10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(5):531-541. doi:10.7150/ijbs.23945 This issue Cite
Research Paper
PARI functions as a new transcriptional target of FOXM1 involved in gastric cancer development
1. Department of Oncology, Shanghai East Hospital, Tongji University School of Medicine, Shanghai 200120, China;
2. Department of Hematology & Oncology, the People's Hospital of Beilun District, Beilun Branch Hospital of the First Affiliated Hospital of Medical School of Zhejiang University, Ningbo 315800, China;
3. Department of Pharmacy, Changhai Hospital, the Second Military Medical University, Shanghai 200433, China.
*These authors contributed equally to this work.
Received 2017-11-20; Accepted 2018-2-16; Published 2018-4-5
Abstract
PARI, an element of the homologous recombination pathway of DNA repair,is involved in the regulation of cell cycle and carcinogenesis in pancreatic cancer. However, little is known about the function and regulatory mechanism of PARI in other cancers. In the present study, we evaluated the expression of PARI in gastric cancer (GC) by immunohistochemical analysis in a tissue microarray and characterized its functions using in vitro assays and in vivo animal models. We found higher expression of PARI protein was shown in GC tissues compared with related adjacent normal gastric mucosa tissues. Knockdown of PARI by RNA inference decreased cell proliferation, migration, and invasion of GC cells in vitro, as well as reduced the xenograft tumor growth and lung metastasis formation in vivo. Quantitative real-time PCR and western blot results revealed that PARI expression was activated by a well-known oncogene FOXM1 and positively correlated with FOXM1 expression at mRNA level in 38 paired of GC samples. Luciferase reporter assay and chromatin immunoprecipitation assay further demonstrated that FOXM1 directly regulated PARI transcription by binding to the specific sequences of PARI promoter. In addition, PARI knockdown blocked the effect of FOXM1 on GC cell migration. Taken together, our results suggest that PARI plays potential oncogenic roles and functions as a transcriptional target and effector of FOXM1 in GC development.
Keywords: PARI, FOXM1, Gastric cancer, Proliferation, Migration
Introduction
Gastric cancer (GC) is the third leading cause of cancer-related mortality worldwide and particularly prevalent in Asian countries [1]. Current treatments, including surgery, chemotherapy and radiotherapy, have been improved at full speed in recent years. However, GC patients remain a pessimistic survival due to the absence of specific biomarkers for early detection and highly effective therapies [2]. Many oncogenes, tumor suppressor genes and microRNAs have been reported to be closely associated with GC progression [3-5], but promising molecules being useful for GC early diagnosis and targeted therapy are still limited to date. As such, it is essential to devote more research to identify new biomarkers, anti-tumor targets and oncogenic processes operative in GC in the near future for this disease therapy.
PARI, also named C12orf48 or PARPBP, is a vital inhibitor of the homologous recombination of human cells during DNA repair [6]. At molecular level, PARI interacts directly with a few of important regulators of DNA repair and cell proliferation, including PARP-1, RAD51 and PCNA [7, 8]. Downregulation of PARI in some cancer cells could improve homologous recombination and preserve genomic stability. Moreover, PARI is also involved in the regulation of cell cycle [9]. Evidence has revealed that PARI could be recruited to replication forks during S-phase through interacting with SUMOylated PCNA. Importantly, aberrant upregulation of PARI has been observed in a variety of malignancies by gene expression profiling analysis, such as breast cancer, colorectal cancer, lung cancer and hepatocellular carcinoma [10-13]. Recently, PARI is also manifested to be overexpressed in pancreatic ductal adenocarcinoma (PDAC) cells [14]. Knockdown of PARI in PDAC cells significantly suppresses cell growth in vitro and in a mouse xenograft model. However, the expression and function of PARI in GC and the relationship between its expression and clinicopathologic significance remain unclear.
In the present study, we provided evidence that PARI has an altered expression profile in GC tissues. We further demonstrated that knockdown of PARI suppressed GC cell proliferation, migration and invasion in vitro, and inhibited the xenograft tumor growth and lung metastasis in vivo. Furthermore, we found that FOXM1, a well-known transcriptional factor that contributes to oncogenesis in a variety of tissues, activated PARI expression by directly binding to the promoter of PARI. These results highlight FOXM1/PARI axis contributes to gastric carcinogenesis and provide new insights on GC progression and metastasis.
Materials and methods
GC tissue microarray and immunohistochemistry
A tissue microarray (Cat#: HStm-Ade180Sur-06, Shanghai Outdo Biotech, Shanghai, China) with 90 matched pairs of primary GC samples and adjacent gastric tissues was applied to evaluate the expression and clinical relevance of PARI. Specific primary antibody against PARI (Cat: #AP10083b, ABGENT, USA) was used for immunostaining. The staining was performed according to the commercial protocol (Outdo Biotech, Shanghai, China). The immunohistochemical score were assessed by two experienced pathologists without knowledge of patients' characteristics. Staining was analyzed based on the percentage of positively stained cells and staining intensity. Briefly, the staining scores graded as follows: no staining, -; weakly positive, +; moderately positive, ++; and strongly positive, +++. For quantification, all stains were assessed at 200× magnifications and at least 3 fields from each core were counted.
