SD of triplicated experiments.
Optimization of enzyme reaction conditions
A series of experiments were carried out to explore the tolerance to extreme temperature and pH of MsPDE. MsPDE exhibited enzyme activity over a broad range of temperatures from 4 to 65 °C and its optimal temperature range was found to be 37-50 °C. On the other hand, both, low (4 °C) and high (65 °C) temperature greatly inhibited its activity (Fig. 3A). As a result, we performed all the following experiments at 37 °C. M. smegmatis is a rapidly growing environmental strain and it is also considered to be an opportunistic human pathogen [32]. Hence, its tolerance to broad range of temperature may contribute to its resilience to the changing environments. However, MsPDE was sensitive to pH change with the optimal pH around 7.5 according to our experiments (Fig. 3B).
We examined metal-ion dependence to identify the physiological metal-ion cofactor for MsPDE. The results indicated that MsPDE was strictly dependent on Co2+, Mn2+, Mg2+ or Fe2+, and showed a maximum activity with Mn2+ as a cofactor (Fig. 3C). Generally, Mg2+ acts as an effective metal cofactor in many PDEs [11, 33], whereas our result showed MsPDE had a strong preference for Mn2+ rather than Mg2+ (Fig. 3D), and this observation is similar to that of YybT [11]. The activity of YybT was strongly inhibited when the concentration of Mn2+ was higher than 1 mM, albeit Mn2+ was found to be its optimum cofactor [11]. By contrast, the activity of MsPDE was enhanced with increasing concentration of Mn2+, and remained stable even at 100 mM Mn2+ (Fig. 3D). In addition, the activity of MsPDE remained relatively low when incubated with Mg2+ and exhibited a maximum activity at 100 mM of Mg2+. We also observed that the activity of MsPDE was inhibited when the reaction mixture contained Ca2+ or Zn2+ (Fig. 3E). The binding site of Mn2+ might be occupied when the reaction mixture contain Ca2+ or Zn2+. In addition, MsPDE was insensitive to ionic strength as its activity remained unchanged with increasing concentration of NaCl (Fig. 3F).
Hence, we formulated “standard reaction mixture” which was composed of 100 mM Tris (pH 7.5), 100 mM NaCl and 10 mM MnCl2.
MsPDE Exhibits Specific PDE Activity toward c-di-AMP
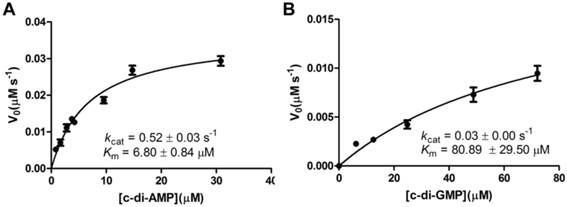
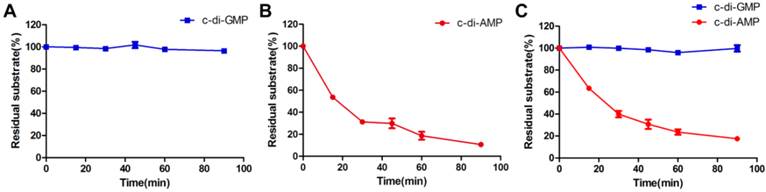
A steady-state kinetic measurement was conducted to explore the substrate specificity of MsPDE (Fig. 4). MsPDE yielded a kcat of 0.52 ± 0.03s-1 and Michaelis-Menten constant (Km) of 6.80 ± 0.84 μM for c-di-AMP and kcat of 0.03 ± 0.00 s-1 and Km of 80.89 ± 29.50 μM for c-di-GMP. Therefore, MsPDE had a 200-fold higher hydrolytic efficiency (kcat/Km) to c-di-AMP than to c-di-GMP, indicating that MsPDE prefers c-di-AMP over c-di-GMP as substrate. The preference of MsPDE for c-di-AMP did not change when incubated with the mixed substrates containing the same amount of c-di-AMP and c-di-GMP (Fig. 5). These observations further confirm that MsPDE is a c-di-AMP- specific PDE.
