10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2016; 12(9):1104-1113. doi:10.7150/ijbs.16064 This issue Cite
Research Paper
An Effective Molecular Target Site in Hepatitis B Virus S Gene for Cas9 Cleavage and Mutational Inactivation
1. Institute of Disease Control and Prevention, Academy of Military Medical Sciences, Beijing, China.
2. Lab of Blood-Borne Viruses, Beijing Institute of Transfusion Medicine, Beijing, China.
3. Transgenic Engineering Research Laboratory, Infectious Disease Center, Guangzhou 458 th Hospital, Guangzhou, China.
4. Research Center for Liver Failure, Beijing 302 nd Hospital, Beijing, China.
5. State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China.
6. Gladcan Consulting Company, Beijing, China.
7. Department of Surgery, University of Michigan, Ann Arbor, MI 48109, USA.
Received 2016-5-4; Accepted 2016-6-28; Published 2016-8-5
Abstract
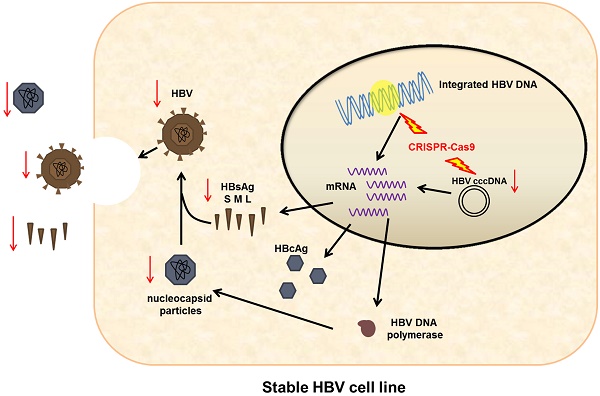
Chronic hepatitis B infection remains incurable because HBV cccDNA can persist indefinitely in patients recovering from acute HBV infection. Given the incidence of HBV infection and the shortcomings of current therapeutic options, a novel antiviral strategy is urgently needed. To inactivate HBV replication and destroy the HBV genome, we employed genome editing tool CRISPR/Cas9. Specifically, we found a CRISPR/Cas9 system (gRNA-S4) that effectively targeted the HBsAg region and could suppress efficiently viral replication with minimal off-target effects and impact on cell viability. The mutation mediated by CRISPR/Cas9 in HBV DNA both in a stable HBV-producing cell line and in HBV transgenic mice had been confirmed and evaluated using deep sequencing. In addition, we demonstrated the reduction of HBV replication was caused by the mutation of S4 site through three S4 region-mutated monoclonal cells. Besides, the gRNA-S4 system could also reduce serum surface-antigen levels by 99.91 ± 0.05% and lowered serum HBV DNA level below the negative threshold in the HBV hydrodynamics mouse model. Together, these findings indicate that the S4 region may be an ideal target for the development of innovative therapies against HBV infection using CRISPR/Cas9.
Keywords: HBV infection, CRISPR/Cas9
Introduction
Chronic infection of hepatitis B virus (HBV) is a major public health problem worldwide, with 350-400 million chronic HBV carriers and 0.5-1 million deaths a year as a result of liver cirrhosis, hepatocellular carcinoma, or HBV related liver failure [1]. China has about 93 million carriers despite the availability of highly effective vaccines against HBV [2], leading to an increased disease burden with treatment costs of approximately USD 16 billion annually[3]. Current antiviral treatment with nucleoside/nucleotide analogs (NAs) or interferon has failed to cure chronic hepatitis B infection, and the cessation of antiviral treatment may cause viremiare bound. Although NAs can effectively suppress HBV replication by suppressing the activity of viral reverse transcriptase, these drugs alone have little or no ability to eliminate replicative HBV templates, i.e., covalently closed circular DNA (cccDNA) [4, 5]. Moreover, NAs also lead to treatment failure because mutant strains are selected and propagate in quasi-species under drug pressure. Seroconversion is a more desirable outcome, because antibody response to HBsAg can lead to virus clearance and a functional cure [6]. Unfortunately, less than 10% of patients exhibit HBsAg loss after HBV-related treatment [7-9], and the rate of HBsAg clearance by interferon treatment is similarly low. Given the incidence of HBV infection and the shortcomings of current therapeutic options, developing new more effective treatments represents a global challenge [10, 11].
Over the last decade, novel genome-editing methods such as ZFNs and TALENs have been developed. Compared with them, CRISPR/Cas9 system requires only a simple design and operation process, which makes it an ideal tool for a wide range of applications [12-14]. The CRISPR/Cas9 system has been used to genome editing in many types of hosts, including human cells [15-17], several model organisms, and proviruses such as HIV [18, 19] and HPV [20], thus this system represents a potential means to radically cure chronic viral infection. Several applications of this technology to HBV research have also been reported [21-25]. In those studies, HBV-specific CRISPR/Cas9 systems effectively mediated gene disruption in HBV templates in expression vectors [22, 23] and HBV cccDNA [21, 24, 25]. It is interesting that a previous study has shown that some target sites on HBV DNA are particularly susceptible to Cas9 cleavage and HBV suppression [21-28]. Zhen et al. showed that gRNAs-S1 exhibited lower efficiency than gRNA-S2 in reducing HBsAg, even though both of them targeted HBV S region [29]. In this study, we found a specific site on HBV genome (S4), whose targeting by the gRNA/Cas9 reduced the levels of HBV surface-antigen and HBV DNA both in vitro and in vivo with minimal off-target effects. Moreover, we established stable S4 site-mutated HBV-infected cells, in which HBV replication was completely suppressed. Our results reveal that the S4 site in HBV genome may be an ideal target for Cas9 in developing innovative therapies against HBV infection.