Tissue samples
Fresh paired gastric cancer tissue samples and adjacent non-cancerous stomach tissues were obtained from 38 patients with GC who underwent primary surgical resection at the Department of General Surgery, Shanghai East Hospital, Tongji University School of Medicine between October 2013 and May 2014. Patients who received preoperative treatment were excluded from the study. All patients in this study provided written informed consent. After resection, tissue samples were flash frozen immediately in liquid nitrogen and stored at -80°C prior to RNA extraction. The use of tissue materials for research was approved by the ethics committee of the Shanghai East Hospital of Tongji University.
Cell lines and culture conditions
The four human GC cell lines MGC803, AGS, BGC823, and SGC7901 were obtained from Shanghai Cell Bank of Chinese Academy of Sciences, China and cultured in MEM medium (Life Technology, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone, USA) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin). All above cells were grown in a humidified incubator with 5% CO2 at 37°C.
RNA interference and PARI overexpression plasmid
The siRNAs targeting PARI were designed as siPARI1 (sense 5- GCUGCUCGAGAGAAACAAAdTdT-3) and siPARI2 (sense 5- CCUUUAAGGACACAUGUAAdTdT-3) and chemically synthesized by GenePharma, Shanghai, China. The FOXM1 siRNA was designed as siFOXM1 (sense 5-CUCUUCUCCCUCAGAUAUAdTdT-3). The irrelevant nucleotides not targeting any annotated human genes were used as negative control: siNC (sense 5-UUCUCCGAACGUGUCACGUdTdT-3). PARI expression construct (pcDNA3.1-PARI-HA) was obtained from Shanghai Tuzhu Biotech, China. The accurate reading frame insertion was verified by DNA sequencing. The plasmid pcDNA3.1-FOXM1 was a gift from Dr. Keping Xie, in the University of Texas MD Anderson Cancer Center, US [15]. The lentivirus knocking down PARI (LV-shPARI) were packaged and purchased from GenePharma, Shanghai using siPARI2 corresponding sequences. Cell transfection with plasmids or siRNAs was conducted using Lipofectamine 2000 (Invitrogen, USA) in accordance with the manufacturer's instructions.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from frozen tissue samples and cultured cell lines using Trizol Reagent (Takara, Japan) according to the manufacturer's protocol. First-strand cDNA was synthesized from 1 μg of total RNA using the PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan). qRT-PCR was performed on the 7500 Real-Time PCR System (Applied Biosystems, USA) using 1μL of cDNA with the SYBR Prime-Script RT-PCR kit (Takara, Japan). β-actin was measured as an internal control for cells and tissues. Specific primers used in the analysis were designed as follows: PARI, forward: 5'-ATAATGAAAAGGTGCAGCTG-3', reverse: 5'-CCCTGCAATGCTTGGGTTTG-3'; FOXM1, forward: 5'-GGGCGCACGGCGGAAGATGAA-3', reverse: 5'-CCACTCTTCCAAGGGAGGGCTC-3'; β-actin, forward: 5'-CCTGGCACCCAGCACAATG-3', reverse: 5'- GGGCCGGACTCGTCATACT-3'. The differential expression level was calculated using 2-ΔCt formula. All the experiments were performed in triplicate.
Western blot assay
Cell lysates from GC cells were prepared using cold lysis buffer (25 mmol/L Tris-Cl [pH7.5], 1% sodium dodecylsulfate [SDS], 5mmol/L ethylenediaminetetraacetic acid, protease inhibitor cocktail [Sigma-Aldrich, St Louis, USA]). Samples were boiled for 3 min, separated by electrophoresis in SDS-PAGE (10% w/v) and transferred to a nitrocellulose membranes. The membrane was blocked in 5% blocking buffer (5% non-fat milk and 0.1% Tween-20 in phosphate-buffered saline [PBS]) for 2 h at room temperature, and then incubated with the primary antibody, anti-PARI (1:500, ABGENT, USA), anti-FOXM1 (1:200, Santa Cruz Biotechnology, USA) and anti-GAPDH (1:20000, Proteintech, China), in 0.1% Tween-20 in PBS overnight at 4°C. Incubation with the secondary antibody was performed for 1 h at room temperature. Detection of proteins was achieved by using the Odyssey Infared Imaging System (Li-COR, USA).
CCK8 and colony formation assays
The cell growth assays were performed as described previously [16]. For proliferation assays, 3×103 cells in 100μl culture medium were plated into a well of 96-well plate. After culturing cell for an appropriate time, 10 μl of Cell Counting Kit 8 (CCK8, Dojindo Laboratories, Japan) was added into each well and cultured for 1-2 h. Then optical density was read at 450 nm wavelength and cell growth curves were determined according to the optical density value. For colony formation assay, GC cells were seeded into 6-well plates (1000 cells/well), and the colonies were stained and counted after culture for 14 days. All experiments were performed in triplicate.