DHH domain is essential for enzymatic activity
Unlike DHH-DHHA1 domain protein of YybT which only converts c-di-AMP to pApA, MsPDE can hydrolyze c-di-AMP to pApA and further to AMP. It means that DHH-DHHA1 domain of MsPDE may contain a special catalytic site to convert pApA to AMP. The proteins of DHH family are predicted to perform a phosphoesterase function, including 5'-3'exonucleases (RecJ) involved in DNA repair, nanoRNases (NrnA) related to RNA processing, cyclic nucleotide PDEs and pyrophosphatases [11, 27, 34, 35]. It is reported that the DHH residues in DHH domain and GGGH residues in DHHA1 domain are highly conserved and responsible for c-di-AMP PDE activity [11, 24]. These key amino acid residues were also conserved in MsPDE (Supplementary Fig. S1). To verify the hypothesis, His-tagged MsPDE with the D134H135H136 residues replaced by AAA, and G313G314G315H316 replaced with AAAA, were expressed and purified, and termed as MsPDEDHH-AAA and MsPDEGGGH-AAAA respectively. Both of the mutant proteins were incubated with c-di-AMP in standard reaction mixture, followed by HPLC analysis of substrate and products. The mutation in DHH domain completely abolished PDE activity of MsPDE, showing that DHH domain was essential for hydrolyzing c-di-AMP (Fig. 6A and 6B). In comparison, the protein harboring the mutant DHHA1 domain still showed a residual PDE activity, partially converting c-di-AMP and pApA to AMP (Fig. 6C and 6D), which suggested that the DHHA1 domain only contributed to fractional activity and its genuine function remained obscure.
Steady-state kinetic analysis of the hydrolysis of c-di-AMP and c-di-GMP catalyzed by MsPDE. The assay conditions used for kinetic analysis were: 0.7 μM MsPDE incubated at 37 °C with 10 μM of c-di-AMP (A) or c-di-GMP (B) in standard reaction mixture. Reactions were stopped at appropriate time intervals and analyzed by HPLC. The errorbars indicates the SD of triplicated experiments.
MsPDE exhibits substrates preference toward c-di-AMP rather than c-di-GMP. (A) and (B) MsPDE was incubated at 37 °C with the same amount of c-di-AMP and c-di-GMP respectively, and then the reaction products were subjected to HPLC analysis. (C) MsPDE was incubated with mixed substrates containing both c-di-AMP and c-di-GMP at the same amount, and the reaction products were analyzed by HPLC. All the reactions were performed in the standard reaction mixture.
Enzymatic activity of MsPDE mutants in hydrolyzing c-di-AMP and pApA. The same amount of MsPDEDHH-AAA (A and B) and MsPDEGGGH-AAAA (C and D) were incubated at 37 °C with 10 μM of c-di-AMP (A and C) or pApA (B and D) in standard reaction mixture.
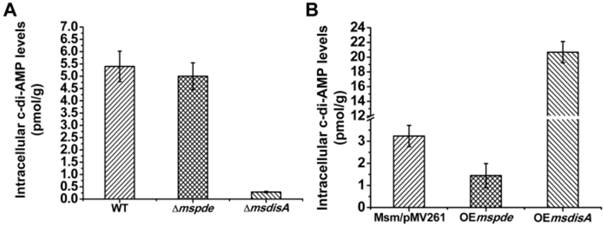
MsPDE and MsdisA altered intracellular c-di-AMP levels of M. smegmatis. (A) Determination of intracellular c-di-AMP concentrations of wild type, Δmspde and ΔmsdisA strains by LC-MS/MS. (B) Determination of intracellular c-di-AMP concentrations of wild type, OEmspde and OEmsdisA strains by LC-MS/MS. 20 μg/ml of kanamycin was added to Msm/pMV261, OEmspde and OEmsdisA cultures to stabilize the overexpression plasmids.
MsPDE exhibits PDE activity toward c-di-AMP in M. smegmatis
To explore the c-di-AMP phosphodiesterase activity of MsPDE in M. smegmatis, intracellular c-di-AMP levels of wild type (WT), Δmspde, ΔmsdisA, OEmspde and OEmsdisA strain were determined by LC-MS/MS. According to our results, intracellular c-di-AMP concentration declined significantly (~2-fold) in OEmspde strain (Fig. 7). Moreover, MsPDE exhibited high c-di-AMP-specific phosphodiesterase activity according to our biochemical studies (Figs. 2, 4 and 5). These results indicated that MsPDE was a functional c-di-AMP-specific phosphodiesterase both in vivo and in vitro. As c-di-AMP levels remained almost unchanged when mspde was deleted (Fig. 7A), but decreased drastically when mspde was overexpressed (Fig. 7B), another un-identified PDE might present in M. smegmatis.
