Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2017; 13(5):615-629. doi:10.7150/ijbs.17051 This issue Cite
Review
Complex Relationship between Obesity and the Fat Mass and Obesity Locus
1. Key Research Center of Liver Regulation for Hyperlipidemia SATCM/Class III Laboratory of Metabolism SATCM, Guangdong TCM Key Laboratory for Metabolic Diseases, Guangdong Pharmaceutical University, Guangzhou, 510006, China;
2. Inorganic Chemistry Laboratory, Oxford University, South Parks Road, OX1 3QR, United Kingdom;
3. Guangzhou Boxabio Technology Ltd, Guangzhou Science City, P R China.
Received 2016-7-31; Accepted 2017-3-24; Published 2017-5-15
Abstract
In the 21st century, obesity has become a serious problem because of increasing obese patients and numerous metabolic complications. The primary reasons for this situation are environmental and genetic factors. In 2007, FTO (fat mass and obesity associated) was the first gene identified through a genome-wide association study (GWAS) associated with obesity in humans. Subsequently, a cluster of single nucleotide polymorphisms (SNPs) in the first intron of the FTO gene was discovered to be associated with BMI and body composition. Various studies have explored the mechanistic basis behind this association. Thus, emerging evidence showed that FTO plays a key role regulating adipose tissue development and functions in body size and composition. Recent prevalent research topic concentrated in the three neighboring genes of FTO: RPGRIP1L, IRX3 and IRX5, as having a functional link between obesity-associated common variants within FTO and the observed human phenotypes. The purpose of this review is to present a comprehensive picture of the impact of FTO on obesity susceptibility and to illuminate these new studies of FTO function in adipose tissue.
Keywords: FTO, obesity, adipogenesis, Rpgrip1l, IRX3, IRX5.
Introduction
According to the definition from the World Health Organization (WHO), overweight and obesity are likely to damage health due to excessive body fat and/or abnormal accumulation. In the clinic, body mass index (BMI) and waist circumference (WC) are measured directly by measuring the external features of the body. BMI is the most important index for the diagnosis of obesity. WC can reflect the degree of accumulation of abdominal fat. The current classification standard of the WHO for obesity is listed below (Table 1). Obesity has become a global problem of epidemic proportions in most emerging economies [1]. The latest figures from the World Health Organization (WHO) provide a global estimate of 1.4 billion overweight individuals. Today, an important concern is the devastating socioeconomic impact of obesity and related metabolic disturbances [2, 3]. A growing number of researchers have devoted themselves to the study of the molecular mechanisms of obesity and obesity-related diseases, with the purpose of ascertaining the real root of obesity to develop reasonable and effective therapies. Our research group has been devoted to exploring the effect and mechanism of chitosan and its degradation products in the treatment of obesity to alleviate the symptoms of obesity [4-12]. According to the relevant literature, approximately 60% of obesity susceptibility is caused by the difference between genotypes [13, 14]. Due to the rapid development of genomics, GWAS has confirmed 300 common human traits and diseases related to approximately 2,000 genes, which contain at least 75 obese genes [15, 16]. Among them, the FTO gene is described as the first identified gene that has a strong association with obesity susceptibility by GWAS [17, 18]. Since 2007, more studies have confirmed the association between SNPs in intron 1 of FTO and BMI in European populations, specifically, Asian populations [19], Africans [20] and Native Americans [21], clearly revealing that the influence of FTO SNPs on obesity is a worldwide epidemiology. This locus is an important gene that contributes to obesity throughout the lifespan and among different ethnic groups. In this paper, we reviewed the recent development of FTO as an obesity gene to illuminate the biological functional mechanisms from many studies that were based on the association between FTO and obesity. We selected the studies on the latest progress in the authoritative journals and others high quality paper with the high number (The data come from Web of Science: the number of citations times is about 21 to 170) of citations in general journals using PubMed database in the past 7 years. At the same time, we also selected the relevant literature about each perspective at home and ±abroad, for horizontal comparison.
Grading standard of WHO on obesity
| Grading standard( Unit: kg/m2) | |
|---|---|
| Underweight | BMI<18.5 |
| Normal Weight | 18.5≤BMI<25.0 |
| Overweight | 25.0≤BMI<30.0 |
| Obesity | 30.0≤BMI<35.0 |
| Severe obesity | 35.0≤BMI<40.0 |
| Morbid obesity | BMI≥40 |
Discovery of FTO
FTO was first identified in Fused toes (Ft) mice in 1999, when it was first cloned by exon trap analysis, which was performed to assess the likelihood a partial deletion in Ft mutants [22]. This gene is expressed throughout embryonic development, and variations in human chromosome 16 in the q12.2 region are associated with the development of anisomastia, dysmorphic facies, mental retardation, obesity and other phenotypes. This area includes a plurality of genes [23]. Subsequently, FTO has been identified as a candidate gene that causes programmed cell death, craniofacial development and the establishment of left-right asymmetry [24]. In 2007, researchers using genomic data from 38,759 European participants, including a population with type 2 diabetes and a control population, found that rs9939609 was representative of some SNPs in the first intron of FTO that were associated with obesity and overweight. In addition, the A allele of rs9939609 exhibited a significant correlation with type 2 diabetes [17]. In this article, approximately 16% of subjects were homozygous for the risk allele of FTO, and their weight was 3 kilograms greater than that of non-risk allele subjects, with a 1.67-fold increased risk for obesity compared with non-risk allele subjects [17]. Obesity is one of the leading causes of type 2 diabetes, so researchers have hypothesized that FTO may be linked to obesity. Subsequent studies have confirmed that a large number of FTO gene polymorphisms are also associated with differences in body mass index (BMI) [25], obesity [26], insulin resistance [27], diabetes (DM), metabolic syndrome [28], hypertension [29], atherosclerosis [30] and C-reactive protein levels [31] (Table 1). Because it was the first gene identified by GWAS to be closely related to obesity, the gene was officially named the fat mass and obesity-associated gene.
FTO in different species
The FTO gene has a long history that can be traced back 450 million years; sequence homology comparisons have revealed that the FTO gene exists in vertebrates, Ostreococcus and Diatoms [45]. FTO genes are widely distributed in the biological world and expressed in a variety of animal tissues. FTO is expressed throughout embryonic and adult tissues, including wide expression in human tissues at different developmental stages. High expression is observed in adipose tissue and skeletal muscle [17], and the highest expression is noted in the area of hypothalamic [46] control of the energy balance, namely, the arcuate nucleus, suggesting that it plays a key role in regulating appetite and energy metabolism.
The nucleotide and amino acid sequences of the FTO gene and protein are highly conserved in different species. The human FTO gene is located on chromosome 16ql2.2 [17], and the mouse FTO gene is located on chromosome 8. Both contain nine exons [22]. Homogeneous analysis demonstrated that these genes exhibit approximately 87% homology at the protein sequence level [24, 47]. In addition, the FTO gene of the pig is located on chromosome 6, which is where many quantitative trait loci for the regulation of fat have been mapped [48]. Homology analysis revealed that the cDNA sequence of FTO in the pig shared 88% and 83% homology with the cDNA sequences of mice and human, respectively [49, 50].
Discovery and structure of the FTO protein
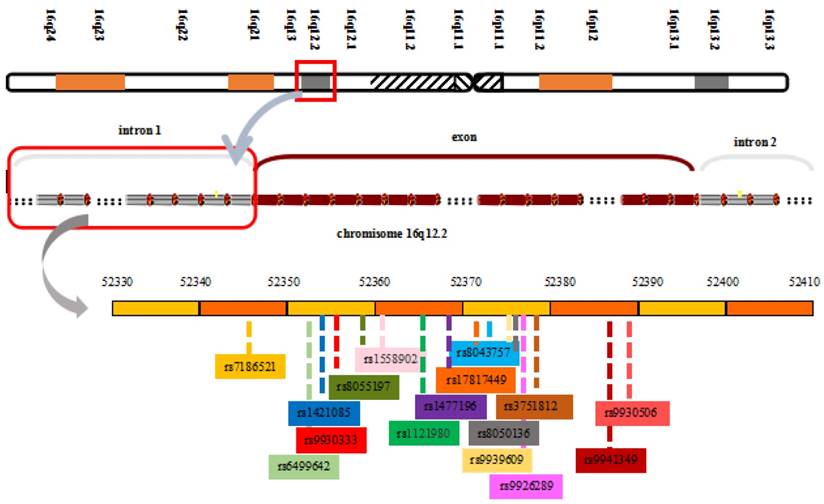
The FTO locus, which spans approximately 417,979 base pairs on human chromosome 16q12.2, from 53,737,875 bp to 54,155,853 bp, contains 9 exons and encodes proteins [17, 46]. The strongest genome-wide association signals lie in introns 1 and 2 of the FTO gene. This sequence includes 89 common variants, which makes confirmation of the causes of variations more difficult [51]. The transcription start site of KIAA1005 lies only 200 base pairs from the 5′ end of the FTO gene and ~61 kb from the 47-kb interval containing the BMI associations (Figure 1) [17]. Based on individual differences in SNPs, the risk alleles in the interval also vary. Some common variants within the first intron of the FTO gene include rs9926289 (G/A), rs9939609, rs8050136, rs9930501 (A/G), rs9930506 (A/G), rs17817449, rs9940646 (C/G), rs9940128 (G/A), rs1421085, and rs8043757 (A/T). These polymorphisms have a strong linkage disequilibrium in all subjects and are associated with increased body weight, body fat, BMI, waist circumference, hip circumference and energy intake [17, 52-56]. The FTO protein belongs to the AlkB family of non-home Fe (II)/dioxygenases, which is part of the Escherichia coli AlkB and human ABH enzyme family [46, 57]. Chinese researchers were the first to analyze the crystal structure of the FTO protein, and the structure further proves that the protein is a type of deoxyribonucleic acid (DNA) demethylase [58]. The crystal structure of FTO reveals an N-terminal AlkB-like catalytic domain and a novel C-terminal domain, which is necessary for FTO catalytic activity. In addition, FTO possesses a specific loop covering one side of the conserved jelly roll motif, which selectively competes with the unmethylated strand of the DNA duplex for binding to FTO. The loop potentially plays an important role in FTO to select against double-stranded nucleic acids [59]. FTO selects 3-meT or 3-meU from other nucleotides through a hydrogen-bonding interaction with the two carbonyl oxygen atoms in 3-meT or 3-meU. A histidine and an aspartate bind tightly to Fe(II) in the catalytic site, which includes five obligate amino acid residues.