Materials and Methods
Selection of HBV-specific gRNA
To find the potential target regions, we compared the HBV genome sequences of subtype's adr (GenBank accession number. HQ638218), ayw (U95551), ayr (NC003977), adw (EF103278), and adr (AB299858), and searched for potential 20-bp target sequences containing a 3′-downstream protospacer adjacent motif (PAM), NGG, in relatively conserved regions (Fig S1). Eight candidate target sequences were selected from them. These target sequences were subsequently inserted into dual-expression vectors with the U6 promoter. GRNA-S3, gRNA-S4, and gRNA-S5 were designed to target the HBV surface-antigen region and the ORF of HBV DNA polymerase; gRNA-SP-I and gRNA-SP-II were designed to target the promoter area of surface-antigen SP-I and SP-II, respectively; gRNA-XP was designed to target the promoter of X protein; and gRNA-CP-BCP and gRNA-CP-URR were designed to target the basic core promoter region (BCP) and upstream regulatory region (URR) at the promoter of the core antigen, respectively.
Plasmids
The gRNA/Cas9 dual-expression vector pSpCas9(BB)-2A-Puro (PX459) was obtained from Addgene (plasmid # 48139) through an MTA, and was constructed according to a previously described protocol [13]. Another gRNA/Cas9 dual-expression plasmid with both EGFP and Puro was ordered from View-solid Biotech (Beijing, China). The HBV-expression vector plasmid pGL3-HBV1.2 was obtained from Beijing Institute of Transfusion Medicine [30]. All plasmids were purified using the EndoFree Plasmid Maxi Kit (Qiagen, Germany).
Cell culture and transfections
HepG2.A64 (CCTCC C 201163), a HBV replicating stable cell line containing 1.1 copies of integrated HBV DNA (GenBank: HQ638218.1) and an entecavir (ETV)-resistance mutation, which was constructed by the 302nd Military Hospital of China [31] and maintained in medium containing 500 μg/ml G418. A total of 2 × 106 HepG2.A64 cells were cultured in a 10 cm plate for 24 h before transfection. The HBV-specific Cas9/gRNA expression vectors were transfected into HepG2.A64 cells by Lipofectamine LTX (Life Technologies). Puromycin was used to increase transfection efficiency (Fig S2). At 72 h after transfection, the transfected cells were inoculated into 24-well plates (4 × 104 cells per well) for subsequent analysis.
CCK-8 assay for cell viability
Viability of cells was assessed using the Cell Counting Kit-8 (Dojindo). Cells were inoculated into 48-well plates at densities ranging from 1 × 104 to 2 × 104 cells/well. CCK-8 and medium were mixed at a ratio of 1:10 per well in a clean 1.5 ml tube. The medium was discarded, and the cells were washed with PBS. Premixed CCK-8 and medium were added to 48-well plates and incubated at 37°C for 0.5-1 h. Absorbance at 450 nm (A450) was measured using an automatic detector (Thermo Scientific).
Immunofluorescence (IF) and Luciferase SSA recombination assay
HepG2.A64 cells were cultured in 24-well chamber slides for 1 day and fixed for 10 min in 4% (wt/vol) paraformaldehyde/PBS. After three rinses with PBS, the cells were treated with 0.5% Triton X-100/PBS for 20 min and blocked in 10% (vol/vol) goat serum for 1 h. Cells were incubated at 4°C overnight with anti-HBsAg antibody (Abcam). After three rinses, the cells were incubated for 1 h with FITC-conjugated secondary antibodies, and then incubated with Hoechst 33258 for 5 min. After three rinses with PBS, the cells were cover slipped with anti-fading aqueous mounting media (Beyotime Biotechnology) and analyzed under an Olympus BX61 fluorescence microscope. The SSA assay was performed as described previously [32]. In brief, a stop codon and a HBV-derived target site were inserted into a luciferase reporter. The insertion site located between two truncated luciferase fragments, and each of them has an 870bp homologous arm. The luciferase activity recovered when double-strand break (DSB) caused by CRISPR/Cas9 was repaired via homology-directed repair (HDR). The CRISPR/Cas9 plasmid with corresponding target, Renilla plasmid (5ng) and SSA reporter plasmid (30 ng) were co-transfected into HEK293T cells. The relative luciferase activity was tested by a dual-luciferase assay system (Promega) at the 2nd day after transfection.
Mice and hydrodynamic injection
pGL3-HBV1.2 (10 μg) and the gRNA/Cas9 dual-expression vector (10 μg) were co-injected into the tail veins of C57BL/6 mice (males, 4-6 weeks old), as described previously [30]. After injection, the C57BL/6 mice were regularly bled at the indicated time points to detect the HBV surface antigen and HBV DNA in serum. HBV transgenic (HBV-Tg) mice (8-10 weeks old) were established by Guangzhou 458th Hospital on BALB/c background. 1.3 copies of the HBV complete genome (subtype ayw) had been integrated into the chromosome of HBV-Tg mice, in which high levels of HBV DNA and HBsAg could be detected stably. Ten micrograms of gRNA-S4 was injected into HBV-Tg mice, and an equal amount of gRNA-empty was used as a control. Two weeks after injection HBV-Tg mice were sacrificed and liver tissue was taken.