Superfluous c-di-AMP boosts the morphologic change of M. smegmatis
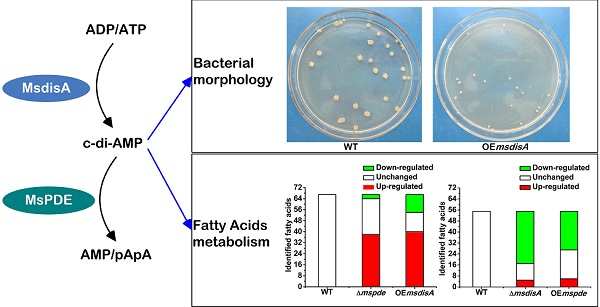
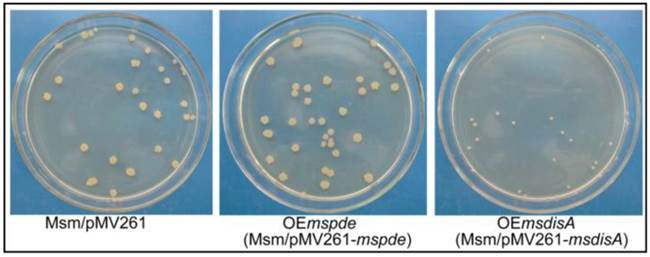
To construct the overexpression strains of msdisA and mspde, pMV261, pMV261-mspde and pMV261-msdisA plasmids were separately transferred into M. smegmatis competent cells by electroporation and cloned cells were plated on 7H10 agar. After three days incubation, an unexpected phenomenon was observed: the OEmsdisA formed small colonies with a convex and smooth appearance, while OEmspde was normal as the control strain (Msm/pMV261) (Fig. 8). The colonial morphology difference in OEmsdisA strain disappeared after continuous culture. This phenomenon was repeatable: after electrotransformation, small colonies appeared in the first generation of OEmsdisA strain and disappeared in continuous culture.
Overexpression of msdisA in M. smegmatis led to a series of abnormal phenotypes including cell expansion, bacterial aggregation and loss of motility [26]. In our study, we also observed the similar cell bulge phenotype through scanning electron microscopy analysis when msdisA was overexpressed (Supplementary Fig. S4). Moreover, the intracellular c-di-AMP level in OEmsdisA strain was sharply increased (~7-fold) (Fig. 7B). These tests indicated that superfluous c-di-AMP could lead to dysfunction of M. smegmatis, resulting in abnormal bacterial morphology.
MsPDE deficiency affects the intracellular concentrations of fatty acids in M. smegmatis
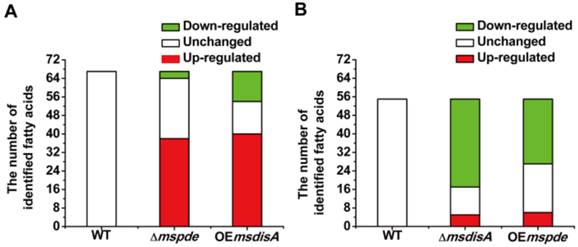
Fatty acids are the essential cellular components of mycobacteria and play an important role in bacterial survival and virulence [36, 37]. Zhang and his co-workers reported that c-di-AMP could regulate metabolism and transport of fatty acids through c-di-AMP-responsive repressor DarR in M. smegmatis [17]. We identified intracellular C7-C20 fatty acids of wild type (WT), △mspde, △msdisA, OEmspde and OEmsdisA strain by GC. Almost half of the identified fatty acids were up-regulated in Δmspde and OEmsdisA strain (Fig. 9A and Supplementary Table S2), while down-regulated in both △msdisA and OEmspde strains (Fig. 9A and Supplementary Table S3). The two group of strains, △mspde/OEmsdisA and △msdisA/OEmspde, exhibited a same tendency in fatty acid changes (Fig. 8 and Supplementary Tables S1-S2), indicating that mspde and msdisA played opposite function in regulating fatty acid concentrations. Among the fatty acids which exhibited significant change, the concentration of C12-C20 fatty acids, including saturated and unsaturated, were more likely to fluctuate in Δmspde and ΔmsdisA strain (Supplementary Tables S2-S3). These results further provided evidences that MsPDE and MsdisA play a cooperative role in controlling fatty acid levels.
c-di-AMP levels affect the colonial morphology of M. smegmatis. The first generation of OEmsdisA strain formed minute colonies with a convex and smooth appearance on 7H9 agar plates, while OEmspde strain was normal as the control strain.