View of the FTO chromosomal location and obesity-associated region with associated common SNPs in the first intron of FTO
FTO variants and BMI modified by physical activity
The obesity epidemic poses a considerable public health threat and is mainly due to one's lifestyle, such as a lack of exercise, sleeplessness and excessive energy intake [60] (Table 3). Numerous preventive strategies have been proposed to control the obesity epidemic [61]. Previous data have consistently demonstrated that the relationship between FTO SNPs and BMI can be modified by physical activity in adults, which has been strongly demonstrated in numerous reported and unreported meta-analyses from 45 studies of 218,166 adults and nine studies of 19,268 children and adolescents [62]. No such interaction was found in children and adolescents. This finding might be attributed to the inaccuracy of BMI as a method of measurement of obesity in young age groups [63]. In another study, adolescents spent at least 1 hour partaking in moderate or intensive physical activity every day, and higher BMI or waist circumference were not affected by the A allele, which is in contrast with that noted in inactive subjects [64]. However, current studies have reported that genetic predisposition for obesity can be modified by lifestyle factors, such as physical activity (PA). A meta-analysis revealed that FTO rs9939609 as a risk allele homozygous SNP can significantly increase the susceptibility to obesity compared with inactive subjects and can lead to a weight reduction of approximately 30% in PA [62]. This meta-analysis of the observations of many individual studies emphasized the association between physical activity and FTO in adults and adolescents. A significant connection between variations in the first intron of FTO and PA has been demonstrated by numerous studies performed with children and adult populations of European, East Asian and African ancestry [65-67]. Another study revealed a significant reduced effect of rs1121980 on both BMI and waist circumference in a group of physically active individuals compared with an inactive group composed of 20,374 European adults [68]. The association with rs1421085 and PA was also observed in a prospective cohort that included approximately 17,400 people from 17 countries in six ethnic groups, including South Asian, European, East Asian, Latin American, African and Native American, who were followed for three years [69]. Fourteen obesity-susceptibility genes were analyzed in this survey, and PA can greatly alleviate the genetic effect of FTO. This interaction was analyzed prospectively using a quantitative measure of PA compared with the recently developed BAI and BMI. Analyzing the impact of obesity-predisposing SNPs on PA revealed novel associations; however, further studies are needed to confirm these effects. The combination of genetic predisposition and environment factors led to variations of the body composition associated with the FTO genotype. An active lifestyle can significantly reduce the genetic effect of FTO on body weight, and this conclusion was applicable to different people. In sum, these data and the results showed that vigorous PA could prevent obesity caused by the powerful gene group, which may be a valuable contribution to the prevention of obesity as well as the global fight against obesity [70]. Moreover, some studies have demonstrated that dietary habits [71] [72, 73] and smoking [74] might also impair the influence of FTO on obesity susceptibility.
Association between FTO SNPs and related diseases
| FTO single nucleotide polymorphisms | Date | Study samples | region | Related diseases | Ref. |
|---|---|---|---|---|---|
| rs1421085 (T/C) | 2013 | 2,077 T2DM patients and 3,085 healthy people (clinic-based) | European | decreased insulin sensitivity index (P = 0.007) T2DM ( P = 0.001) | [32] |
| 2013 | 10447 men and women (population-based) | African American and European | decline in cognitive function, assessed by performance on the Delayed Word Recall Test (P = 0.002) | [33] | |
| rs9939609(T/A) | 2011 | 3,919 T2D patients and 4,172 healthy people (clinic-based) | South Asian | increased chances of T2DM ( P = 10-8) | [34] |
| 2012 | 2,858 PCOS patients and 2,358 healthy people (clinic-based) | Asian | T2DM ( P = 6 x 10-8) polycystic ovary syndrome( P = 1.3 x 10-11) | [35] | |
| 2009 | 37,927 men and women (population-based) | European | increased systolic ( P = 4 x 10-5) increased diastolic blood pressure (P = 0.003) increased chances of hypertension ( P = 2 x 10-4) | [36] | |
| 2012 | 2,816 patients and 2,932 healthy people (clinic-based) | European and Asian | increased chances of pancreatic cancer | [37] | |
| rs8050136(C/A) | 2012 | 6,835 men and women (population-based) | American Indian | lower eGFR (P = 0.002) | [38] |
| 2010 | 21,674 women (population-based) | European | increased risk of cardiovascular disease | [39] | |
| rs8044769 | 2013 | 9764 OA patients and 5362 healthy people (clinic-based) | European | increased chances of knee osteoarthritis risk and body mass index | [40] |
| 2016 | 196 OA cases and 442 healthy people (clinic-based) | Chinese | not modify individual susceptibility to osteoarthritis (P = 0.791) | [41] | |
| rs11075995 rs9939609 rs17817449(T/G) rs8050136(C/A) rs1477196 rs6499640 rs16953002 rs1121980(T/C) | 2015 | an overview | African American and European | increased chances of ER-negative breast cancer | [42, 43] |
| rs16953002 | 2013 | 12,313 patients and 55,667 healthy people (clinic-based) | European | Increased chances of melanoma (P = 4 x 10-12) | [44] |
abbreviations: e-GFR, estimated glomerular filtration rate; ER, estrogen receptor; T2DM, type 2 diabetes mellitus
Physical Activity Attenuates the Effect of the FTO Genotype on fat parameters
| FTO single nucleotide polymorphisms | date | Study design | The risk alleles | participants | N | region | Subjects BMI(kg/m2) | measurement parameters | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| rs9939609 | 2011 | a literature-based meta-analysis | A/T | Adults (46±8 year) | 237,434 | Europe North America Asia | Risk of overweight and obesity: 25.0≤BMI<30.0; 30.0≤BMI<35.0 | PA significantly (beta interaction = 20.14 kg/m2 per allele, P-interaction = 0.005) attenuated the association between the FTO variant and BMI of adults, but No such interaction was found in children and adolescents. | [62] |
| rs9939609 | 2010 | Cross-sectional study from October 2006 to December 2007 | A/T | Adolescents (15 year) | 752 (339 Boys; 413 Girls ) | Europe | Risk of overweight 24.0≤BMI | weight, height, WC( P =0.1), BMI( P =0.02) body fat percentage( P =0.06) | [63] |
| rs9939609 | 2008 | Clinical Trials | A/T | adults | 17,508 | Danes | subjects with BMI<25 and BMI≥25 | physically inactive homozygous risk A-allele carriers had an increase in BMI level by 1.95±0.3 kg/m2 compared with homozygous T-allele carriers. ( P =0.007) | [75] |
| rs1421085 | 2016 | a longitudinal multi-ethnic study (clinical trial) | T/C | the ages of 18-85 years | 17,423 (10,608 women and 6799 men) | six ethnic groups | a mean BMI of 30.2 (SD= 6.22) kg/m2 a mean BAI of 33.0 (SD= 7.49) | BMI Reduced by 57%( P = 0.01) BAI Reduced by 56%( P =0.04) | [69] |
| rs9939609 | 2011 | a prospective cohort study | A/T | aged 45-66 years | 12,523(6,894 women and 5,611 men) | African & European Americans | subjects with 25.0≤BMI<30.0 | P ≤0.04 for all traits( BMI, WC) only in AA men and EA men | [76] |
| rs9939609 | 2016 | randomized controlled trial | A/T | Adults (aged ≥18 years) | 1,280(537 women and 743 men) | European | a mean BMI of 25.5(SD= 4.80) | Attenuated WC( P =0.02) and BMI(P= 0.005) | [67] |
| rs9939609 | 2015 | randomized controlled trial | A/T | 18-36 years old | 550(231 males and 319 females) | Portuguese | Men: 22.9% were overweight and 4.3% obese, Women: 12.2% were overweight and 4.4% obese | decreased in the body-fat percentage( P =0.0002) an increased in BMI | [66] |
| rs1121980 | 2009 | prospective population-based cohort | T/C | Adults (39-79 y of age) | 20,374(10,059 males and 10,315 females) | European | Men BMI: (26.5±6.33) Women BMI: (26.1± 6.42) | Attenuated WC( P =0.02) and BMI( P = 0.004) | [68] |
| rs1121980 | 2013 | 11 cohorts study | T/C | Adults (aged ≥30 years) | 111,421(More than 50% females) | European | subjects with 20.0≤BMI<35.0 | attenuated BMI ( P =0.003) almost 10-fold larger in North American than in European cohorts | [70] |
| rs9939609 | 2015 | randomized trial | A/T | children and adolescents (aged 10-18 years) | 135 (77 males and 61 females) | Chinese | BMI≥25 | BMI, insulin, TC, TG, HDL, RBC, HB and HCT were significantly declined by the combined intervention(P < 0.05) | [77] |
Abbreviations: BMI: body mass index, BAI: body adiposity index, SD: standard deviation, N: sample size, PA: physical activity, PAI: physical activity index, EA: European Americans; AA: African Americans, TC: total cholesterol, TG: triglyceride, HDL: high-density lipoprotein, RBC: red blood cell, HB: Hemoglobin, HCT: hematocrit
FTO variants with macronutrient intake
An increasing number of reports suggest that the FTO SNPs are associated with satiety, appetite ratings and overeating. Several studies have probed FTO SNPs associated with food intake to elucidate the mechanisms for variation in FTO and the risk of obesity (Table 4). A study of the link between rs1421085 and protein intake was observed by using the frequency of foods questionnaire in 7,724 subjects [78], and another study found that the A allele of rs8050136 was positively correlated with the percentage of fat in dairy foods and negatively correlated with energy from carbohydrates in 36,973 subjects [79]. Moreover, a meta-analysis across 37 studies in 177,330 subjects confirmed that FTO rs9939609 played an important role in carbohydrate intake and total energy, especially when the risk allele carrier had a significantly reduced fiber intake and increased protein intake [80]. A similar association was noted between the A allele of rs8050136 and total energy intake as measured using food diaries in Asian populations [56]. In addition, children who were carriers of the A allele (AA or AT) exhibited a significantly increased energy intake compared with children who were TT allele carriers across the three conditions. However, only a very small effect on protein intake and no effect on carbohydrate intake were noted [81]. A recent study of feeding behavior was conducted in 64 FTO SNPs. Overall, only rs1292170 and rs12446047 play an important role on total energy and among all macronutrients [82]. The findings of the study indicate that the FTO gene has a potential role in dietary traits [83, 84]. A report of macronutrient intake in more than 70,000 individuals illustrated that the BMI-increasing allele of FTO SNPs was also positively correlated with protein intake (P=10-9), which might explain the effect of FTO on BMI [78]. The milestone study reported a mechanistic link between FTO SNPs and gut peptides [85]. Researchers performed a considerable amount of work based on studying the mechanism of FTO in animals. Then, studies were performed in humans. Young and healthy participants were divided into risk-allele carriers (AA) and low risk-allele carriers (TT). After a test meal, comparisons between the TT group and AA group were made; the latter group was discovered to have a subtle increase in acyl ghrelin and a weak inhibitory effect on the visual analog hunger score. Compared with the TT group, the risk-allele carriers had an increase in both FTO expression and ghrelin precursor GHRL mRNA and, conversely, a decrease in m6A methylation of ghrelin mRNA from peripheral blood cells after two hours of fasting. In in vitro studies in which FTO was overexpressed, an increase in ghrelin and ghrelin O-acyltransferase mRNA, as well as total and acyl ghrelin concentrations in cell lysates, and a reduction in m6A methylation of ghrelin mRNA were noted. These results confirmed the suggestion that the association of the FTO risk allele with food intake might be regulated through ghrelin by increasing m6A ghrelin mRNA demethylation.