Detection of HBV DNA and HBV surface antigen
HBV DNA inside the viral nucleocapsid was extracted as described previously [33]. To extract HBV cccDNA, total DNA was subjected to ATP-dependent DNase (PSAD). Then, rolling circle amplification (RCA) was performed to amplify circular DNA selectively. To rule out the possibility of genomic DNA contamination, specific primers of HBsAg and A1AT gene were used (Fig S3). Then, HBV cccDNA was amplified and quantitated using cccDNA-selective primers and a specific probe targeting the gap region of two direct repeat (DR1 and DR2 in HBV cccDNA[34]. HBsAg in cell supernatants and HBsAg in serum were determined using Elecsys HBsAg II (Roche) or enzyme linked immunosorbent assay (ELISA); data are reported in IU/ml or S/CO. The amounts of HBV DNA in serum and cell culture supernatants were measured using the Quantitative PCR Diagnostic Kit for HBV DNA (SinoMD).
Statistical analysis
Quantitative data were shown as the mean ± standard deviation of usually 3-5 independent experiments. Student's t-test was processed using the SAS software suite. P-values <0.05 or 0.01 were considered statistically significant.
Ethics Statement
All animal studies were performed in strict accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People's Republic of China. This study was approved by the ethics committee of the National Beijing Center for Drug Safety Evaluation and Research. All mice used for experiments were housed in cages in a controlled environment (22-25°C, 50% humidity, 12 h light/dark cycle) and were sacrificed by ether anesthesia.
Deep sequencing of CRISPR/Cas9 modified target S4 region
The S4 region in HBV genome was amplified by Phusion high-fidelity polymerase. The DNA samples and genomic DNA libraries were prepared by the NGS facility at the AUWIGENE Company (Beijing). Deep sequencing libraries were made as the Nextera protocol and were sequenced with paired-end 150-bp reads on Illumina MiSeq machine. The raw reads were mapped against the S4 region of HBV DNA (GenBank: HQ638218.1) using Burrows-Wheeler Aligner (BWA).
Results
Screening and optimization of an effective HBV-specific CRISPR/Cas9 system
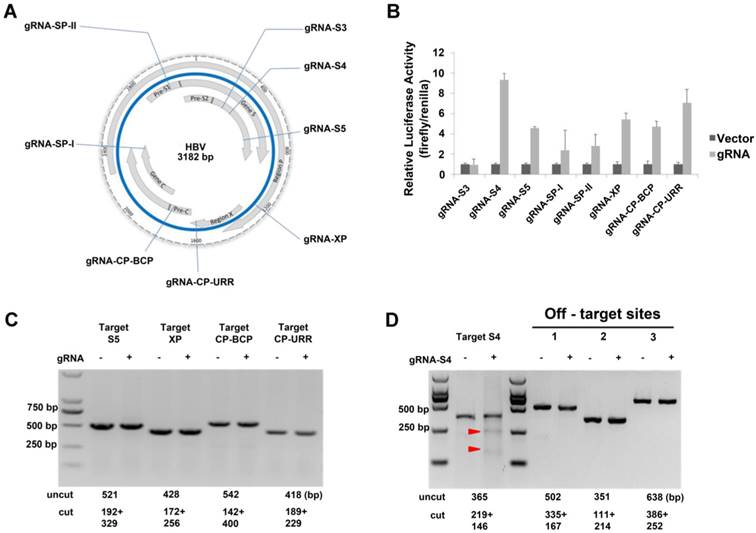
The HBV virion has four overlapping open reading frames (ORFs) that encode seven proteins: core protein, precore protein, viral polymerase, three forms of S antigen, and the X protein. We chose eight target sequences from conserved regions as gRNAs (Table 1, Fig S1) to construct the CRISPR/Cas9 systems. In some cases, these gRNAs targeted overlapping ORFs of the virus so that multiple viral RNAs would be simultaneously affected by a single gRNA system (Fig 1A). We examined the efficiency of the dual-expression vectors in 293T cells using the luciferase SSA recombination assay and it indicated that the gRNA-S4 gRNA-S5, gRNA-XP, gRNA-CP-BCP, and gRNA-CP-URR system could generated double-strand breaks (DSBs) efficiently (Fig 1B). To determine whether those cassettes could efficiently target HBV DNA in a stable HBV cell line, we conducted a T7EI assay[13] after transfecting the selected gRNAs into HepG2.A64, a stable HBV cell line containing 1.1 copies of integrated HBV DNA with an entecavir tablets (ETV)-resistance mutation. The result showed that gRNA-S4 system successfully introduced DSBs into the HBV genomes in HepG2.A64 (Fig 1D), whereas the other systems did not (Fig 1C, D). In addition, to analysis the off-target effect of gRNA-S4, we predicted 20 potential off-target sites for gRNA-S4 (Table S2) using a published prediction tool [35]. Then we amplified the top three potential off-target sites and the target-S4 locus and measured the DNA cleavage effects using the T7EI assay [16]. The result revealed ~11.3% indels at the target S4 locus in the gRNA-S4-treated cells. By contrast, there were no detectable indels in the 3 top-ranking off-target sites (Fig 1D), indicating the off-target effect of gRNA-S4 was below the detection limit of the assay.