Cell migration and invasion assays
3×104 transfected GC cells were cultured in 400 μl serum-free media and seeded in the upper part of each transwell chamber in a 24-well format (Corning, USA). 800 μl of normal MEM medium containing 10% FBS was added as a chemoattractant in the bottom chamber. Similarly, approximately 6×104 cells suspended in 400μl serum-free media were seeded independently in the top chamber coated with matrigel for invasion assays. After incubation for 24-48 h, the cells on the upper part of the chamber were removed with a cotton applicator, whereas the cells migrated through the membrane were stained with crystal violet. The migrated cells on the bottom surface of the membrane were photographed and counted on an inverted microscope. The migrated cells were counted in five random fields under a light microscope (200×magnification) and the average number of five fields was calculated. All assays were performed in triplicate and repeated three times.
Tumorigenicity and lung metastasis models
The protocol for animal experiments was viewed and approved by the Institutional Animal Care and Use Committee of Shanghai East Hospital, Tongji University. Four-week-old Male athymic nude mice were obtained from SLAC Laboratories Animal, Shanghai, China and maintained in sterile laminar flow cabinets. 1.5×106 of SGC7901 or MGC803 cells was inoculated bilaterally and subcutaneously into the flanks of nude mice. Bidimensional tumor measurements were taken with vernier calipers every 4 days, and the mice were euthanized after 4 weeks. The volume of the implanted tumor was calculated using the formula: volume = (π*width2*length)/6. In addition, the subcutaneous tissues were fixed in 4% paraformaldehyde, stored in 70% ethanol, and then treated with paraffin-embedding, sectioning, H&E staining and immunohistochemistry examination with PARI antibody (1:100, ABGENT, USA) and PCNA antibody (1:100, Proteintech, China).
For lung metastasis assay (n=6 for each group), the mice at the age of 6 weeks old were injected with 1×106 cells of SGC7901-Lv-shNC, SGC7901-Lv-shPARI through tail vein, respectively. After 4 weeks, the groups were sacrificed, the lungs were removed and photographed, obvious lung metastatic tumors on the surface were calculated.
Promoter Reporters and Dual-luciferase Assay
The PARI promoter (-1500, +76) was amplified and the fragment was cloned into the luciferase reporter plasmids pGL3-basic vector (Promega, USA), designated as pGL3-PARI. Mutant construct pGL3-PARI-MU was generated as shown in Figure 6C by site-directed mutagenesis. For luciferase assay, 5×104 cells per well in 24-well plates were cultured without antibiotics overnight and then transfected with pGL3-PARI or pGL3-PARI-MU and pcDNA3.1-FOXM1 or negative vector. In the meantime, all cells were co-transfected with pRL-SV40 Renilla luciferase construct for normalization. After 24 h, cells were washed with PBS, subjected to lysis, and their luciferase activities measured by using a dual luciferase assay kit (Promega, USA). All transfections were performed in triplicate and repeated three times.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was performed using a ChIP assay kit (Millipore, CA, USA) according to the manufacturer's instructions. In brief, SGC7901 cells were lysed using SDS lysis buffer and DNA was sheared by sonication. Protein-DNA complexes were precipitated by control immunoglobulin G and anti-FOXM1 antibody (Proteintech, Wuhan, China) respectively, followed by eluting the complex from the antibody. PCR was carried out with primers specific for PARI promoter. The primer pair 5′-ACACAAGGGTTGTCCTGAA-3′ and 5′- CCTTCAAAAGTCTCCAGTC-3′ were used to amplify a 147-bp region (-637 to -490 bp) of the PARI promoter. The PCR products were resolved electrophoretically on a 2% agarose gel and visualized using ethidium bromide staining.
Statistical analysis
Statistical analyses were performed using SPSS (Statistical Package for the Social Sciences) 17.0 software. Results obtained from in vitro and in vivo experiments were presented as mean ± standard deviation (SD) and analyzed by two-tailed Student's t-test. P < 0.05 was considered significant.
Results
PARI is frequently upregulated in GC
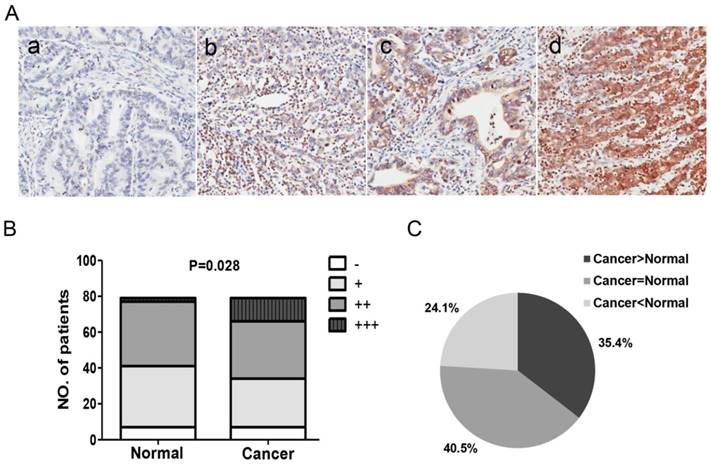
To assess the potential effect of PARI on GC, we first evaluated the expression pattern of PARI in GC samples by immunohistochemical staining. A GC tissue microarray that contains 90 primary gastric cancer specimens and paired adjacent normal gastric mucosa tissues was stained with PARI specific antibody. Among them, 11 paired samples were failed in staining. The rest 79 paired of samples were classified into -, +, ++ and +++ level expression as described in methods section (Figure 1A). The staining level of PARI was significantly higher in GC tissue group compared with the adjacent noncancerous mucosa control group (Figure 1B). When PARI expression was evaluated in GC samples and their matched adjacent normal gastric mucosa tissues, we found PARI was upregulated in 35.4% of GC tissues, whereas downregulated in 24.1% of GC samples (Figure 1C). We next analyzed the relationship between some clinicopathological features and PARI expression levels in gastric cancer cases. However, due to the insufficiency of sample capacity, the clinicopathological features such as age, gender, TNM stage and tumor differentiation between the two groups showed no-statistical significance. These results indicated that PARI expression is upregulated in GC tissues and may play important roles in gastric carcinogenesis.
