10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(1):85-99. doi:10.7150/ijbs.17390 This issue Cite
Research Paper
Metadata Analysis of Phanerochaete chrysosporium Gene Expression Data Identified Common CAZymes Encoding Gene Expression Profiles Involved in Cellulose and Hemicellulose Degradation
Department of Biology, Lakehead University, 955 Oliver Road, Thunder Bay, Ontario, P7B 5E1, Canada.
Abstract
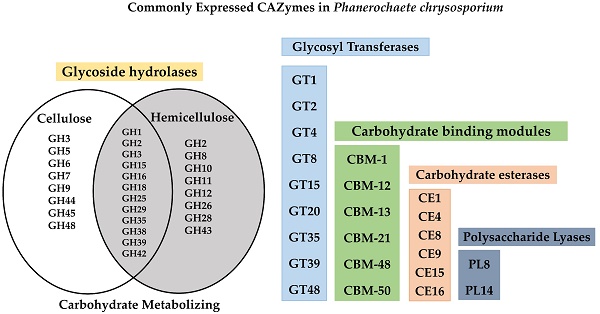
In literature, extensive studies have been conducted on popular wood degrading white rot fungus, Phanerochaete chrysosporium about its lignin degrading mechanisms compared to the cellulose and hemicellulose degrading abilities. This study delineates cellulose and hemicellulose degrading mechanisms through large scale metadata analysis of P. chrysosporium gene expression data (retrieved from NCBI GEO) to understand the common expression patterns of differentially expressed genes when cultured on different growth substrates. Genes encoding glycoside hydrolase classes commonly expressed during breakdown of cellulose such as GH-5,6,7,9,44,45,48 and hemicellulose are GH-2,8,10,11,26,30,43,47 were found to be highly expressed among varied growth conditions including simple customized and complex natural plant biomass growth mediums. Genes encoding carbohydrate esterase class enzymes CE (1,4,8,9,15,16) polysaccharide lyase class enzymes PL-8 and PL-14, and glycosyl transferases classes GT (1,2,4,8,15,20,35,39,48) were differentially expressed in natural plant biomass growth mediums. Based on these results, P. chrysosporium, on natural plant biomass substrates was found to express lignin and hemicellulose degrading enzymes more than cellulolytic enzymes except GH-61 (LPMO) class enzymes, in early stages. It was observed that the fate of P. chrysosporium transcriptome is significantly affected by the wood substrate provided. We believe, the gene expression findings in this study plays crucial role in developing genetically efficient microbe with effective cellulose and hemicellulose degradation abilities.
Keywords: Phanerochaete chrysosporium, Transcriptome, Lignocellulose, Gene Expression Omnibus (GEO), GEO2R, Bioconductor, Carbohydrate Active Enzyme database (CAZy).

 Global reach, higher impact
Global reach, higher impact