Sequences of CRISPR gRNAs used in this study.
| Name | gRNA sequence (5'-3') | PAM | Nucleotide position (in U95551) |
|---|---|---|---|
| gRNA-S3 | GGACTTCTCTCAATTTTCTA | GGG | 263-285 |
| gRNA-S4 | GCTATCGCTGGATGTGTCTG | CGG | 368-390 |
| gRNA-S5 | CCATTTGTTCAGTGGTTCGT | AGG | 688-710 |
| gRNA-SP-I | TCGCAGAAGATCTCAATCTC | GGG | 2418-2440 |
| gRNA-SP-II | GGGTGGAGCCCTCAGGCTCA | GGG | 3042-3064 |
| gRNA-XP | CCTCTGCCGATCCATACTG | CGG | 1257-1278 |
| gRNA-CP-BCP | AAGCCTCCAAGCTGTGCCTT | GGG | 1868-1890 |
| gRNA-CP-URR | ATGTCAACGACCGACCTTG | AGG | 1682-1703 |
Identification of an effective HBV-specific CRISPR/Cas9 system. (A) Illustration of eight target sequences on the HBV genome. (B) SSA recombination assay of gRNA-Cas9 systems in 293T cells. The cleavage activity of the gRNA-S4 system was ~9-fold higher than that of the vector control. Error bars indicate SD, n = 3. (C) DNA extracted from HepG2.A64 cells transfected with gRNA-S5, gRNA-XP, gRNA-CP-BCP, and gRNA-CP-URR was analyzed by T7EI assay. None of these constructs could generate an apparent cleavage in target DNA. (D) Assessment of off-target cutting by gRNA-S4. T7EI assay in HepG2.A64 cells treated with gRNA-empty (-) and gRNA-S4 (+). gRNA-S4 and gRNA-S4 off-target sites 1, 2, and 3 were amplified using PCR. T7EI nuclease-cleaved gRNA PCR products are indicated by Red arrow heads. The sizes of cut and uncut bands are indicated below.
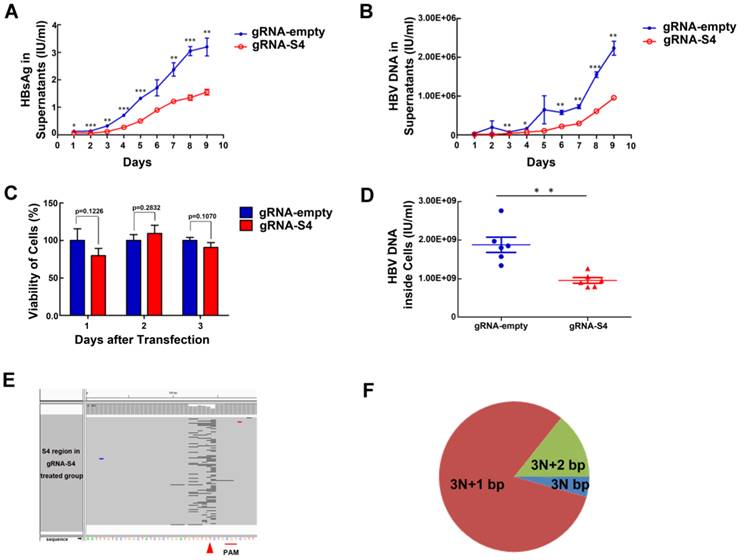
Inhibition of both HBV antigen expression and HBV replication by gRNA-S4 in HepG2.A64 cells. (A, B) Inhibition of HBsAg and HBV DNA in cell culture supernatant at the indicated times points after transfection of HepG2.A64 cells with gRNA-S4. (C) CCK-8 assay performed at the indicated time points after transfection (absorbance at 450 nm, A450); cell viability did not significantly differ between the gRNA-empty and gRNA-S4 samples on 3 successive days after transfection. (D) qPCR-based determination of concentrations of HBV DNA inside viral nucleocapsids on the 9th day after transfection. (E) Representative views of target S4 region deletion in gRNA-S4-treated cells. Black bars represent deletions. Arrowhead indicates the predicated cleavage site of Cas9. (F) The percentage of each indel type which calculated by the length of indels modulo 3.
GRNA-S4 system specifically inactivates HBV replication without affecting cell viability
GRNA-S4 expression cassettes target the region encoding the HBV surface antigens and DNA polymerase. To determine the knockdown efficiency of gRNA-S4 in HepG2.A64 immediately after efficient transfection, we used the Elecsys HBsAg IIsystem (Roche) to determine the amount of HBsAg in supernatants at various time points. HBsAg levels were significantly reduced after transfection with gRNA-S4 (Fig 2A). Also, compared with cells transfected with gRNA-empty vectors, the level of HBV DNA in HepG2.A64 cell culture supernatants decreased after transfection with the gRNA-S4 system (Fig 2B). In order to determine whether gRNA-S4 affected viral production via an effect on cell viability, we performed the CCK-8 assay at various time points after transfection. The results indicated that gRNA-S4 had no significant effect on cellular proliferation (Fig 2C). Next, to more sensitively detect HBV replication, we measured the amount of HBV DNA inside the viral nucleocapsid at the 9th day post-transfection by quantitative real-time PCR, which revealed that gRNA-S4 reduced HBV replication by 47.52 ± 10.78% relative to the gRNA-empty-treated control cells (Fig 2D). These results indicated that the HBV-specific gRNA-S4 system could reduce overall viral load in HBV stable cell lines. Also, to confirm that the mutation of the S4 region was caused by gRNA-S4, we performed deep sequencing of the S4 region in total DNA from cells transfected with HepG2.A64. Sequencing revealed that 10.76 ± 0.19% of sequencing reads from gRNA-S4-treated cells had insertion (Fig S4) or deletion (Fig 2E) (indel) mutations in the S4 region, versus 0.00 ± 0.00% in gRNA-empty-treated cells (n = 3, P < 0.001). In the gRNA-S4-treated cells, most of the mutations were predicted to lead to frame shifts, as inferred from the length of indels (Fig 2F).