According to the description, the FTO risk allele carriers can increase weight, through the movement of fat mass, so we can exercise to prevent obesity and other low calorie diet is the key to prevent obesity, in general, a healthy lifestyle to prevent obesity is essential.
The FTO variants on food intakes and related parameters
| FTO single nucleotide polymorphisms | date | Study design | The risk alleles | subjects | N(sex) | region | Types of food intake and related parameters | Ref. |
|---|---|---|---|---|---|---|---|---|
| rs1421085 | 2013 | 12 prospective cohorts study | T/C | adults | 7,724 | European descent | the BMI-increasing minor allele was associated with higher protein intake (P=9.96×10-10) | [78] |
| rs8050136 | 2013 | prospective cohorts study | A/T | Multiethnic adult (23-74y) | 36,973 | 5 racial/ ethnic groups | Macronutrient intake (carbohydrate, protein, ethanol, and fat) (P <0.01) positive correlation with protein (P =4×10-5) negatively correlated with carbohydrate (P=4×10-5) | [79] |
| rs1421085 | 2012 | The Look AHEAD trial | C/T | aged 45-76 y | 2,075 (56% women) | African American | eating episodes per day (P =0.001 ) total energy intake (P =0.031, P =0.067) FTO may bias meal patterning and perhaps total caloric intake and consumption of sweet or high-fat foods | [80] |
| 68 FTO SNPs | 2014 | The randomized trial | many | Adults (34.5±10.6 y) | 237 (117 males/120 females) | Mixed ethnic | moderated the relationship between RRV food and energy intake to predict an additional 4.9-7.4% of variance in energy intake(p < 0.001) moderated the relationship between RRVfood and protein intake, sugar intake (p < 0.001) | [82] |
| rs9939609 | 2014 | Prospective cohort study | A/T | aged 30-75 | 177,330 (51, 529 males/121, 700 females) | Mixed ethnic | Total energy A allele lower ( P = 0.001) across all ethnicities higher Protein (P = 2.4×10-16) lower Carbohydrate A allele (P =0.004) no associated with Fat (P =0.24) associated with fried food consumption (p<0.001) | [86] |
| rs1421085 rs17817449 | 2014 | The randomized trial | C/T | the ages of 25 and 40 years | 133 (21 males/ 112 females) | Dutch, French, German, British | associated with poorer eating behaviors (p < 0.05), a higher intake of high fat foods and refined starches and more depressive symptoms (p < 0.05) | [87] |
| rs8050136 | 2009 | The randomized trial | A/T | Adults (35 to 45 years of age) | 380 (171 males/ 209 females) | German | significantly associated with higher energy intake( P =0.01) associated with fibre intake ( P =0.31) | [88] |
| rs9939609 | 2008 | The randomized trial | A/T | Children (4 to 10 years of age) | 2726 | Scottish | A allele was associated with increased energy intake( P =0.006) with 30% higher fat intake after the low-energy premeal load in the A allele carriers (P = 0.004) with 16% greater energy density of food ingested by carriers ( P = 0.03) | [89] |
| rs8050136 rs11076023 | 2016 | case-control study | A/T | Individuals aged 30-65 | 1,618 (916 males/ 702 females) | Asian Indians | interaction between SNP rs8050136 and carbohydrate intake (% energy) (P = 0.04) SNP rs11076023 and dietary fiber intake (P=0.0008) the highest tertile of carbohydrate intake (P=1.0×10-5) | [90] |
FTO and energy metabolism
Despite the fact that FTO deficiency causes some serious developmental deformities, there are increasing reports that FTO might also affect energy homeostasis. Early experiments reported that FTO-/- and FTO+/- mice resist obesity induced by a high-fat diet and exhibited an obvious phenomenon of hyperphagia [91]. Three mouse models with different additional copies of the FTO gene exhibited a positive correlation with body mass and fat mass measurements regardless of whether mice were fed a standard or high-fat diet [92]. In a similar global knockout model, food intake was unaffected, but power consumption was elevated, despite the fact that physical activity was diminished. Thus, the lack of the FTO gene results in a lean phenotype [93]. Mice with a point mutation in the FTO gene that reduced FTO activity were not developmentally compromised. They developed a lean phenotype and an increased rate of energy expenditure and respiratory exchange ratio (RER). However, no change in physical activity levels and food intake were noted [94-96]. The food intake was significantly increased in mice with a knockout of the FTO gene (FTO-/-), and FTO+/- mice did not necessarily become obese with a high-fat diet [97]. A mouse model with additional copies of FTO promoted increases in body weight and fat mass in a dose-dependent manner regardless of whether the mice were fed a standard or high-fat diet [98]. However, other report indicated that mice deficient in Fto significantly increased their fat mass after exposure to a HFD and that FTO+/-and FTO-/- mice exhibited an obvious anorexigenic phenomenon due to the increase of leptin [99]. According to these different mouse models, a complex connection is noted between FTO and food intake. In summary, changes in the content of FTO in mice regulate the energy changes in mice and subsequently affect the phenotype regarding body mass. It has been hypothesized that the effect of FTO on energy may be due to FTO being a 2-oxoglutarate sensor and the effect of the 2-oxoglutarate level, which is a member of the citric acid cycle. FTO enhanced TCA cycle activity by regulating 2-oxoglutarate levels to promote differences in body energy metabolism [100]. However, at present, there is another hypothesis that FTO may be a sensor of circulating amino acids rather than a sensor of 2-oxoglutarate [101-106]. This action mainly occurs in FTO-deficient mice that exhibit developmental deformities. Thus, presumably, some association between FTO and the translation of mRNA exists that impacts the multi-tRNA synthetase complex, which is composed of aminoacyl-tRNA [101, 107].
Deficiency and mutation of FTO in human/mice
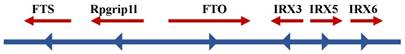
Many genes that control the same phenotypes with lipid storage and obesity have been identified, such as PCSK1 [108, 109], MCR4 [110-112], BDNF [113], and POMC [114-116]. Therefore, it was reasonable to determine the phenotype of FTO deficiency. FTO was one of six consecutive genes, including Ftl, Ftm, Irx3, Irx5, and Irx6, detected on the 1.6-Mb chromosome segment, which is missing in fusedtoe mice (Figure 2). The mutant homozygous mice died as embryos and exhibited underdeveloped facial structures, whereas heterozygous mutants had severe developmental malformations with damaged hypothalamic development, defective left-right asymmetry and hyperplasia of the thymus and fused digits [93]. Compared with the above-mentioned facts, a specific targeted deletion of FTO in mice did not exhibit the above-mentioned features, but exhibited high postnatal mortality, decreased fat mass on a normal diet, and increased food consumption [93]. Analogously, mice with a global germline loss of FTO displayed a high perinatal mortality as well as reduced body length and body weight after birth, whereas their weight increased with feeding and strengthen metabolism [117]. Subsequently, two independent studies of FTO loss also confirmed the phenotypic characteristic of postnatal growth retardation [96, 118]. One of the experiments used a conditional allele to delete FTO in the nervous system [96], and phenotypes were produced that were similar to those described if much of the research on a global germline loss. This finding suggested that FTO has a crucial role in the central nervous system to regulate growth development.
Schematic diagram of FTO and its neighboring genes on chromosome 16q12.2
The deletion of the FTO gene in humans also leads to a severe syndrome that is mainly characterized by postnatal growth retardation, facial dysmorphology, microcephaly, severe psychomotor delay and functional brain deficits [119]. In some patients, the syndrome also includes structural abnormalities in the brain, cardiac defects, genital abnormalities and cleft palate. More severe symptoms may lead to infant death within 3 years [119]. It is also possible that some individuals will carry a R316Q mutation in FTO, in which the arginine residue at position 316 is replaced by a glutamine. This mutation prevents 2oxyglutarate binding, resulting in a loss of FTO demethylase activity [120]. An important study reported on a family with FTO loss with an unreported malformation syndrome that was characterized by poly-malformation and childhood mortality due to a homozygous R316Q mutation in the FTO gene at cDNA position 947 [121]. Other researchers have comprehensively investigated whether nonsynonymous variants of FTO might be represented in either obese or lean individuals [119]. Complete sequencing of the FTO coding regions in obese participants and lean individuals revealed that the ratio of nonsynonymous mutations is uniform. In addition, heterozygous mutations were identified that severely damaged the enzymatic activity of FTO in both obese and lean cohorts [121]. Recently, researchers investigated a 1-year-9-month-old girl who presented failure to thrive, distinctive facial features, left ventricular cardiac hypertrophy, global developmental delay, bilateral hearing loss and reduced vision. A novel FTO homozygous missense mutation was identified that affected a highly conserved residue in the functional domain (c.956C>T; p.(Ser319Phe)) in the patient and her parents upon sequencing 4,813 clinically relevant genes. The results of the biochemical experiments showed that p.(Ser319Phe) FTO had impaired N-methyl-nucleoside demethylase activity and 2-oxoglutarate accumulation [122], which were consistent with previous reports. These studies revealed that FTO homozygous mutations could cause developmental delay syndromes and rare growth retardation and further demonstrated that FTO plays a significant role in the development of the cardiovascular and central nervous systems in infants. The above studies in humans demonstrated that rare variants in FTO are distributed, with different proportions between lean or obese individuals, but no evidence has reported any particular variant in individuals. Thus, although many problems remain unresolved, it seems plausible that FTO plays an important role in the growth and development of humans and mice.