Both mspde and msdisA affect the concentrations of intracellular fatty acids in M. smegmatis. (A) Determination of intracellular fatty acid concentrations of wild type, △mspde and OEmsdisA strains by GC. (B) Determination of intracellular fatty acid concentrations of wild type, △msdisA and OEmspde strains by GC.
DISCUSSION
MsPDE is a genuine c-di-AMP-specific PDE in M. smegmatis
In this study, we identified MsPDE as a genuine c-di-AMP specific-PDE in M. smegmatis. MsPDE is an ortholog of B. subtilis YybT. Comparing with YybT, which contains an extra transmembrane domain and a modified GGDEF domain involved in signal sensing, MsPDE only contains DHH-DHHA1 domain which is conserved in all the reported c-di-AMP specific-PDEs [7, 11, 33, 38-40]. Recently, a structural study on PDE revealed the molecular mechanism of its flexibility on substrates recognition [27]. Our study suggested that MsPDE could degrade c-di-AMP, pApA, c-di-GMP and pGpG, but not cAMP or cGMP. Moreover, c-di-AMP appears to be the preferred substrates as the MsPDE has a 200-fold higher hydrolytic efficiency (kcat/Km) to c-di-AMP than to c-di-GMP. We also provided evidences that overexpession of mspde in M. smegmatis resulted in direct modulation of c-di-AMP levels (Fig. 7B). Our results showed that in M. smegmatis, deletion of msdisA partially abolished bacterial C12-C20 fatty acid production, whereas deletion of mspde significantly enhanced C12-C20 fatty acid accumulation. Moreover, the fatty acid profile of Δmspde strain was similar to OEmsdisA strain, which was significantly up-regulated. It suggested that MsPDE and MsdisA had inverse function in fatty acid regulation.
Collectively, MsPDE is likely to function as the c-di-AMP PDE in vivo, albeit the substrates of MsPDE are various in vitro.
The function of DHH family proteins are versatile and various
All the reported c-di-AMP-specific PDEs belong to DHH family [7, 11, 33, 38-40], and the function of DHH family proteins is versatile and various. B. subtilis possesses four DHH family proteins: BsYybT, BsYtqI, BsRecJ and BsPPase. Only BsYybT was cyclic nucleotide PDE, hydrolyzing both c-di-AMP and c-di-GMP, and DHH domain was responsible for the catalytic activity [11]. BsYtqI and BsRecJ have both oligoribonuclease and pAp-phosphatase activities [41, 42], and BsPPase is a pyrophosphatase which provides thermal energy for many biosynthetic reactions [43]. B. thuringiensis, one of the relatives of B. subtilis, possesses three DHH family proteins (BtYybT, BtYtqI and BtRecJ). According to our research, both BtYybT and BtYtqI exhibited a c-di-AMP PDE activity (Supplementary Fig. S5A and B), but RecJ could not hydrolyze c-di-AMP (Supplementary Fig. S5C). Moreover, the hydrolysis products of BtYybT and BtYtqI were different when they were incubated with c-di-AMP. BtYybT hydrolyzed c-di-AMP into pApA (Supplementary Fig. S5A), but BtYtqI converted c-di-AMP directly into AMP (Supplementary Fig. S5B). Other than Bacillus, Mycobacterium species usually contain one DHH family protein. As a result, mycobacterial DHH proteins tend to be potential multifunctional proteins. DHH family proteins also function differently among Mycobacterium species. Although MsPDE shares significant identity with M. tuberculosis PDE, and both have cyclic nucleotide PDE activity, MsPDE appeared to be much more active (Supplementary Fig. S3). The wide substrates and functional diversity of DHH proteins contribute to rapid environmental adaptability of bacteria, and the overwhelming cyclic nucleotide PDE activity is likely to play a predominant role.