PARI significantly promotes GC cell proliferation
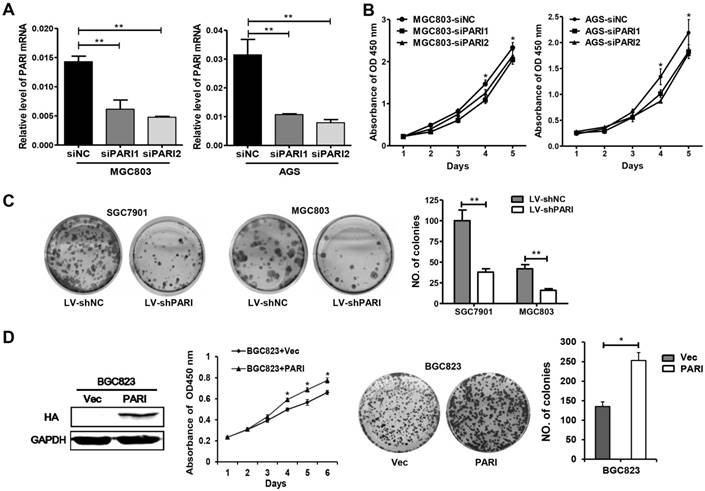
To characterize the function of PARI in GC, we first investigated the effect of PARI silencing on cellular growth. Two specific small interference RNAs, siPARI1, siPARI2 and negative control siNC were transiently transfected into MGC803 or AGS cells, respectively. The efficiency of PARI knockdown was analyzed by qRT-PCR (Figure 2A). Then, cell viability assays were performed to monitor the growth of transfected cells. As shown in Figure 2B, both MGC803 and AGS cells transfected with siPARIs slowed cell growth compared with the negative control group, indicating PARI knockdown inhibited GC cell growth. Furthermore, we conducted colony formation assays to evaluate the long-term effects of PARI on cell proliferation. Two lentivirus particles containing shPARI and shNC were packaged and infected into SGC7901 and MGC803 cells. After puromycin screening, the stable cells were seeded on 6-well plates and allowed to grow for two weeks. The final data exhibited that PARI knockdown also decreased the ability of colony formation of GC cells (Figure 2C). Conversely, the proliferation ability of PARI-overexpressed BGC823 cells was increased significantly relative to the control group as indicated in Figure 2D. These observations above strongly suggested that PARI has a positive effect on GC cell growth in vitro.
Knockdown of PARI suppresses xenograft tumor growth
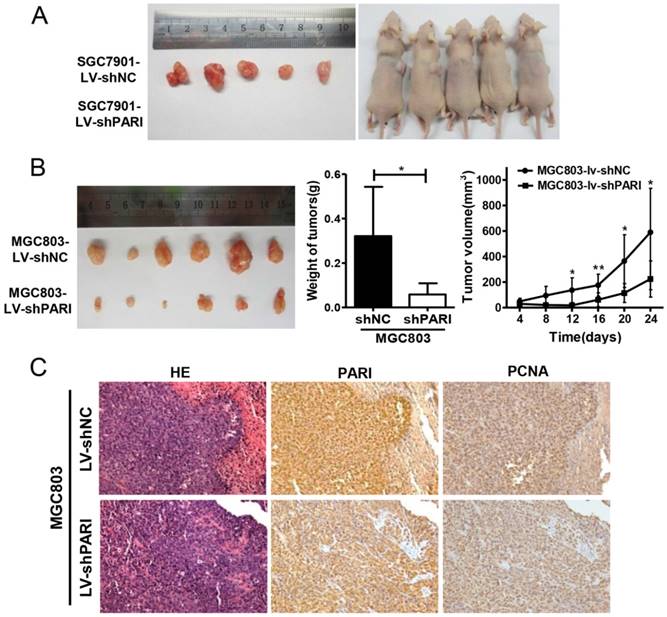
To determine the role of PARI on GC cell growth in vivo, SGC7901-LV-shPARI cells were subcutaneously inoculated into a side of nude mice to form xenograft, while SGC7901-LV-shNC cells were used as controls to inoculate into the opposite side of the same mice. As a result, no visible tumors were observed in the PARI silencing group after 4 weeks, whereas tumors were markedly generated in the control group (Figure 3A), implicating that knockdown of PARI abolished tumor formation ability of SGC7901 cells in nude mice. Meanwhile, MGC803-LV-shPARI cells and MGC803-LV-shNC cells were also employed to detect the tumorigenicity. Consistently, the tumors formed from PARI silenced MGC803 cells were significantly smaller in size and lower in weight than those formed from the control cells (Figure 3B). Moreover, immunostaining of the xenograft tumors with an anti-PARI antibody were performed. PCNA is a cell cycle promotive protein, the expression of which implicates cell proliferation rate. In line with reduced tumor growth, the expression of PCNA was found to be substantially decreased in PARI downregulated tumor tissues (Figure 3C), suggesting PARI expression is critical for tumor cell proliferation in vivo.