FTO and m6A demethylation
Similar to other members of the AlkB family, the FTO protein contains both an Fe2+ binding motif and a 2-oxyglutarate (2OG) AlkB dioxygenases demethylase and leads to non-syndromic human obesity [46, 59]. The FTO protein is closely related to the bacterial DNA demethylase AlkB and the mammalian AlkB homologs ABH1 and ABH2 [58, 123]. However, FTO has a much weaker binding affinity to RNA molecules and a very different sensitivity to RNase treatment at the cellular level than ALKBH5 [124, 125]. FTO is mainly localized in the nucleoplasm, and it was originally shown to catalyze the oxidative demethylation of 3-methylthymidine (m3T) or 3-methyluracil (m3U) in single strand DNA/RNA, suggesting that nuclear RNAs are the main substrates of FTO. However, FTO has relatively reduced activities compared with other AlkB family enzymes that catalyze an extensive range of oxidative reactions [126], demonstrating another role for FTO in nucleic acid repair or modification [127]. FTO overexpression results in a decrease in m6A levels in cultured cells, suggesting that m6A is a physiological substrate of FTO [128]. A link between FTO-dependent m6A demethylation and gene expression was demonstrated in two independent studies. FTO-knockout mice exhibited changes in m6A levels in the midbrain and striatal, with an increased m6A methylation level of key genes in the dopaminergic pathway and reduced expression of these genes [98, 129]. Hydrogen bonds between R96 and 3-meT as well as between Glu234 and 3-meT form the main structural interactions of the FTO protein for the specific identification of 3-methylation of U and T [130]. The structure of the complex described above also provides a basis for the design of small-molecule inhibitors that interfere with the enzyme activity of FTO by interfering with fat metabolism [131]. The FTO protein possesses strong demethylating activity for m6A in single-stranded RNA, both in vitro and in vivo, due to partial localization in nuclear speckles. It is positioned to demethylate m6A on nuclear RNA [132]. Interestingly, m6A demethylation by FTO involves 2 intermediate products: N6-hydroxylmethyladenosine (hm6A) and N6-formyladenosine (f6A), which is a more oxidized product of hm6A. Moreover, m6A RNA decoration during adipogenesis is inversely correlated with FTO gene expression, and these findings are consistent with the recently demonstrated m6A-dependent mRNA decay mediated by YTHDF2. Recently, a pioneering study identified FTO as an important regulator of mRNA splicing by regulating the RNA-binding ability of SRSF2. FTO-regulated m6A demethylation is the regulatory mechanism by which RNA splicing is controlled, explaining its important role during the regulation of adipogenesis [133]. Yet, the role of m6A demethylation by FTO in the regulation of energy homeostasis and body composition remains unclear.
In addition, in neuron-specific FTO-deficient mice, a subset of mRNAs that are involved in dopaminergic (DA) signaling exhibits increased m6A levels [83, 134]. In a mouse model in which FTO was knocked out only in DA neurons, FTO demethylates an mRNA subset that is involved in neuronal function, which provides more evidence for the critical role of FTO in RNA metabolism [135, 136]. This research applied diazirine photo-cross-linking to m6A recognition mediated by the FTO demethylase. The α-KG co-substrate and Fe(II) cofactor formed activated FTO complexes that contributed to capturing dioxygen and recognizing m6A. In addition, the demethylation activity of FTO is related to the physiological process [137]. A recent report showed that FTO overexpression might regulate 3T3-L1 differentiation by increasing Wnt signaling, suppressing PPARγ activity and decreasing m6A[138]. Collectively, the above studies provide valuable evidence for the role of FTO in modulating preadipocyte differentiation through m6A demethylation.
Novel mechanism of FTO
The role of RUNX1T1 and FTO
As a nucleic acid demethylase, FTO removes methyl groups from both DNA and RNA [134, 135, 139, 140]. There is a prevalent belief that the most significant function of FTO is demethylating N6-methyladenosine (m6A), which could adjust the alternative splicing of mRNAs [141-143]. Studies using 3T3-L1 cell culture verify that FTO regulates mRNA splicing by controlling the binding activity of the splicing factor SRSF2 to mRNA [138, 143]. Runt-related transcription factor 1 (RUNX1T1), an adipogenesis-related transcription factor, is spliced by SRSF2 into two variants: a short (S) and a long (L) isoform. In 3T3-L1 cells, S isoform overexpression of RUNX1T1 promoted adipogenesis, indicating that FTO might enhance adipocyte formation by affecting RUNX1T1. However, previous studies investigated the role of FTO in regulating adipogenesis in mice primordial cells with FTO overexpression or deletion [118]. Next, they demonstrated that FTO works early in adipogenesis to increase fat cell numbers during mitotic clonal expansion (MCE). The prerequisites for adipose differentiation that occurs within 48 hours is Mitotic clonal expansion (MCE). Mouse embryonic fibroblasts (MEFs) in which FTO was either deleted (FTOKO) or overexpressed (FTOO) were treated with a cocktail containing IBMX, insulin and dexamethasone to induce differentiation into mature adipocytes [144]. The transcription factor C/EBPβ binds to DNA, resulting in the elevation of C/EBPα and PPARγ, which are the most important master regulators of adipogenesis. The results of the experiment validated that FTO knockdown suppresses adipogenesis immediately prior to MCE and that FTO overexpression enhances the expression of PPARγ and C/EBPα. These results indicate that FTO regulated RUNX1T1 and then affected adipogenesis. Mice with FTOKO in MEFs decreased RUNX1T1-S expression, and mice with FTOO in MEFs exhibited high RUNX1T1-S expression. WT MEFs with knockdown of RUNX1T1 aided in the further discovery of the reduction of the expression of cell cycle genes and cell proliferation during MCE. Previous studies in 3T3-L1 cells suggested that adipogenesis was impaired due to overexpression of RUNX1T1-L and that overexpression of RUNX1T1-S enhanced adipogenesis. These findings illustrated that adipocyte differentiation and proliferation depend on the relative abundances of the S and L isoforms of RUNX1T1 binding to C/EBPβ, but differences in the expression between L and S isoforms were not observed [145]. RUNXT1-L exhibited higher expression levels than RUNXT1-S in 3T3-L1 cells. The reduction in adipogenesis is potentially due to the primary role of RUNXT1-L in WT MEFs with RUNX1T1 knockdown. Moreover, research now demonstrates that FTO acts as a transcriptional coactivator of C/EBPβ [146]. These studies demonstrated that FTO acts on adipogenesis and that the fat mass (Figure 3) and body weight were further controlled.
Roles of IRX3 and IRX5
A follow-up study in mice showed that FTO expression influences weight and the body composition. However, the following experiment found that FTO expression and FTO intron variation in the area were not directly related. Irx3 within the FTO gene region was initially not remarkable. Then, a report by Ragvin et al sought to identify the relationship between the FTO gene [147]. This team found that the FTO LD block seemingly contained a large number of highly conserved noncoding elements (HCNEs) and determined that potential regulatory elements may affect target genes. GFP reporter gene analysis confirmed the base sequence of zebra fish linkage disequilibrium with FTO, but the discovery and expression pattern matched those of IRX3. Given that Irx3 is highly expressed in the pancreas, it is possible that at least some of the effects ascribed to the FTO gene, such as susceptibility to type 2 diabetes, might be attributed to Irx3. Studies in murine and human genomes established that the in vitro development of Irx3 is involved in the association between human obesity and FTO. Chromatin conformation capture was used in both embryonic tissue and brain tissue from adult mice to investigate cis-regulatory interactions between FTO and IRX3 [148]. The results clearly indicate that IRX3 relied on long-range regulation from FTO, and the expression of this region in the remote enhancer and IRX3 protein expression followed the same trend. These findings indicate that the obesity-related area in FTO commonly regulated FTO and IRX3. Consistent with this finding, in human brain cells, the expression of obesity-related variability was associated with IRX3, but the expression of FTO was not relevant. The relations between IRX3 expression and body weight as well as weight composition in a mouse model have also been confirmed. The body weight of Irx3 deficient mice was at least 25 to 30% reduced compared with control mice, and the weight reduction was mainly due to the reduction in adipose tissue and the increase in basal metabolic rate. Finally, the hypothalamic expression of a dominant negative Irx3 gene resulted in a phenotype similar to that of Irx3-deficient mice. The study showed that the IRX3 gene is a functional long-range target of FTO SNPs and is one of the decisive factors of body mass and composition. Previous studies demonstrated a relation between FTO gene regulation associated with obesity and the hypothalamus, the area of the human brain that controls appetite or physical activity [149, 150]. By contrast, new research has found that the FTO polymorphisms exhibited the strongest effect in preadipocytes, and this pathway is independent of the brain. Thermogenesis production is a method of energy consumption in adipocyte cells that is triggered by movement, diet or temperature [151]. New pathways control the browning of white adipose and can affect the body's energy balance [152, 153].