Suppression of HBV replication in the mutated S4 region cell line
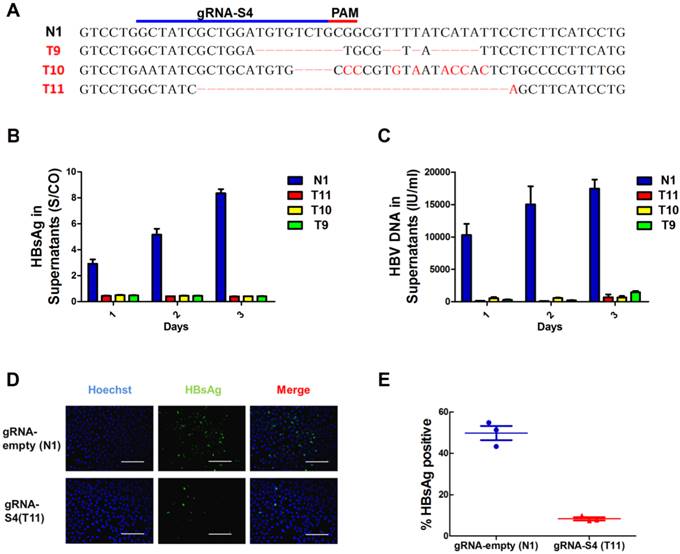
Next, to determine whether mutation of the S4 region could lead to inactivation of HBV replication, we established 4 monoclonal cell lines from HepG2.A64 transfected with gRNA-S4 and gRNA-empty, 3 of which—T9, T10 and T11contained different mutations in the target S4 region (Fig 3A) and monoclonal cell line N1 was established from HBV stable cell line transfected with gRNA-empty. The HBsAg levels were assessed by ELISA. As shown in Figure 3B and 3C, the supernatant HBsAg levels and the amount of supernatant HBV DNA levels markedly maintained below negative threshold (HBsAg < 1 S/CO, HBV DNA < 1000IU/ml) in both 3 mutated monoclonal cell lines during 3 days continuous cultivation. In order to measure a more detailed assessment, 9 consecutive days observation had been conducted on monoclonal cell T11 (Fig S5). The level of supernatant HBsAg in T11 cell clone was under the detection threshold (0.05 IU/ml) (Fig S5A), and the amount of supernatant HBV DNA level in gRNA-S4-treated group (T11) kept a very low level (792.67±504.11 IU/ml) during all those days (Fig S5B). Compared with gRNA-empty-treated group (N1) at the 9th day, the amount of supernatants HBV DNA in gRNA-S4-treated group decreased 99.95±0.01% (Fig S5B), HBV cccDNA decreased 95.37±0.64% (Fig S5C), replicable HBV DNA in cell decreased 99.87±0.25% (Fig S5D). Besides, immunofluorescence (IF) staining of T11 and N1 at the 9rd days revealed that gRNA-S4 exhibited an antiviral effect with a reduction of HBsAg in the cytoplasm (Fig 3D, E).
Suppression of HBV expression and replication in S4 region mutated cell lines. (A)Sanger sequencing results of S4 region of 4 individual monoclonal cell lines, which were separated from transfected HepG2.A64 cell lines, showing indels (red) introduced by gRNA-S4. (B, C) Titers of supernatants HBsAg and HBV DNA in those 4 monoclonal cell lines during three consecutive days' cultivation, the test results of HBsAg were always negative (<1S/CO) and the amount of HBV DNA is around or under the negative critical value in these three S4 site mutated monoclonal cells (T9, T10, T11, n=3). (D) Immunofluorescence analyses with anti-HBsAg antibody showed the high expression level of HBsAg in a gRNA-empty-treated cell clone (N1) and low level in a gRNA-S4-treated cell clone (T11). Scale bars represent 200 μm. (E) Results from a quantitative analysis of HBsAg-positive cells from three different micros.