PARI knockdown decreases the ability of GC cell migration and invasion
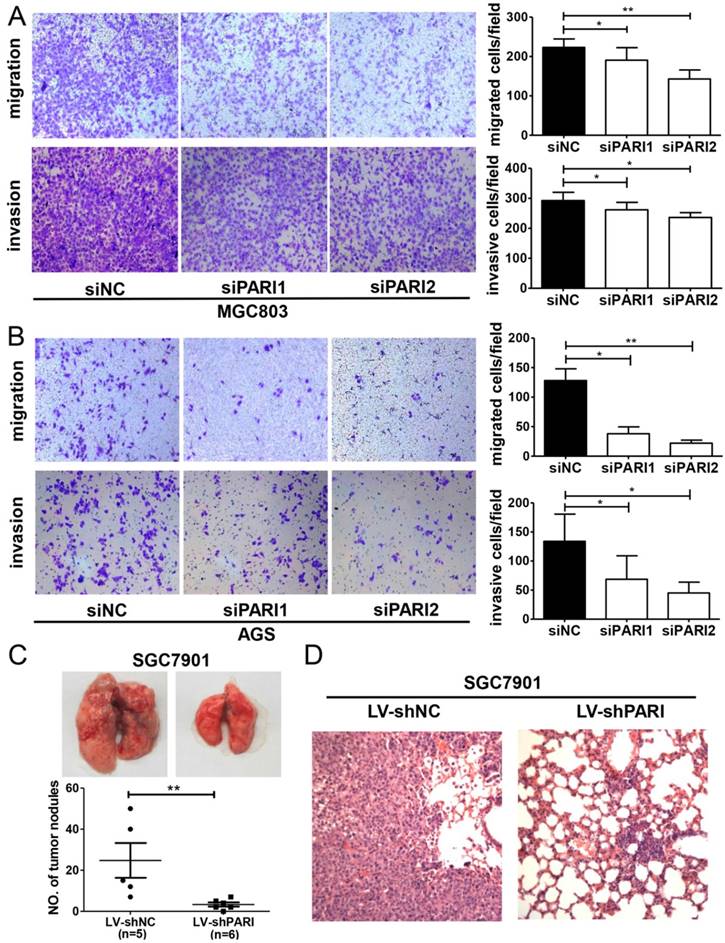
Next, we assessed the effects of PARI on cell migration and invasion, which are key determinants of malignant progression and metastasis of GC. The results showed that cell migration and invasion of MGC803 and AGS cells were significantly attenuated by PARI knockdown in the Transwell assay (Figure 4A and 4B). Consistent with the in vitro assay, the in vivo pulmonary metastasis model result also demonstrated that PARI downregulation suppressed GC cell metastasis, since fewer and smaller tumors formed in the lungs of those mice injected with SGC7901-LV-shPARI cells than those of mice infected with SGC7901-LV-shNC cells. Histological analysis confirmed the presence of metastatic tumors in the lungs of these mice (Figure 4D). These collective data suggested that PARI silencing may be involved in the suppression of metastasis of GC.
PARI expression is regulated by FOXM1 in GC
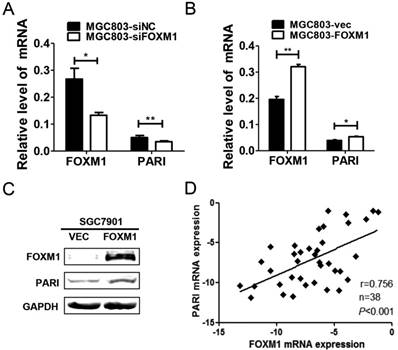
Previous studies have shown that overexpression of FOXM1 contributes to gastric carcinogenesis [17, 18], while the underlying mechanisms remain to be elucidated. Interestingly, a Meta-analysis using oncomine cancer microarray database implicated that PARI is co-expressed with FOXM1 [9]. Therefore, to identify the molecular correlation between PARI and FOXM1, we transfected FOXM1 siRNA into MGC803 cells and detected the expression of PARI and FOXM1 by qRT-PCR. The results indicated that the mRNA level of PARI was significantly decreased with FOXM1 knockdown (Figure 5A). On the contrary, when FOXM1 was overexpressed in this cell line by transiently transfection with FOXM1 expression plasmid, PARI was increased in mRNA level (Figure 5B). Furthermore, PARI was also upregulated in protein level with FOXM1 overexpression in SGC7901 cells (Figure 5C). These results demonstrated that PARI expression is activated by FOXM1. To verify the relationship between FOXM1 and PARI in GC tissue samples, 38 pairs of gastric cancer tissues and neighboring non-cancerous stomach tissues were analyzed by qRT-PCR. Using linear regression analysis, we found there was a significant positive correlation between PARI and FOXM1 expression in GC tissues (Figure 5D), supporting the conclusion that PARI expression is regulated by FOXM1.