Compared with risk-allele carriers, non-risk-allele carriers exhibited reduced IRX3 and IRX5 expression. These results showed that FTO gene mutations promote increased expression of IRX3 and IRX5. Further experiments revealed that the obesity risk of the FTO area depends on a key nucleotide. When thymine (T) is replaced by cytosine (C), which leads to the derepression of a potent preadipocyte enhancer and a doubling of IRX3 and IRX5 expression during preadipocyte differentiation, there is a reduction in mitochondrial thermogenesis. This action results in body energy imbalance, lipid accumulation and subsequent obesity without a change in physical activity or appetite. When C is converted to T, IRX3 and IRX5 expression is repressed. Then, the thermogenesis process returns to normal, and the storage of excess lipids is halted. Researchers used CRISPR-Cas9 editing [154-156] in human cells and mice to manipulate these new pathways to reverse obesity. These results showed that regulation of IRX3 and IRX5 expression can alter the function of adipocytes, which shift from energy-dissipating beige adipocytes [153, 157] to energy-storing white adipocytes. Knockdown of IRX3 or IRX5 in primary adipocytes of mice significantly affects the energy balance of the entire body, leading to weight loss and a reduction in lipid storage. Above all, mice were completely resistant to high-fat diet. The researchers also summed these findings vividly using the phrases to "solve the case". First, they found that the direct "perpetrator" is not the FTO gene, but is IRX3 and IRX5. Second, the "crime locations" are preadipocytes. Third, the "crime" is that they inhibit lipid burning and energy metabolism. This pathway for the treatment of obesity offers new hope (Figure 3). The future of obesity treatment can be edited on the basis of an individual's risk for obesity by changing the genome; manipulating the body's metabolic master switch is expected to offset the environmental, lifestyle or genetic factors. However, the researchers also noted that although we understand the principle by which these genes lead to obesity, there is no simple method to develop a medication to control obesity. These discoveries will be crucial to move us toward the obesity treatment that we crave.
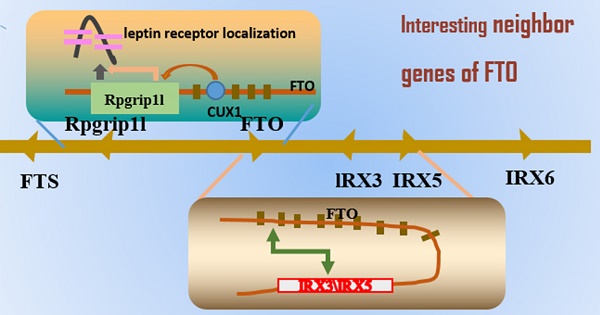
Role of Rpgrip1l
As mentioned earlier, originally of the FTO gene research, followed by the association of single nucleotide polymorphisms in intron 3 and intron 8, the formation of a large number of studies on the biology of the FTO gene [158-160]. However, the Leibel group found another direction, choosing to focus on Rpgrip1l as being the potential target point [161, 162]. They originally mainly studied two intronic single nucleotide polymorphisms, rs17817449 and rs8050136, and they predicted that these SNPs were located in a binding site for the transcription factor CUTL1, which acts as a transcriptional repressor by displacing activators and by recruitment of histone deacetylase 1. Preliminary in vitro data showed that siRNA-mediated reduction in cutl1 of 70% led to a decrease in FTO expression of 90% and a decrease in rpgrip1l of 65%; they are able to act as cux1 protein binding sites, and the FTO variation indirectly affects RPGRIP1L expression. Therefore, they determined that the two genes by cutl1/cux1 through the FTO gene first intron of a single adjustment possibility. Subsequently, researchers confirmed that RPGRIP1L plays a role in obesity. They used hemizygous RPGRIP1L mice, which reduced but did not eliminate the function of the gene (the loss of both copies of the gene in mice creates several serious defects, masking effects on food intake itself). The Rpgrip1l+/- mice had higher food intake and gained significantly more weight, with a higher percentage of body fat, than the control group. In subsequent experiments, the researchers found that RPGRIP1L deficient mice also had deficient leptin receptor signaling. The receptor was not properly set to the cell surface, so it was speculated that RPGRIP1L plays a role in the formation of leptin receptor clustering, which is important for efficient signaling. This model showed that decreased leptin receptor aggregation, negative energy balance, resulted in reduced CUX1 P200 and CUX1 P110 protein levels, showing that the cilia molecular abnormalities affect leptin receptor (a hormone that suppresses appetite), resulting in weight gain (Figure 3). Tests on mice show that is expected through the adjustment of intervention ciliary function so as to improve obesity. To understand the molecular mechanism of obesity, will help researchers to manipulate this mechanism, achieve the purpose of weight loss.
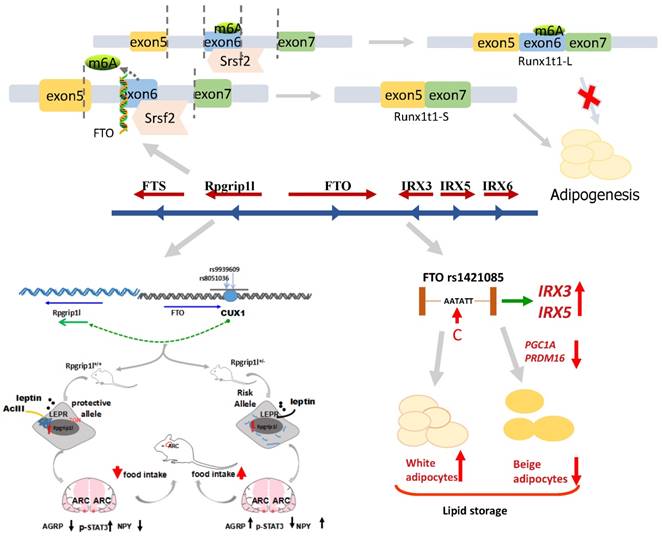
The following Figure 3 shows the above three mechanisms of FTO locus association with the formation of fat cells and weight gain. It refers to FTO in relation to m6A demethylation, RUNX1T1, IRX3, IRX5, and Rpgrip1l, and the role of each.
Conclusions
Currently, we have a good understanding that multiple single nucleotide polymorphisms (SNPs) in intron 1 of FTO are strongly associated with obesity. More experiments on the mechanism of FTO SNPs on obesity should be performed due to the different alleles and different effects on different ethnic populations. The identification of a mechanistic basis for the association between the FTO locus and obesity in humans is now favorable; the relevant cell types and target genes have been partially identified. We also know that FTO is a DNA/RNA nucleic acid demethylase that localizes to the nucleus [139]. Many studies have demonstrated that demethylation modification of m6A by FTO is required for inhibiting preadipocyte differentiation. However, there is still a need for further investigation into the molecular mechanisms underlying the involvement of FTO during adipogenesis. In addition, scientists have confirmed that FTO and mature fat cells interact, but the connection between these mechanisms and the FTO gene must be elucidated. Thus, additional studies are still required to explore the direct relevance of FTO SNPs and obesity. Moreover, understanding the interaction effects between FTO and its downstream partners could potentially offer new therapeutic directions to reverse the global trends towards fatness in combination with an FTO inhibitor. These bold visions will be essential to move us toward the world of precision medicine to which we aspire as well as the traditional treatment concept of transformation and innovations.
The mechanism of FTO locus association with obesity, suggesting that the transition from T to C in FTO rs1421085 may regulate the expression of IRX3 and IRX5, reducing the mitochondrial heat production and increasing white adipocytes mass. The binding site of FTO intron 1 interacts with transcription factor CUX1 and then regulates the neighboring group RPGRIP1L to control the fat mass. The removal of m6A by FTO demethylation leads to a short product of Runx1t1, which enhances pre-adipocyte differentiation. Conversely, if no FTO exists, this results in a long product of Runx1t1, which inhibits pre-adipocyte differentiation.
Acknowledgements
This work was supported by the National Science Foundation of China (no. 81173107); and the Science and Technology Planning Project of Guangdong, China (2013B021100018, 2013B090600050, 2015A010101318); and the Industry-university-research Collaborative Innovation Major Projects of Guangzhou Science Technology Innovation Commission (no.201604020164), China.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Malik VS, Willett WC, Hu FB. Global obesity: trends, risk factors and policy implications. Nature Reviews Endocrinology. 2013;9:13-27
2. Anis AH, Zhang W, Bansback N, Guh D, Amarsi Z, Birmingham C. Obesity and overweight in Canada: an updated cost-of-illness study. Obes Rev. 2010;11:31-40
3. Laurson KR, Welk GJ, Eisenmann JC. Diagnostic performance of BMI percentiles to identify adolescents with metabolic syndrome. Pediatrics. 2014;133:e330-8
4. Su Z, Wu S, Zhang H, Feng Y. Development and validation of an improved Bradford method for determination of insulin from chitosan nanoparticulate systems. Pharm Biol. 2010;48:966-73
5. Zhang H-l, Wu S-h, Tao Y, Zang L-q, Su Z-q. Preparation and characterization of water-soluble chitosan nanoparticles as protein delivery system. J Nanomaterials. 2010;2010:1
6. Tao Y, Zhang H, Gao B, Guo J, Hu Y, Su Z. Water-Soluble chitosan nanoparticles inhibit hypercholesterolemia induced by feeding a high-fat diet in male Sprague-Dawley rats. J Nanomaterials. 2011;2011:6
7. Wu S, Tao Y, Zhang H, Su Z. Preparation and characterization of water-soluble chitosan microparticles loaded with insulin using the polyelectrolyte complexation method. J Nanomaterials. 2011;2011:2
8. Zhang H-l, Tao Y, Guo J, Hu Y-m, Su Z-q. Hypolipidemic effects of chitosan nanoparticles in hyperlipidemia rats induced by high fat diet. Int Immunopharmacol. 2011;11:457-61
9. Zhang H-l, Zhong X-b, Tao Y, Wu S-h, Su Z-q. Effects of chitosan and water-soluble chitosan micro-and nanoparticles in obese rats fed a high-fat diet. Int J Nanomed. 2012;7:4069
10. Chen J, Huang G-D, Tan S-R, Guo J, Su Z-Q. The preparation of capsaicin-chitosan microspheres (CCMS) enteric coated tablets. Int J Mol Sci. 2013;14:24305-19
11. Tao Y, Zhang H-L, Hu Y-M, Wan S, Su Z-Q. Preparation of chitosan and water-soluble chitosan microspheres via spray-drying method to lower blood lipids in rats fed with high-fat diets. Int J Mol Sci. 2013;14:4174-84