The regulatory mechanism of c-di-AMP in M. smegmatis is quite different from other bacteria
Our study also demonstrated that the regulatory mechanism of c-di-AMP in M. smegmatis is quite different from other bacteria. c-di-AMP is dominated in the regulation of various physiological processes in bacteria. So c-di-AMP is essential for many bacteria, and bacteria having DAC deletion cannot survive [11]. Like most bacteria, M. smegmatis contains only one DAC. MsdisA was reported to be the only functional DAC in M. smegmatis, sharing 84% identity to M. tuberculosis DAC MtdisA [13]. We further confirmed its DAC activity through HPLC analysis. MsDisA could synthesize c-di-AMP from two molecules of ATP or ADP in vitro (Supplementary Fig. S6). Interestingly, we successfully knocked-out msdisA from M. smegmatis genome using homologous recombination strategy. The msdisA mutant showed normal growth trend, as well as the same cell morphology with wild type (Supplementary Figs. S4 and S7). This indicated that msdisA deficiency only had a weak effect on survival of M. smegmatis.
In M. smegmatis, msdisA and the adjacent gene radA are in the same operon. RadA physically interacts with MsDisA and inhibits c-di-AMP synthesis activity of MsDisA [26]. The expression of radA was relatively high at logarithmic and stationary phases but down-regulated in death phase according to our RNA-seq data (data not shown). Furthermore, the activity of MtDisA, the homolog of M. tuberculosis MsDisA, is strongly inhibited by 1 mM ATP [13]. The intracellular concentration of ATP in mycobacteria is about 1 mM [44]. All the evidences suggest that the activity of MsDisA is suppressed to guarantee the normal growth of M. smegmatis. On the contrary, the high concentration of c-di-AMP will induce abnormal morphological phenotypes. However, what induces the re-activation of MsDisA, is still unknown.
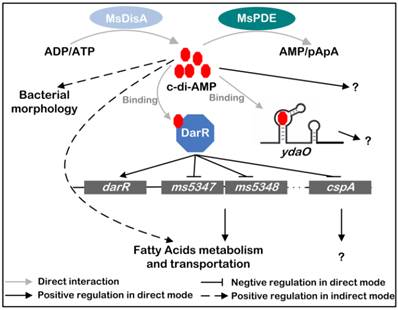
A summary of the regulatory network mediated by c-di-AMP in M. smegmatis
Based on previous and present studies, we delineate the regulatory network mediated by c-di-AMP in M. smegmatis (Fig. 10) [17, 24]. c-di-AMP is an important regulator which maintains normal growth of M. smegmatis and regulates the metabolism and transportation of fatty acids through DarR or some other undiscovered pathways. The predicted ydaO riboswitch, a newly identified c-di-AMP receptor [18, 20], is located in the 5'-UTR of rpfA gene coding for resuscitation-promoting factor A (RpfA) [45]. However, the link between c-di-AMP and ydaO, as well as the function of mycobacterial ydaO, is also obscure. The regulatory mechanism mediated by c-di-AMP should be further expanded in the future research. Moreover, tetrameric c-di-GMP was identified and reported to regulate Streptomyces development by controlling the dimerization of transcription factor BldD [46]. In a study of DAC in M. tuberculosis, multimeric c-di-AMP was identified by LC-ESI-MS in assay mixture [13], however, its presence and function within cell is still not reported. Moreover, a crystal structure of LmPC tetramer in complex with two c-di-AMP molecules was reported, and another c-di-AMP that bridged two c-di-AMP molecules bound to neighboring LmPC in crystal was observed [47]. If the bridging c-di-AMP is an artifact in that crystal form or naturally exists in cells is unknown. So it is worth to re-examine the binding strategy of c-di-AMP to its receptors in the future study.
Signaling network mediated by c-di-AMP in M. smegmatis. c-di-AMP is synthetized by DAC and hydrolyzed by PDE. It binds to its responsive proteins or RNAs and regulates various physiological processes in M. smegmatis. MsDisA, M. smegmatis DAC [26]; MsPDE, M. smegmatis c-di-AMP PDE; DarR, cyclic di-AMP receptor regulator [17]; ydaO, predicted c-di-AMP riboswitch [18, 20].