PARI expression pattern in gastric cancer samples. (A) Representative immunohistochemical analysis pictures for PARI in clinical samples. (-), representative sections of negative staining; (+), slight positive; (++), moderate positive; (+++), strong staining. Magnification: ×200. (B) Distribution of each level of PARI expression in gastric cancer and normal tissues. (C) The percentage of PARI expression alteration in 79 paired gastric cancer samples.
Effects of PARI knockdown on cell proliferation in vitro. (A) Synthesized siPARIs knocked down the endogenous PARI in MGC803 and AGS cells, as indicated by quantitative reverse transcription polymerase chain reaction. Data represent as means ± SD of three independent experiments. **P<0.01. (B) PARI knockdown inhibited the cellular proliferation of MGC803 and AGS cells, where siNC was used as control. Data represent as means ± SD of three independent experiments. *P<0.05. (C) Stained colonies in a typical well by infecting lentivirus into SGC7901 and MGC803 cells. The number of colonies was calculated and analyzed. The data came from three independent experiments. **P<0.01. (D) PARI overexpression promoted BGC823 cell growth and colony formation. The protein level of PARI overexpression was detected by western blot analysis. *P<0.05.
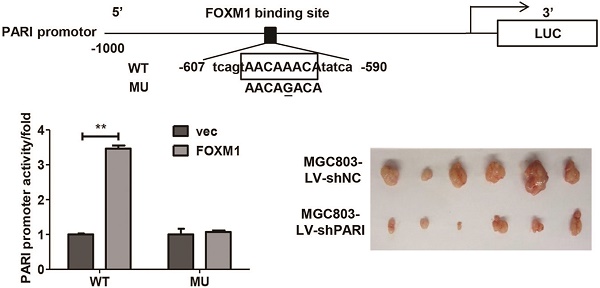
FOXM1 directly binds to the PARI promoter and regulates its activity in GC cells
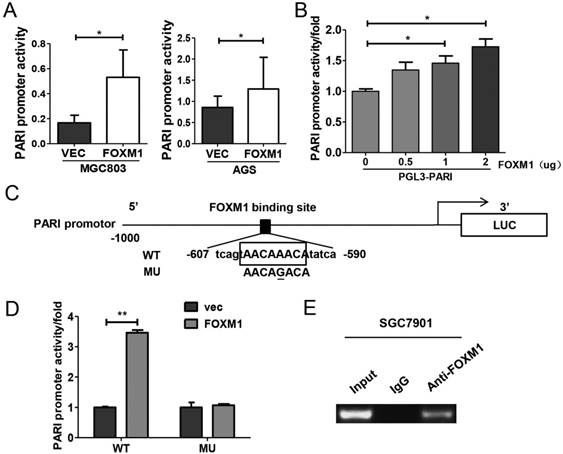
Since FOXM1 is a transcriptional activator, we explored whether FOXM1 regulates PARI promoter activity. The PARI promoter luciferase construct pGL3-PARI and FOXM1 expression plasmid or control vector were co-transfected into MGC803 or AGS cells. As shown in Figure 6A, the luciferase activity was higher in FOXM1 overexpressed cells than the control cells. More importantly, the luciferase activity of PARI promoter was regulated by FOXM1 expression in a dose-dependent manner (Figure 6B). When we analyzed the sequence of PARI promoter using the FOXM1 consensus sequence 5'-AC/ TAAAC/TAA-3', we found a putative FOXM1 binding site in the region of -602 to -595 bp. By site-specific mutagenesis, we generated a mutated PARI promoter construct, pGL3-PARI-MU, as shown in Figure 6C. As expected, disruption of the FOXM1 binding site significantly attenuated the effect of FOXM1 expression on PARI promoter activity (Figure 6D). To address whether FoxM1 may directly bind to endogenous PARI promoter region, we performed chromatin immunoprecipitation assay with SGC7901 cells. The data showed that endogenous FOXM1 could bind to the putative binding site of FOXM1 in PARI promoter region (Figure 6E). All these results above suggested that FOXM1 directly regulates PARI expression in GC cells.