Inhibition of HBV expression and replication in vivo by gRNA-S4
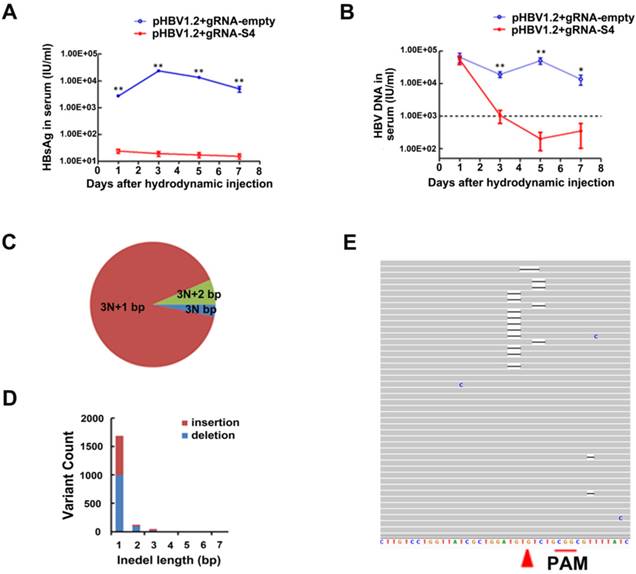
As a model of HBV persistence, HBV hydrodynamics mouse has been widely used [21-23, 26, 27, 30]. In this study, the HBV-expression vector pGL3-HBV1.2 and CRISPR/Cas9 expressing vector gRNA-S4 were co-injected into the tail veins of C57BL/6 mice by hydrodynamics. In the sera of gRNA-empty-treated group, the average concentration of HBsAg was 2.35×104 IU/ml at the 3rd day post injection (dpi) and continued to stay at high level during 7 days post injection. Notably, we found the level of HBsAg in serum were continually significantly lower (18.93 IU/ml on 3th dpi) among five independent mice receiving HBV-specific gRNA-S4 system after hydrodynamics (Fig 4A). The level of serum HBV DNA was quantified using qPCR and the average titer between two groups was 6.25×104 IU per milliliter at 1st dpi. However, serum HBV DNA level in gRNA-S4-treated group rapidly decreased to the negative value (<1000 IU/ml) and maintained below negative threshold since the 3rd dpi while the serum HBV DNA in gRNA-empty-treated group maintained a high level (>10000 IU/ml) during the 7 days (Figure 4B). To confirm the effect of the gRNA-S4 system in HBV transgenic mice (HBV-Tg mice), the CRISPR/Cas9 system was delivered via hydrodynamic tail-vein injection into the liver of adult HBV-Tg mice [36] and we also injected the gRNA-empty plasmid into an equal number of mice as a control. Afterward, liver genomic DNA was isolated two weeks later and the result of deep sequencing revealed 1.33 ± 0.31% indels in the S4 region of gRNA-S4-treated mice, versus 0.63 ± 0.01% in the gRNA-empty-treated group (n = 3, P < 0.05) (Fig 4C-E and Fig S4).
GRNA-S4-mediated mutations and inhibition of HBV replication in vivo. (A, B) The reduced efficiency of gRNA-S4 on serum HBsAg in HBV hydrodynamics mouse (A) The difference in serum HBsAg between the gRNA-empty- and gRNA-S4-treated groups. The differences between the two groups are significant (**P < 0.01, n = 5) on all days. (B) The level of serum HBV DNA in mice after injection of gRNA-empty control or the gRNA-S4 system (*P < 0.05, **P < 0.01, n = 5). The negative critical value is represented by a dashed line. (C-E) Representative target S4 indels in the liver of HBV-Tg mice two weeks after gRNA-S4/Cas9 injection. (C) The percentage of each indel type which calculated by the length of modulo 3. (D) Distribution of S4 indel length. (E) Representative views of target S4 region deletion in gRNA-S4-treated mice. Black bars indicate deletions. Arrowhead indicates the predicated cleavage site of Cas9.
Discussion
Many novel and efficient antiviral drugs are undergoing clinical trials, such as non-nucleoside antiviral ARC-520 from Arrowhead Research and immune-based drugs GS-9620 from Gilead Science. Although RNAi gene silencer ARC-520 [37] and TLR7-agonist GS-9620 [38] are able to reduce serum levels of HBsAg, HBeAg and HBV DNA, they fail to disrupt HBV cccDNA and HBV integrated DNA, both being the roots of HBV replication. The third genome-editing method, CRISPR/Cas9 system has been predicated to be a promising genome-editing tool to radically cure chronic hepatitis B viral infections because this system could disrupt HBV cccDNA with favourable effects [21, 24, 25]. In this study, we developed an effective CRISPR/Cas9 system (gRNA-S4) targeting the region of overlap between ORF S and P, which effectively suppressed markers of viral replication by disrupting HBV DNA both in HBV stable cell lines and in a HBV hydrodynamic mouse model. Using monoclonal cell lines, we further demonstrated that mutation of the S4 region could cause the inactivation of HBV replication. Moreover, we confirmed and evaluated the mutation caused by gRNA-S4 in the HBV stable cell line and HBV-Tg mice. Particularly worth mentioning was that we reduced serum surface-antigen levels by 99.91 ± 0.05%, significantly more than the largest reduction (93%) reported in previous studies [21]. Thus, the CRISPR/Cas9 system targeting the surface antigen region (S4) could be used as a gene therapy to reduce the inflammation and necrosis associated with HBV.
HBV is transmitted through the blood and infects hepatocytes in the liver. The sustained production of viral antigens especially HBsAg could cause inflammation and elevation of liver enzymes, which may result in hepatitis, cirrhosis, hepatocellular carcinoma, and liver failure [39]. Thus, the resultant reduction in surface-antigen production would also prevent inflammation and necrosis [39]. At present, no NA treatment is available to reduce HBsAg expression in patients [7-9], and the rate of HBsAg clearance by interferon treatment is similarly low. Besides, HBV DNA must be destroyed if a definitive and long lasting cure for HBV infection is to be obtained. Fortunately, three forms of the envelope protein HBsAg and the viral polymerase were encoded by the overlapping ORF where designed S4 target right located. Therefore, in this study, targeting S4 region could affect not only the synthesis of those three envelope proteins but the viral polymerase and it is plausible that the inflammation and necrosis may not be induced because of lacking of surface antigen [39]. Besides, generating mutation in polymerase region by gRNA-S4 could also affect the synthesis of viral polymerase and eliminates polymerase activity which may result in reduction of HBV replication. Interestingly, although the rate of indel at S4 site was 10.76 ± 0.19%, the percentage declines in HBV antigens was much more than it. It possibly because that HBV cccDNA had been effectively cleaved into a high percentage of linear DNAs which is not repaired but destroyed [26]. Moreover, the cell model we use in this study has ETV resistance mutation, thus, it also indicates that the CRISPR/Cas9 system is not only a perfect strategy for eradicating the HBV but also a simple pathway to overcome drug resistance.