12. Sirong T, Bing G, Yi T, Jiao G, Zheng-Quan S. Antiobese effects of capsaicin-chitosan microsphere (CCMS) in obese rats induced by high fat diet. J Agric Food Chem. 2014;62:1866-74
13. Elks CE, Den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos R. et al. Variability in the heritability of body mass index: a systematic review and meta-regression. Front Endocrinol (Lausanne). 2012;3:1-16
14. Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325-51
15. Lu Y, Loos R. Obesity genomics: assessing the transferability of susceptibility loci across diverse populations. Genome Med. 2013;5:55
16. Day FR, Loos RJF. Developments in obesity genetics in the era of genome-wide association studies. Journal of Nutrigenetics & Nutrigenomics. 2011;4:222-38
17. Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM. et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889-94
18. Scuteri A, Sanna S, Chen W-M, Uda M, Albai G, Strait J. et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115
19. Fawwad A, Siddiqui IA, Basit A, Zeeshan NF, Shahid SM, Nawab SN. et al. Common variant within the FTO gene, rs9939609, obesity and type 2 diabetes in population of Karachi, Pakistan. Diabetes & Metabolic Syndrome Clinical Research & Reviews. 2015;10:43-7
20. Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA. et al. A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet. 2013;45:690-6
21. Keaton JM, Bailey JNC, Palmer ND, Freedman BI, Langefeld CD, Ng MC. et al. A comparison of type 2 diabetes risk allele load between African Americans and European Americans. Hum Genet. 2014;133:1487-95
22. Peters T, Ausmeier K, Rüther U. Cloning of Fatso (Fto), a novel gene deleted by the Fused toes (Ft) mouse mutation. Mamm Genome. 1999;10:983-6
23. Stratakis CA, Lafferty A, Taymans SE, Gafni RI, Meck JM, Blancato J. Anisomastia Associated with Interstitial Duplication of Chromosome 16, Mental Retardation, Obesity, Dysmorphic Facies, and Digital Anomalies: Molecular Mapping of a New Syndrome by Fluorescent in Situ Hybridization and Microsatellites to 16q13 (D16S419-D16S503) 1. J Clin Endocrinol Metab. 2000;85:3396-401
24. Peters T, Ausmeier K. et al. mouse Fused toes (Ft) mutation is the result of a 1.6-Mb deletion including the entire Iroquois B gene cluster. Mamm Genome. 2002
25. Hubacek J, Dlouha D, Lanska V, Adamkova V. Strong Gender-Specific Additive Effects of the NYD-SP18 and FTO Variants on BMI Values. Physiol Res. 2015;64:S419
26. Church C, Moir L, McMurray F, Girard C, Banks GT, Teboul L. et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42:1086-92
27. de Luis DA, Aller R, Izaola O, Pacheco D. Role of rs9939609 FTO gene variant in weight loss, insulin resistance and metabolic parameters after a high monounsaturated vs a high polyunsaturated fat hypocaloric diets. Nutr Hosp. 2015;32:175-81
28. Elouej S, Belfki-Benali H, Nagara M, Lasram K, Attaoua R, Sallem OK. et al. Association of rs9939609 Polymorphism with Metabolic Parameters and FTO Risk Haplotype Among Tunisian Metabolic Syndrome. Metab Syndr Relat Disord. 2016
29. He D, Fu M, Miao S, Hotta K, Chandak GR, Xi B. FTO gene variant and risk of hypertension: A meta-analysis of 57,464 hypertensive cases and 41,256 controls. Metabolism. 2014;63:633-9
30. M Ä, Ronkainen J, Huusko T, Malo E, Savolainen ER, Savolainen MJ. et al. The fat mass and obesity-associated (FTO) gene variant rs9939609 predicts long-term incidence of cardiovascular disease and related death independent of the traditional risk factors. Ann Med. 2015:1-9
31. Ligthart S, de Vries PS, Uitterlinden AG, Hofman A, Franco OH, Chasman DI. et al. Pleiotropy among common genetic loci identified for cardiometabolic disorders and C-reactive protein. PLoS One. 2015;10:e0118859
32. Robiou-du-Pont S, Bonnefond A, Yengo L, Vaillant E, Lobbens S, Durand E. et al. Contribution of 24 obesity-associated genetic variants to insulin resistance, pancreatic beta-cell function and type 2 diabetes risk in the French population. Int J Obes (Lond). 2013;37:980-5
33. Bressler J, Fornage M, Demerath EW, Knopman DS, Monda KL, North KE. et al. Fat mass and obesity gene and cognitive decline: the Atherosclerosis Risk in Communities Study. Neurology. 2013;80:92-9
34. Rees SD, Islam M, Hydrie MZI, Chaudhary B, Bellary S, Hashmi S. et al. An FTO variant is associated with Type 2 diabetes in South Asian populations after accounting for body mass index and waist circumference. Diabet Med. 2011;28:673-80
35. Li H, Kilpeläinen TO, Liu C, Zhu J, Liu Y, Hu C. et al. Association of genetic variation in FTO with risk of obesity and type 2 diabetes with data from 96,551 East and South Asians. Diabetologia. 2012;55:981-95
36. Timpson NJ, Harbord R, Davey SG, Zacho J, Tybjaerg-Hansen A, Nordestgaard BG. Does greater adiposity increase blood pressure and hypertension risk?: Mendelian randomization using the FTO/MC4R genotype. Hypertension. 2009;54:84-90
37. Li G, Chen Q, Li W, Ke D, Yuan Z. Association between FTO gene polymorphism and cancer risk: evidence from 16,277 cases and 31,153 controls. Tumor Biol. 2012;33:1237-43
38. Franceschini N, Shara NM, Wang H, Voruganti VS, Laston S, Haack K. et al. The association of genetic variants of type 2 diabetes with kidney function. Kidney Int. 2012;82:220-5
39. Ahmad T, Chasman DI. et al. The Fat-Mass and Obesity-Associated (FTO) Gene, Physical Activity, and Risk of Incident Cardiovascular Events in Caucasian Women. Am Heart J. 2010;160:1163-9
40. Panoutsopoulou K, Metrustry S, Doherty SA, Laslett LL, Maciewicz RA, Hart DJ. et al. The effect of FTO variation on increased osteoarthritis risk is mediated through body mass index: a mendelian randomisation study. Geochim Cosmochim Acta. 2013;148:34-49
41. Wang Y, Chu M, Rong J, Xing B, Zhu L, Zhao Y. et al. No association of the single nucleotide polymorphism rs8044769 in the fat mass and obesity-associated gene with knee osteoarthritis risk and body mass index. Bone & Joint Research. 2016;5:169-74
42. Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN. et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat Genet. 2013;45:392-8 8e1-2
43. Hernández-Caballero ME, Sierra-Ramírez JA. Single nucleotide polymorphisms of the FTO gene and cancer risk: an overview. Mol Biol Rep. 2015;42:1-6
44. Iles MM, Law MH, Stacey SN, Han J, Fang S, Pfeiffer R. et al. A variant in FTO shows association with melanoma risk not due to BMI. Nat Genet. 2013;45:428-32
45. Robbens S, Rouzé P, Cock JM, Spring J, Worden AZ, Peer YVD. The FTO gene, implicated in human obesity, is found only in vertebrates and marine algae. J Mol Evol. 2008;66:80-4
46. Gerken T, Girard CA, Tung Y-CL, Webby CJ, Saudek V, Hewitson KS. et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469-72
47. van der Hoeven F, Schimmang T, Volkmann A, Mattei M-G, Kyewski B, Ruther U. Programmed cell death is affected in the novel mouse mutant Fused toes (Ft). Development. 1994;120:2601-7
48. Fan B, Du Z-Q, Rothschild MF. The fat mass and obesity-associated (FTO) gene is associated with intramuscular fat content and growth rate in the pig. Anim Biotechnol. 2009;20:58-70
49. Fontanesi L, Scotti E, Buttazzoni L, Davoli R, Russo V. The porcine fat mass and obesity associated (FTO) gene is associated with fat deposition in Italian Duroc pigs. Anim Genet. 2009;40:90-3
50. Sanchez-Pulido L, Andrade-Navarro MA. The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 2007;8:1-6
51. Stratakis CA, Lafferty A, Taymans SE, Gafni RI, Meck JM, Blancato J. Anisomastia associated with interstitial duplication of chromosome 16, mental retardation, obesity, dysmorphic facies, and digital anomalies: molecular mapping of a new syndrome by fluorescent in situ hybridization and microsatellites to 16q13 (D16S419-D16. J Clin Endocrinol Metab. 2000;85:3396-401
52. González JR, González-Carpio M, Hernández-Sáez R, Vargas VS, Hidalgo GT, Rubio-Rodrigo M. et al. FTO Risk Haplotype Among Early Onset and Severe Obesity Cases in a Population of Western Spain. Cardiovasc J Afr. 2012;20:909-15
53. David A, Clévio N, Licínio M. Association of FTO polymorphisms with obesity and obesity-related outcomes in Portuguese children. J Thermoplast Compos Mater. 2011;24:767-76
54. Zermeno-Rivera J, Astocondor-Perez J, Valle Y, Padilla-Gutierrez J, Orozco-Castellanos R, Figuera L. et al. Association of the FTO gene SNP rs17817449 with body fat distribution in Mexican women. Gen Mol Res. 2014;13:8561-7
55. Prakash J, Srivastava N, Awasthi S, Agarwal C, Natu S, Rajpal N. et al. Association of FTO rs17817449 SNP with obesity and associated physiological parameters in a north Indian population. Ann Hum Biol. 2011;38:760-3
56. Chuenta W, Phonrat B, Tungtrongchitr A, Limwongse C, Chongviriyaphan N, Santiprabhob J. et al. Common variations in the FTO gene and obesity in Thais: a family-based study. Gene. 2015;558:75-81
57. Bleijlevens B, Shivarattan T, van den Boom KS, de Haan A, van der Zwan G, Simpson PJ. et al. Changes in protein dynamics of the DNA repair dioxygenase AlkB upon binding of Fe2+ and 2-oxoglutarate. Biochemistry. 2012;51:3334-41
58. Han Z, Niu T, Chang J, Lei X, Zhao M, Wang Q. et al. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464:1205-9
59. Jia G, Yang C-G, Yang S, Jian X, Yi C, Zhou Z. et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313-9
60. Simmons AL, Schlezinger JJ, Corkey BE. What are we putting in our food that is making us fat? Food additives, contaminants, and other putative contributors to obesity. CurrObesRep. 2014;3:273-85