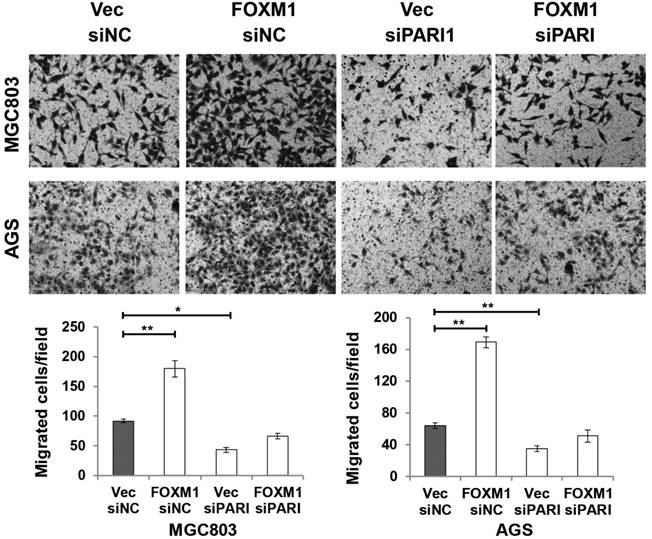
To elucidate the functional link between PARI and FOXM1, we transiently transfected FOXM1 expression plasmid into PARI silenced MGC803 and AGS cells, respectively. Transwell assays were performed to analyze the effect on cell migration. The resulting data indicated that PARI knockdown suppressed the effect of FOXM1 on cell migration (Figure 7), suggesting that PARI may act as a functional downstream effector of FOXM1.
Discussion
Tumor relapse and metastasis are thought of as the major causes of death in GC patients. Unfortunately, because of the lack of knowledge regarding the mechanism underlying tumorigenesis, no effective therapies that block the development and progression have been identified to date. In the present study, we provided evidence that PARI, an element of the homologous recombination pathway of DNA repair, exhibits an altered expression profile in GC that has functional impact in cellular proliferation, migration and metastasis. Our findings may help to understand the tumor development and metastasis and provide a new promising therapy target for GC.
As known, tumorigenesis may originate from the dysregulation of normal developmental genes [19, 20]. Homologous recombination promotes error-free DNA repair of strand breaks in S-phase and G2 by using the sister chromatid as repair template [21]. Inappropriate homologous recombination often causes genomic instability and cancer [22-24]. PARI has been known as a factor involved in homologous recombination pathway of DNA repair. Reports have revealed that PARI downregulation improves homologous recombination and genomic stability in HR-deficient Fanconi Anemia/BRCA pathway- inactivated cancer cells [8]. However, it is found overexpressed in PDAC cell lines and leads to homologous recombination inhibition, DNA damage hypersensitivity, and genomic instability [14]. Furthermore, knockdown of PARI significantly suppresses human HeLa cell growth in another study [25]. They put forward that PARI is an oncogene. A meta-analysis indicates that PARI is co-expressed with a number of mitosis related genes such as AurkA, Plk-1, Cdc20, Cdk1,Nek2, Top2A and CENP family members. This suggests it may be an important molecule for both interphase and mitosis and accordingly regulate cell cycle progression [9]. Therefore, these experimental and bioinformatic findings led us to hypothesize that PARI contributes to tumor progression in GC. Our present data demonstrate that knockdown of PARI decreases the proliferation and metastasis of human gastric cancer cells in vitro and in vivo. All of the results support our hypothesis that PARI plays a stimulative role in progression of gastric cancer and is a potential therapeutic strategy for gastric cancer.
Knockdown of PARI inhibited tumorigenicity of gastric cancer cells in vivo. (A) The picture of xenograft tumors isolated from nude mice (left) and image of nude mice (right) inoculated subcutaneously with SGC7901-LV-shNC and SGC7901-LV-shPARI cells. SGC7901 cells infected with LV-shPARI showed significantly reduced tumor formation ability in the left flank of nude mice. Cells infected with LV-shNC were used as a negative control and were injected into the opposite flank of the same mice (n = 5). (B) MGC803 cells infected with LV-shPARI showed significantly reduced tumor formation and tumor volume in nude mice, where cells infected with LV-shNC was used as control. Tumor weight also remarkably decreased compared to the control group (n=6). *P<0.05, **P<0.01. (C) The expression levels of PARI and PCNA protein in the implanted tumors above were evaluated by immunohistochemistry assays. Tumor tissues were processed by H&E, anti-PARI and anti-PCNA staining. Original magnification, ×200.
Effects of PARI knockdown on cell migration, invasion and metastasis. Silencing PARI inhibited migration and invasion of MGC803 cells (A) and AGS cells (B). Cells were plated in 6-well plates, transfected with siPARIs and control siNC for 48 h. Cell invasion and migration were examined by transwell assay with or without Matrigel. Results are expressed as the mean ± SD of at least three independent experiments. *P<0.05, **P<0.01. (C) Knockdown of PARI reduced gastric cancer cell metastasis in vivo. Lung metastatic tumor nodules observed in lung surface of nude mice received tail vein injection with SGC7901-LV-shNC or SGC7901-LV-shPARI cells. The tumor nodules from each experimental group were shown. **P<0.01. (D) Pulmonary metastases were processed by H&E staining.