In a previous study, a HBV-specific CRISPR/Cas9 system effectively mediated gene disruption on HBV templates in an expression vector [22]. In this study, we designed eight different anti-HBV gRNA systems; however, most of them were insufficiently effective in the HBV stable cell line HepG2.A64 using the T7EI assay (although they were effective in 293T cells, as demonstrated by the SSA assay). It indicates that not all HBV-target gRNAs can disrupt and inactivate the HBV genome efficiently as reported in previous studies [21-28]. Besides, considering most of the less effective targets are located in the promoter regions and conserved regions, we hypothesize that disruption of RNA polymerase-binding sites, or some structures within conserved regions, can lead to competitive inhibition of gRNA/Cas9. Thus, it indicates that the S4 region is an effective CRISPR/Cas9 target site on HBV genome. Specifically, we found that the cell lines carrying the CRISPR/Cas9 mediated S4-mutation had much lower levels of supernatant HBsAg (Fig 3B), supernatant HBV DNA (Fig 3C), HBV cccDNA (Fig S5C) and replicable HBV DNA in the viral nucleocapsid (Fig S5D), compared with the wild type. One potential mechanism is that the transcription of mutated HBV DNA into pre-genomic RNA, its export to the cytoplasm, and its packaging into virions could result in cccDNA that is defective being a template for the HBV expression. Still, more efficient cleavage targets on HBV genome are expected. Fortunately, a gDNA guided NgAgo system has been found suitable for genome editing in human cells [40]. This system has lower off-target effect and does not require any PAM sequence, which can be used to select more efficient targets in the future.
In HBV-Tg mice, we delivered the gRNA-S4 system to the liver of adult HBV-Tg mice using hydrodynamic tail-vein injection and found only 1.33 ± 0.31% indels in the corresponding target site in mice liver at 14 dpi using deep sequencing. This could have been due to the low transfection efficiency, which may not significantly lowered the levels of surface antigen in serum; thus, for HBV gene therapy using CRISPR/Cas9 system, it will be necessary to develop a more efficient delivery system [41]. As HBV infected liver cells in HCC patients, the stable HBV cell line also contains many different HBV replication templates such as HBV cccDNA and many integrated subgenomic HBV DNA fragments. Although we introduced mutations in HBV genome and suppressed the expression of HBV, there are still many integrated HBV DNA fragments on the chromosomes which play an important role in carcinogenesis [42] and we still could not remove them all. It largely limited the prospect of developing a fundamental therapeutic method of chronic HBV infection through CRISPR/Cas9. Therefore, more researches about the mechanisms of HBV integration and HBV pathogenesis are still needed.
Abbreviations
HBV: hepatitis B virus; NAs: nucleoside/nucleotide analogs; cccDNA: covalently closed circular DNA; CRISPR: clustered regularly interspaced short palindromic repeat; gRNA: guide RNA; PAM: protospacer adjacent motif; BCP: basic core promoter region; URR: upstream regulatory region; SSA: single-strand annealing; DSBs: double-strand breaks; HBS: hepatitis B surface antigen; indel: insertion or deletion; dpi: days post-injection; HBV-Tg: HBV transgenic; SD: standard deviation.
Supplementary Material
Supplementary tables and figures.
Competing Interests
The authors declare that they have no competing interests.
Acknowledgements
This work was supported by grants from the Mega-Projects of Science and Technology Research (nos. 2013ZX10004607, 2012ZX10004503), the National Nature Science Foundation of China (nos. 81371854), Beijing Nature Science Foundation (7162145), the Beijing Science and Technology Nova program (no.xx2013061) and the Innovation Foundation of AMMS (no. 2015CXJJ26). We thank Dayang Zou, Zhihui Xu, Qi Li, and Chenchen Ji for sharing reagents and for discussions. We thank the Research Center for Liver Failure, Beijing 302nd Hospital, Lab of Blood-Borne Viruses, Beijing Institute of Transfusion Medicine, and the Transgenic Engineering Research Laboratory, Infectious Disease Center, 458th Hospital for technical support.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Kao JH, Chen DS. Global control of hepatitis B virus infection. The Lancet Infectious diseases. 2002;2:395-403
2. Liaw YF, Chu CM. Hepatitis B virus infection. Lancet. 2009;373:582-92
3. Ren JJ, Liu Y, Ren W, Qiu Y, Wang B, Chen P. et al. Role of general practitioners in prevention and treatment of hepatitis B in China. Hepatobiliary & pancreatic diseases international: HBPD INT. 2014;13:495-500
4. Dandri M, Burda MR, Will H, Petersen J. Increased hepatocyte turnover and inhibition of woodchuck hepatitis B virus replication by adefovir in vitro do not lead to reduction of the closed circular DNA. Hepatology. 2000;32:139-46
5. Moraleda G, Saputelli J, Aldrich CE, Averett D, Condreay L, Mason WS. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. Journal of virology. 1997;71:9392-9
6. Yuen MF, Lai CL. Treatment of chronic hepatitis B: Evolution over two decades. Journal of gastroenterology and hepatology. 2011;26(Suppl 1):138-43
7. Heathcote EJ, Marcellin P, Buti M, Gane E, De Man RA, Krastev Z. et al. Three-year efficacy and safety of tenofovir disoproxil fumarate treatment for chronic hepatitis B. Gastroenterology. 2011;140:132-43
8. Funk ML, Rosenberg DM, Lok AS. World-wide epidemiology of HBeAg-negative chronic hepatitis B and associated precore and core promoter variants. Journal of viral hepatitis. 2002;9:52-61
9. Gish RG, Chang TT, Lai CL, de Man R, Gadano A, Poordad F. et al. Loss of HBsAg antigen during treatment with entecavir or lamivudine in nucleoside-naive HBeAg-positive patients with chronic hepatitis B. Journal of viral hepatitis. 2010;17:16-22