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31270105 and 31421064) and the National High-tech R&D Program of China (863 Program, grant 2011AA10A205). We especially thank Prof. Zhengguo He from Huazhong Agricultural University for providing us the M. smegmatis strain, the expression vector pMV261 and the vectors involved in gene knockout experiments.
SUPPLEMENTARY MATERIAL
Figures S1-S7 and Tables S1-S3.
CONFLICT OF INTERESTS
The authors have declared that no conflict of interest exists.
References
1. Gomelsky M. cAMP, c-di-GMP, c-di-AMP and now cGMP: bacteria use them all!. Mol Microbiol. 2011;79:562-5
2. Kalia D, Merey G, Nakayama S. et al. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev. 2013;42:305-41
3. Corrigan RM, Grundling A. Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 2013;11:513-24
4. Witte G, Hartung S, Buttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30:167-78
5. Witte CE, Whiteley AT, Burke TP, Sauer JD, Portnoy DA, Woodward JJ. Cyclic di-AMP is critical for Listeria monocytogenes growth, cell wall homeostasis, and establishment of infection. MBio. 2013;4:e00282-13
6. Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7:e1002217
7. Cho KH, Kang SO. Streptococcus pyogenes c-di-AMP phosphodiesterase, GdpP, influences SpeB processing and virulence. PLoS One. 2013;8:e69425
8. Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12:594-601
9. Zheng C, Wang J, Luo Y, Fu Y, Su J, He J. Highly efficient enzymatic preparation of c-di-AMP using the diadenylate cyclase DisA from Bacillus thuringiensis. Enzyme Microb Technol. 2013;52:319-24
10. Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stulke J. Cyclic di-AMP homeostasis in bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem. 2013;288:2004-17
11. Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285:473-82
12. Rao F, Ji Q, Soehano I, Liang ZX. Unusual heme-binding PAS domain from YybT family proteins. J Bacteriol. 2011;193:1543-51
13. Manikandan K, Sabareesh V, Singh N, Saigal K, Mechold U, Sinha KM. Two-step synthesis and hydrolysis of cyclic di-AMP in Mycobacterium tuberculosis. PLoS One. 2014;9:e86096
14. Ye M, Zhang JJ, Fang X. et al. DhhP, a cyclic di-AMP phosphodiesterase of Borrelia burgdorferi, is essential for cell growth and virulence. Infect Immun. 2014;82:1840-9
15. Huynh TN, Luo S, Pensinger D, Sauer JD, Tong L, Woodward JJ. An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc Natl Acad Sci U S A. 2015
16. Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc Natl Acad Sci U S A. 2013;110:9084-9
17. Zhang L, Li W, He Z-G. DarR, a TetR-like Transcriptional Factor, Is a Cyclic Di-AMP-responsive Repressor in Mycobacterium smegmatis. Journal of Biological Chemistry. 2013;288:3085-96
18. Ren A, Patel DJ. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat Chem Biol. 2014;10:780-6
19. Gao A, Serganov A. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat Chem Biol. 2014;10:787-92
20. Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9:834-9
21. Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328:1703-5
22. Jin L, Hill KK, Filak H. et al. MPYS is required for IFN response factor 3 activation and type I IFN production in the response of cultured phagocytes to bacterial second messengers cyclic-di-AMP and cyclic-di-GMP. J Immunol. 2011;187:2595-601
23. Parvatiyar K, Zhang Z, Teles RM. et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat Immunol. 2012;13:1155-61
24. Yang J, Bai Y, Zhang Y, Gabrielle VD, Jin L, Bai G. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol Microbiol. 2014;93:65-79
25. Bai Y, Yang J, Zhou X, Ding X, Eisele LE, Bai G. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS One. 2012;7:e35206
26. Zhang L, He ZG. Radiation-sensitive gene A (RadA) targets DisA, DNA integrity scanning protein A, to negatively affect cyclic Di-AMP synthesis activity in Mycobacterium smegmatis. J Biol Chem. 2013;288:22426-36