61. Totura CM, Figueroa HL, Wharton C, Marsiglia FF. Assessing implementation of evidence-based childhood obesity prevention strategies in schools. PrevMedRep. 2015;2:347-54
62. Kilpeläinen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E. et al. Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8:e1001116
63. Ruiz JR, Labayen I, Ortega FB, Legry V, Moreno LA, Dallongeville J. et al. Attenuation of the effect of the FTO rs9939609 polymorphism on total and central body fat by physical activity in adolescents: the HELENA study. Arch Pediatr Adolesc Med. 2010;164:328-33
64. Hakanen M, Raitakari OT, Lehtimäki T, Peltonen N, Pahkala K, Sillanmaki L. et al. FTO genotype is associated with body mass index after the age of seven years but not with energy intake or leisure-time physical activity. J Clin Endocrinol Metab. 2009;94:1281-7
65. Marsaux CF, Celis-Morales C, Livingstone KM, Fallaize R, Kolossa S, Hallmann J. et al. Changes in Physical Activity Following a Genetic-Based Internet-Delivered Personalized Intervention: Randomized Controlled Trial (Food4Me). J Med Internet Res. 2016:18
66. Muc M, Padez C, Manco L. Influence of physical activity on the association between the FTO variant rs9939609 and adiposity in young adults. Am J Hum Biol. 2015;27:734-8
67. Celis-Morales C, Marsaux CFM, Livingstone KM, Navas-Carretero S, San-Cristobal R, O'Donovan CB. et al. Physical activity attenuates the effect of the FTO genotype on obesity traits in European adults: The Food4Me study. Obesity. 2016;24:962-9
68. Vimaleswaran KS, Li S, Zhao JH, Luan Ja, Bingham SA, Khaw K-T. et al. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin Nutr. 2009;90:425-8
69. Reddon H, Gerstein HC, Engert JC, Mohan V, Bosch J, Desai D. et al. Physical activity and genetic predisposition to obesity in a multiethnic longitudinal study. Sci Rep. 2016:6
70. Ahmad S, Rukh G, Varga TV, Ali A, Kurbasic A, Shungin D. et al. Gene× physical activity interactions in obesity: combined analysis of 111,421 individuals of European ancestry. PLoS Genet. 2013;9:e1003607
71. Dougkas A, Yaqoob P, Givens DI, Reynolds CK, Minihane AM. The impact of obesity-related SNP on appetite and energy intake. Br J Nutr. 2013;110:1151-6
72. Livingstone K, Celis-Morales C, Lara J, Ashor A, Lovegrove J, Martinez J. et al. Associations between FTO genotype and total energy and macronutrient intake in adults: a systematic review and meta-analysis. Obes Rev. 2015;16:666-78
73. Qi Q, Kilpeläinen TO, Downer MK, Tanaka T, Smith CE, Sluijs I. et al. FTO genetic variants, dietary intake, and body mass index: insights from 177,330 individuals. Hum Mol Genet. 2014 ddu411
74. Fesinmeyer MD, North KE, Lim U, Bůžková P, Crawford DC, Haessler J. et al. Effects of smoking on the genetic risk of obesity: the population architecture using genomics and epidemiology study. BMC Med Genet. 2013;14:1
75. Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G. et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57:95-101
76. Demerath EW, Lutsey PL, Monda KL, Linda-Kao WH, Bressler J, Pankow JS. et al. Interaction of FTO and Physical Activity Level on Adiposity in African-American and European-American Adults: The ARIC Study. Obesity. 2011;19:1866-72
77. Zou ZC, L JM, Shi YY, Chen JH, Wang LS, Cai W. Effect of exercise combined with dietary intervention on obese children and adolescents associated with the FTO rs9939609 polymorphism. Eur Rev Med Pharmacol Sci. 2015:19
78. Tanaka T, Ngwa JS, Van Rooij FJ, Zillikens MC, Wojczynski MK, Frazier-Wood AC. et al. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 2013;97:1395-402
79. Park SL, Cheng I, Pendergrass SA, Kucharskanewton AM, Lim U, Ambite JL. et al. Association of the FTO Obesity Risk Variant rs8050136 With Percentage of Energy Intake From Fat in Multiple Racial/Ethnic Populations. Am J Epidemiol. 2013;178:780-90
80. McCaffery JM, Papandonatos GD, Peter I, Huggins GS, Raynor HA, Delahanty LM. et al. Obesity susceptibility loci and dietary intake in the Look AHEAD Trial. Am J Clin Nutr. 2012;95:1477-86
81. Cecil J, Dalton M, Finlayson G, Blundell J, Hetherington M, Palmer C. Obesity and eating behaviour in children and adolescents: contribution of common gene polymorphisms. International Review of Psychiatry. 2012;24:200-10
82. Scheid JL, Carr KA, Lin H, Fletcher KD, Sucheston L, Singh PK. et al. FTO polymorphisms moderate the association of food reinforcement with energy intake. Physiol Behav. 2014;132:51-6
83. Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Brönneke HS. et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16:1042-8
84. Xiang L, Wu H, Pan A, Patel B, Xiang G, Qi L. et al. FTO genotype and weight loss in diet and lifestyle interventions: a systematic review and meta-analysis. Am J Clin Nutr. 2016;103:1162-70
85. Karra E, O'Daly OG, Choudhury AI, Yousseif A, Millership S, Neary MT. et al. A link between FTO, ghrelin, and impaired brain food-cue responsivity. The Journal of clinical investigation. 2013;123:3539-51
86. Qi Q, Chu AY, Kang JH, Huang J, Rose LM, Jensen MK. et al. Fried food consumption, genetic risk, and body mass index: gene-diet interaction analysis in three US cohort studies. BMJ. 2014;348:g1610-g
87. Harbron J, Van dML, Zaahl MG, Kotze MJ, Senekal M. Fat mass and obesity-associated (FTO) gene polymorphisms are associated with physical activity, food intake, eating behaviors, psychological health, and modeled change in body mass index in overweight/obese Caucasian adults. Nutrients. 2014;6:3130-52
88. Haupt A, Thamer C, Staiger H, Tschritter O, Kirchhoff K, Machicao F. et al. Variation in the FTO gene influences food intake but not energy expenditure. Experimental and clinical endocrinology & diabetes: official journal, German Society of Endocrinology [and] German Diabetes Association. 2009;117:194-7
89. Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359:2558-66
90. Vimaleswaran KS, Bodhini D, Lakshmipriya N, Ramya K, Anjana RM, Sudha V. et al. Interaction between FTO gene variants and lifestyle factors on metabolic traits in an Asian Indian population. Nutr Metab (Lond). 2016;13:39
91. Julia F, Linda K, Christian E, Jeanette V, Thomas P, Brüning JC. et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:123-9
92. Chris C, Lee M, Fiona MM, Christophe G, Banks GT, Lydia T. et al. Overexpression of Fto leads to increased food intake and results in obesity. Nat Genet. 2010;42:1086-92
93. McMurray F, Church CD, Larder R, Nicholson G, Wells S, Teboul L. et al. Adult onset global loss of the fto gene alters body composition and metabolism in the mouse. PLoS Genet. 2013;9:e1003166
94. Tschöp MH, Speakman JR, Arch JRS, Auwerx J, Brüning JC, Chan L. et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2012;9:57-63
95. Church C, Lee S, Bagg EA, McTaggart JS, Deacon R, Gerken T. et al. A mouse model for the metabolic effects of the human fat mass and obesity associated FTO gene. PLoS Genet. 2009;5:e1000599
96. Gao X, Shin YH, Li M, Wang F, Tong Q, Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PLoS One. 2010;5:e14005
97. Ronkainen J, Huusko TJ, Soininen R, Mondini E, Cinti F, Mäkelä KA. et al. Fat mass-and obesity-associated gene Fto affects the dietary response in mouse white adipose tissue. Sci Rep. 2015:5
98. Merkestein M, McTaggart JS, Lee S, Kramer HB, McMurray F, Lafond M. et al. Changes in gene expression associated with FTO overexpression in mice. PLoS One. 2014;9:e97162
99. Tung YL, Gulati P, Liu C-H, Rimmington D, Dennis R, Ma M. et al. FTO is necessary for the induction of leptin resistance by high-fat feeding. Mol metab. 2015;4:287-98
100. Ma M, Harding HP, O'rahilly S, Ron D, Yeo GS. Kinetic analysis of FTO (fat mass and obesity-associated) reveals that it is unlikely to function as a sensor for 2-oxoglutarate. Biochem J. 2012;444:183-7
101. Cheung M, Gulati P, O'Rahilly S, Yeo G. FTO expression is regulated by availability of essential amino acids. Int J Obes. 2013;37:744-7
102. Huang T, Qi Q, Li Y, Hu FB, Bray GA, Sacks FM. et al. FTO genotype, dietary protein, and change in appetite: the Preventing Overweight Using Novel Dietary Strategies trial. Am J Clin Nutr. 2014;99:1126-30
103. Daoud H, Zhang D, McMurray F, Yu A, Luco SM, Vanstone J. et al. Identification of a pathogenic FTO mutation by next-generation sequencing in a newborn with growth retardation and developmental delay. J Med Genet. 2015 jmedgenet-2015-103399
104. Pitman RT, Fong JT, Billman P, Puri N. Knockdown of the fat mass and obesity gene disrupts cellular energy balance in a cell-type specific manner. PLoS One. 2012;7:e38444
105. Lin L, Hales CM, Garber K, Jin P. Fat mass and obesity-associated (FTO) protein interacts with CaMKII and modulates the activity of CREB signaling pathway. Hum Mol Genet. 2014;23:3299-306
106. Caruso V, Bahari H, Morris MJ. The beneficial effects of early short-term exercise in the offspring of obese mothers are accompanied by alterations in the hypothalamic gene expression of appetite regulators and FTO (fat mass and obesity associated) gene. J Neuroendocrinol. 2013;25:742-52
107. Gulati P, Yeo GS. The biology of FTO: from nucleic acid demethylase to amino acid sensor. Diabetologia. 2013;56:2113-21
108. Benzinou M, Creemers JW, Choquet H, Lobbens S, Dina C, Durand E. et al. Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat Genet. 2008;40:943-5
109. Jackson RS, Creemers JW, Ohagi S, Raffin-Sanson M-L, Sanders L, Montague CT. et al. Obesity and impaired prohormone processing associated with mutations in the human prohormone convertase 1 gene. Nat Genet. 1997;16:303-6