10. Sharon A, Chu CK. Understanding the molecular basis of HBV drug resistance by molecular modeling. Antiviral research. 2008;80:339-53
11. Shaw T, Bartholomeusz A, Locarnini S. HBV drug resistance: mechanisms, detection and interpretation. Journal of hepatology. 2006;44:593-606
12. Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: progress, implications and challenges. Human molecular genetics. 2014;23:R40-6
13. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281-308
14. Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173-83
15. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE. et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823-6
16. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-23
17. Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013;31:230-2
18. Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F. et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11461-6
19. Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Scientific reports. 2013;3:2510
20. Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochemical and biophysical research communications. 2014;450:1422-6
21. Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY. et al. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene therapy. 2015
22. Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC. et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Molecular therapy Nucleic acids. 2014;3:e186
23. Liu X, Hao R, Chen S, Guo D, Chen Y. Inhibition of Hepatitis B Virus by CRISPR/Cas9 System via Targeting the Conserved Regions of Viral Genome. The Journal of general virology. 2015
24. Kennedy EM, Bassit LC, Mueller H, Kornepati AV, Bogerd HP, Nie T. et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2015;476:196-205
25. Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Molecular therapy Nucleic acids. 2014;3:e216
26. Dong C, Qu L, Wang H, Wei L, Dong Y, Xiong S. Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral research. 2015;118:110-7
27. Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A. et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Scientific reports. 2015;5:10833
28. Wang J, Xu ZW, Liu S, Zhang RY, Ding SL, Xie XM. et al. Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World journal of gastroenterology: WJG. 2015;21:9554-65
29. Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY. et al. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene therapy. 2015;22:404-12
30. Liang SQ, Du J, Yan H, Zhou QQ, Zhou Y, Yuan ZN. et al. A mouse model for studying the clearance of hepatitis B virus in vivo using a luciferase reporter. PloS one. 2013;8:e60005
31. Wei-ming Y, Yan L, Lan-lan S, Zhi-hui X, Peng-gao L, Lv-ping B. et al. Inhibitory effect of herbal concoction Suduxing on hepatitis B virus replication in vitro (in Chinese). Med J Chin PLA. 2014;39:4
32. Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nature biotechnology. 2011;29:699-700
33. Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. Journal of virology. 1995;69:6158-69
34. Zhong Y, Han J, Zou Z, Liu S, Tang B, Ren X. et al. Quantitation of HBV covalently closed circular DNA in micro formalin fixed paraffin-embedded liver tissue using rolling circle amplification in combination with real-time PCR. Clinica chimica acta; international journal of clinical chemistry. 2011;412:1905-11
35. Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology. 2013;31:827-32
36. Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene therapy. 1999;6:1258-66
37. Wang XY, Chen HS. Emerging antivirals for the treatment of hepatitis B. World journal of gastroenterology: WJG. 2014;20:7707-17
38. Lanford RE, Guerra B, Chavez D, Giavedoni L, Hodara VL, Brasky KM. et al. GS-9620, an oral agonist of Toll-like receptor-7, induces prolonged suppression of hepatitis B virus in chronically infected chimpanzees. Gastroenterology. 2013;144:1508-17 17 e1-10
39. Wooddell CI, Rozema DB, Hossbach M, John M, Hamilton HL, Chu Q. et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Molecular therapy: the journal of the American Society of Gene Therapy. 2013;21:973-85
40. Gao F, Shen XZ, Jiang F, Wu Y, Han C. DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nature biotechnology. 2016
41. Schiffer JT, Swan DA, Stone D, Jerome KR. Predictors of hepatitis B cure using gene therapy to deliver DNA cleavage enzymes: a mathematical modeling approach. PLoS computational biology. 2013;9:e1003131
42. Brechot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:S56-61
Author contact
![]() Corresponding authors: Prof. Hongbin Song, Institute of Disease Control and Prevention, Academy of Military Medical Science, No. 20 East Street, Fengtai district, Beijing 100071, China. E-mail address: songhbac.cn; Phone/Fax: +86-10-66948475; Dr. Shaofu Qiu, Institute of Disease Control and Prevention, Academy of Military Medical Science, No. 20 East Street, Fengtai district, Beijing 100071, China; e-mail address: qiushf0613com; or Prof. Qiao Li, Department of Surgery, University of Michigan, Ann Arbor, MI 48109, USA. Phone:734-615-1977, Email: qiaoliedu
Corresponding authors: Prof. Hongbin Song, Institute of Disease Control and Prevention, Academy of Military Medical Science, No. 20 East Street, Fengtai district, Beijing 100071, China. E-mail address: songhbac.cn; Phone/Fax: +86-10-66948475; Dr. Shaofu Qiu, Institute of Disease Control and Prevention, Academy of Military Medical Science, No. 20 East Street, Fengtai district, Beijing 100071, China; e-mail address: qiushf0613com; or Prof. Qiao Li, Department of Surgery, University of Michigan, Ann Arbor, MI 48109, USA. Phone:734-615-1977, Email: qiaoliedu

 Global reach, higher impact
Global reach, higher impact