27. Srivastav R, Kumar D, Grover A. et al. Unique subunit packing in mycobacterial nanoRNase leads to alternate substrate recognitions in DHH phosphodiesterases. Nucleic Acids Res. 2014;42:7894-910
28. Yang M, Gao C, Cui T, An J, He ZG. A TetR-like regulator broadly affects the expressions of diverse genes in Mycobacterium smegmatis. Nucleic Acids Res. 2012;40:1009-20
29. Li W, He ZG. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res. 2012;40:11292-307
30. Burhenne H, Kaever V. Quantification of cyclic dinucleotides by reversed-phase LC-MS/MS. Methods Mol Biol. 2013;1016:27-37
31. Tighe SW, de Lajudie P, Dipietro K, Lindstrom K, Nick G, Jarvis BD. Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium species using the Sherlock Microbial Identification System. Int J Syst Evol Microbiol. 2000;50(Pt 2):787-801
32. Wallace RJ Jr, Nash DR, Tsukamura M, Blacklock ZM, Silcox VA. Human disease due to Mycobacterium smegmatis. J Infect Dis. 1988;158:52-9
33. Bai Y, Yang J, Eisele LE. et al. Two DHH subfamily 1 proteins in Streptococcus pneumoniae possess cyclic di-AMP phosphodiesterase activity and affect bacterial growth and virulence. J Bacteriol. 2013;195:5123-32
34. Yuan H, Liu XP, Han Z, Allers T, Hou JL, Liu JH. RecJ-like protein from Pyrococcus furiosus has 3'-5' exonuclease activity on RNA: implications for proofreading of 3'-mismatched RNA primers in DNA replication. Nucleic Acids Res. 2013;41:5817-26
35. Uemura Y, Nakagawa N, Wakamatsu T. et al. Crystal structure of the ligand-binding form of nanoRNase from Bacteroides fragilis, a member of the DHH/DHHA1 phosphoesterase family of proteins. FEBS Lett. 2013;587:2669-74
36. Lee W, VanderVen BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem. 2013;288:6788-800
37. Kowalski K, Trzepinski P, Druszczynska M, Boratynski J. [Mycolic acids--biological role and potential application in Mycobacterium detection and differentiation]. Postepy Hig Med Dosw (Online). 2014;68:350-8
38. Smith HE, Damman M, van der Velde J. et al. Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67:1750-6
39. Smith WM, Pham TH, Lei L. et al. Heat resistance and salt hypersensitivity in Lactococcus lactis due to spontaneous mutation of llmg_1816 (gdpP) induced by high-temperature growth. Appl Environ Microbiol. 2012;78:7753-9
40. Du B, Ji W, An H. et al. Functional analysis of c-di-AMP phosphodiesterase, GdpP, in Streptococcus suis serotype 2. Microbiol Res. 2014;169:749-58
41. Mechold U, Fang G, Ngo S, Ogryzko V, Danchin A. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007;35:4552-61
42. Wakamatsu T, Kim K, Uemura Y, Nakagawa N, Kuramitsu S, Masui R. Role of RecJ-like protein with 5'-3' exonuclease activity in oligo(deoxy)nucleotide degradation. J Biol Chem. 2011;286:2807-16
43. Halonen P, Tammenkoski M, Niiranen L. et al. Effects of active site mutations on the metal binding affinity, catalytic competence, and stability of the family II pyrophosphatase from Bacillus subtilis. Biochemistry. 2005;44:4004-10
44. James BW, Williams A, Marsh PD. The physiology and pathogenicity of Mycobacterium tuberculosis grown under controlled conditions in a defined medium. J Appl Microbiol. 2000;88:669-77
45. Rickman L, Scott C, Hunt DM. et al. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol Microbiol. 2005;56:1274-86
46. Tschowri N, Schumacher MA, Schlimpert S. et al. Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell. 2014;158:1136-47
47. Sureka K, Choi PH, Precit M. et al. The cyclic dinucleotide c-di-AMP is an allosteric regulator of metabolic enzyme function. Cell. 2014;158:1389-401
Author contact
![]() Corresponding author: Jin He, State Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, PR China. Phone: +86-27-87280670; Fax: +86-27-87280670. E-mail: hejinhzau.edu.cn
Corresponding author: Jin He, State Key Laboratory of Agricultural Microbiology, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, Hubei 430070, PR China. Phone: +86-27-87280670; Fax: +86-27-87280670. E-mail: hejinhzau.edu.cn

 Global reach, higher impact
Global reach, higher impact