110. Doulla M, McIntyre AD, Hegele RA, Gallego PH. A novel MC4R mutation associated with childhood-onset obesity: A case report. Paediatr Child Health. 2014;19:515-8
111. Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, Froguel P. et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716-8
112. Vaisse C, Clement K, Guy-Grand B, Froguel P. A frameshift mutation in human MC4R is associated with a dominant form of obesity. Nat Genet. 1998;20:113-4
113. Reyesizquierdo T, Argumedo R, Shu C, Nemzer B, Pietrzkowski Z. Stimulatory Effect of Whole Coffee Fruit Concentrate Powder on Plasma Levels of Total and Exosomal Brain-Derived Neurotrophic Factor in Healthy Subjects: An Acute Within-Subject Clinical Study. Food & Nutrition Sciences. 2013;4:984-90
114. Koch M, Varela L, Kim JG, Kim JD, Hernández-Nuño F, Simonds SE. et al. Hypothalamic POMC neurons promote cannabinoid-induced feeding. Nature. 2015;519:45-50
115. Langdon DR. Severe early-onset obesity, adrenal insufficiency and red hair pigmentation caused by POMC mutations in humans. Clin Pediatr (Phila). 1999;38:251
116. Challis BG, Pritchard LE, Creemers JW, Delplanque J, Keogh JM, Luan Ja. et al. A missense mutation disrupting a dibasic prohormone processing site in pro-opiomelanocortin (POMC) increases susceptibility to early-onset obesity through a novel molecular mechanism. Hum Mol Genet. 2002;11:1997-2004
117. Fischer J, Koch L, Emmerling C, Vierkotten J, Peters T, Brüning JC. et al. Inactivation of the Fto gene protects from obesity. Nature. 2009;458:894-8
118. Mcmurray F, Church CD, Larder R, Nicholson G, Wells S, Teboul L. et al. Adult onset global loss of the fto gene alters body composition and metabolism in the mouse. PLoS Genet. 2012;9:e1003166-e
119. Boissel S, Reish O, Proulx K, Kawagoe-Takaki H, Sedgwick B, Yeo GS. et al. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am J Hum Genet. 2009;85:106-11
120. Meyre D, Proulx K, Kawagoe-Takaki H, Vatin V, Gutiérrez-Aguilar R, Lyon D. et al. Prevalence of loss-of-function FTO mutations in lean and obese individuals. Diabetes. 2010;59:311-8
121. Meyre D, Delplanque J, Chèvre J-C, Lecoeur C, Lobbens S, Gallina S. et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157-9
122. Daoud H, Dong Z, Mcmurray F, Yu A, Luco SM, Vanstone J. et al. Identification of a pathogenic FTO mutation by next-generation sequencing in a newborn with growth retardation and developmental delay. BMC Genomics. 2015;64:1-14
123. Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB family of Fe (II)/α-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem. 2015;290:20734-42
124. Shen F, Huang W, Huang J-T, Xiong J, Yang Y, Wu K. et al. Decreased N 6-Methyladenosine in Peripheral Blood RNA From Diabetic Patients Is Associated With FTO Expression Rather Than ALKBH5. J Clin Endocrinol Metab. 2014;100:E148-E54
125. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H. et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2014 gku1276
126. Aik W, Demetriades M, Hamdan MK, Bagg EA, Yeoh KK, Lejeune C. et al. Structural basis for inhibition of the fat mass and obesity associated protein (FTO). J Med Chem. 2013;56:3680-8
127. Zdżalik D, Vågbø CB, Kirpekar F, Davydova E, Puścian A, Maciejewska AM. et al. Protozoan ALKBH8 oxygenases display both DNA repair and tRNA modification activities. PLoS One. 2014;9:e98729
128. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-7
129. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
130. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635-46
131. McMurray F, Demetriades M, Aik W, Merkestein M, Kramer H, Andrew DS. et al. Pharmacological Inhibition of FTO. PLoS One. 2015;10:e0121829
132. Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ. et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. NatCommun. 2013;4:1798
133. Zhao X, Yang Y, Sun B-F, Shi Y, Yang X, Xiao W. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403-19
134. Sevgi M, Rigoux L, Kühn AB, Mauer J, Schilbach L, Hess ME. et al. An Obesity-predisposing variant of the FTO gene regulates D2R-dependent reward learning. J Neurosci. 2015;35:12584-92
135. Davis W, van Rensburg SJ, Cronje FJ, Whati L, Fisher LR, van der Merwe L. et al. The fat mass and obesity-associated FTO rs9939609 polymorphism is associated with elevated homocysteine levels in patients with multiple sclerosis screened for vascular risk factors. Metab Brain Dis. 2014;29:409-19
136. Bravard A, Veilleux A, Disse E, Laville M, Vidal H, Tchernof A. et al. The expression of FTO in human adipose tissue is influenced by fat depot, adiposity, and insulin sensitivity. Obesity (Silver Spring). 2013;21:1165-73
137. Jeong HS, Hayashi G, Okamoto A. Diazirine photocrosslinking recruits activated FTO demethylase complexes for specific N(6)-methyladenosine recognition. ACS Chem Biol. 2015;10:1450-5
138. Zhang M, Zhang Y, Ma J, Guo F, Cao Q, Zhang Y. et al. The Demethylase Activity of FTO (Fat Mass and Obesity Associated Protein) Is Required for Preadipocyte Differentiation. PLoS One. 2015;10:e0133788
139. G J, Y F, X Z, Q D, G Z, Y Y. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-7
140. Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z. et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313-9
141. Meyer K, Saletore Y, Zumbo P, Elemento O, Mason C, Jaffrey S. Comprehensive Analysis of mRNA Methylation Reveals Enrichment in 3′ UTRs and near Stop Codons. Cell. 2012;149:1635-46
142. Dan D, Sharon MM, Schraga S, Mali SD, Lior U, Sivan O. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
143. Xu Z, Ying Y, Bao-Fa S, Yue S, Xin Y, Wen X. et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403-19
144. Merkestein M, Laber S, Mcmurray F, Andrew D, Sachse G, Sanderson J. et al. FTO influences adipogenesis by regulating mitotic clonal expansion. NatCommun. 2015:6
145. Rochford JJ, Semple RK, Matthias L, Boyle KB, Constantinos C, Claire M. et al. ETO/MTG8 is an inhibitor of C/EBPbeta activity and a regulator of early adipogenesis. Mol Cell Biol. 2004;24:9863-72
146. Qiong W, Saunders RA, Maria SM, Serna IDL, Khew-Voon C. The obesity-associated Fto gene is a transcriptional coactivator. Biochem Biophys Res Commun. 2010;401:390-5
147. Anja R, Enrico M, David F, Pavla N, ?yvind D, Engstr?M PRG. et al. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci U S A. 2010;107:775-80
148. Scott S, Tena JJ, Kyoung-Han K, Gamazon ER, Sakabe NJ, Carlos GM. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371-5
149. Tao H, Qibin Q, Yanping L, Hu FB, Bray GA, Sacks FM. et al. FTO genotype, dietary protein, and change in appetite: the Preventing Overweight Using Novel Dietary Strategies trial. American Journal of Clinical Nutrition. 2014;99:1126-30
150. Muc M, Padez C, Manco L. Influence of physical activity on the association between the FTO variant rs9939609 and adiposity in young adults. Am J Hum Biol. 2015;00:734-8
151. Melina C, Dankel SN, Kyoung-Han K, Gerald Q, Wouter M, Christine H. et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373:895-907
152. Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T. et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518-25
153. Labros S, Shingo K. Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. J Clin Invest. 2015;125:478-86
154. Tönjes A, Zeggini E, Kovacs P, Böttcher Y, Schleinitz D, Dietrich K. et al. Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Sadhana. 2009;18:104-10
155. Li C, Cao W. Advances in CRISPR/Cas9-mediated gene editing. Sheng wu gong cheng xue bao= Chinese journal of biotechnology. 2015;31:1531-42
156. Steyer B, Carlson-Stevermer J, Angenent-Mari N, Khalil A, Harkness T, Saha K. High content analysis platform for optimization of lipid mediated CRISPR-Cas9 delivery strategies in human cells ☆. Acta Biomater. 2015;34:143-58
157. Adebowale A, Guanjie C, Jie Z, Daniel S, Ayo D, Hanxia H. et al. FTO genetic variation and association with obesity in West Africans and African Americans. Diabetes. 2010;59:1549-54
158. Tonjes A, Ekovacs Z. Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Sadhana. 2009;18:104-10
159. Díaz-Anzaldúa A, Ocampo-Mendoza Y, Hernández-Lagunas JO, Díaz-Madrid FA, Romo-Nava F, Juárez-García F. et al. Differences in body mass index according to fat mass- and obesity-associated (FTO) genotype in Mexican patients with bipolar disorder. Bipolar Disorders. 2015;17:662-9
160. Mailly F, Bérubé G, Harada R, Mao PL, Phillips S, Nepveu A. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16:5346-57
161. George S, Leduc CA, Cremona ML, Chung WK, Leibel RL. Cut-like homeobox 1 (CUX1) regulates expression of the fat mass and obesity-associated and retinitis pigmentosa GTPase regulator-interacting protein-1-like (RPGRIP1L) genes and coordinates leptin receptor signaling. J Biol Chem. 2011;286:2155-70
162. Stratigopoulos G, Martincarli J, O'Day D, Wang L, Leduc C, Lanzano P. et al. Hypomorphism for RPGRIP1L, a Ciliary Gene Vicinal to the FTO Locus, Causes Increased Adiposity in Mice. Cell Metabolism. 2014;19:767-79
Author contact
![]() Corresponding authors: suzhqedu.cn (Z-Q.S.) and wshxalbcom (J.G.). Tel.: +86-20-3935-2067 (Z-Q.S.). Fax: +86-20-3935-2067 (J.G.) and +86-20-3935-2067 (Z-Q.S.).
Corresponding authors: suzhqedu.cn (Z-Q.S.) and wshxalbcom (J.G.). Tel.: +86-20-3935-2067 (Z-Q.S.). Fax: +86-20-3935-2067 (J.G.) and +86-20-3935-2067 (Z-Q.S.).

 Global reach, higher impact
Global reach, higher impact