Previous studies have demonstrated that FOXM1 is a member of the Forkhead box (Fox) transcription factor family, and is generally highly expressed in several aggressive human carcinomas and related to oncogenesis in many tissue types, including GC [26-31]. In accordance with FOXM1, PARI has the similar function in GC based on our study. Additionally, a meta-analysis has implicated that PARI is co-expressed with FOXM1 in oncomine cancer microarray database. Importantly, we found a putative binding site of FOXM1 in the promoter of PARI. Subsequent molecular biology experiments confirmed PARI transcription is modulated by FOXM1 directly. Furthermore, a strong correlation of the expression of FOXM1 and PARI were observed in patients with GC. Enforced FOXM1 expression did not restore the suppressive phenotype of PARI knockdown on cell migration. Therefore, abnormal expression of FOXM1 during the initiation and development of gastric cancer contributes to abnormal PARI expression and activation; the latter may be a novel molecular marker for poor prognosis and contribute to gastric cancer pathogenesis.
In conclusion, our study is the first to demonstrate the expression pattern of PARI and its potential roles in human GC, as well as the involvement in the FOXM1 mediated oncogenesis. Further study is still required to clarify the downstream pathway of PARI in the future.
PARI expression is related with FOXM1 expression in human GC cell line and specimens. (A) Downregulation of FOXM1 by transfecting siFOXM1 reduced the mRNA level of PARI by qRT-PCR. (B) Upregulation of FOXM1 by transfecting FOXM1 expression plasmid increased the mRNA level of PARI. The results of qRT-PCR were analyzed by using the 2-ΔCt method. *P<0.05, **P<0.01. (C) Upregulation of FOXM1 by transfecting FOXM1 expression plasmid increased the protein expression of PARI in SGC7901 cells by western blot analysis. (D) A positive correlation between PARI and FOXM1 expression in 38 paired gastric cancer samples was measured by linear regression (r = 0.756, P< 0.001).
FOXM1 positively regulates PARI transcription by binding to the PARI promoter. (A) The PARI promoter reporter plasmid pGL3-PARI was co-transfected into MGC803 and AGS cells in triplicate with pcDNA3.1-FOXM1 or empty vector. Luciferase activity was normalized to Renilla luciferase activity. The relative PARI promoter activities were measured 24 h after transfection. Data represents means ± SD of at least three independent experiments. *P < 0.05. (B) Luciferase activities of PARI promoter reporter were measured in MGC803 cells transfected with different dose of pcDNA3.1-FOXM1 using dual luciferase assay kit. *P < 0.05. (C) Schematic of the PARI promoter reporter and its putative FOXM1-binding site. Both wild-type (WT) and mutant (MU) forms are shown. (D) Luciferase reporter assay in MGC803 cells transfected with luciferase reporter constructs containing wild-type (WT) or mutant (MU). Data represent means ± SD of at least three independent experiments. **P < 0.01. (E) A ChIP assay was performed using a specific anti-FOXM1 antibody and oligonucleotides flanking the PARI promoter region containing putative FOXM1-binding site. The oligonucleotides of the cells were immunoprecipitated with anti-FOXM1 antibody or negative control IgG and subjected to PCR on a 2% agarose gel.
PARI knockdown suppresses the effect of FOXM1 on cell migration. MGC803 and AGS cells were co-transfected with siRNAs and plasmids as indicated. Representative photographs of migratory cells on the transwell membrane (upper, magnification, ×200). The average GC cell numbers of triplicate were shown (bottom, *P<0.05, **P<0.01.)
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 81302064, No. 81472576 and No. 81272917) and Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (No. PWZxq2017-13)
Competing Interests
The authors have declared that no competing interest exists.
References
1. Herrero R, Parsonnet J, Greenberg ER. Prevention of gastric cancer. Jama. 2014;312:1197-8
2. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M. et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer. 2015;136:E359-86
3. Zhang Y, Guan DH, Bi RX, Xie J, Yang CH, Jiang YH. Prognostic value of microRNAs in gastric cancer: a meta-analysis. Oncotarget. 2017;8(33):55489-510
4. Zhang M, Du X. Noncoding RNAs in gastric cancer: Research progress and prospects. World journal of gastroenterology. 2016;22:6610-8
5. Kim J, Yum S, Kang C, Kang SJ. Gene-gene interactions in gastrointestinal cancer susceptibility. Oncotarget. 2016;7:67612-25
6. Piao L, Nakagawa H, Ueda K, Chung S, Kashiwaya K, Eguchi H. et al. C12orf48, termed PARP-1 binding protein, enhances poly(ADP-ribose) polymerase-1 (PARP-1) activity and protects pancreatic cancer cells from DNA damage. Genes, chromosomes & cancer. 2011;50:13-24
7. Burkovics P, Dome L, Juhasz S, Altmannova V, Sebesta M, Pacesa M. et al. The PCNA-associated protein PARI negatively regulates homologous recombination via the inhibition of DNA repair synthesis. Nucleic acids research. 2016;44:3176-89
8. Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, Boulton SJ. et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Molecular cell. 2012;45:75-86
9. Varisli L. Meta-analysis of the cell cycle related C12orf48. Biocell. 2013;37:11-6
10. Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A. et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer cell. 2006;9:121-32
11. Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D. et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PloS one. 2010:5
12. Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M. et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938-47
13. Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H. et al. Transcriptome profile of human colorectal adenomas. Molecular cancer research: MCR. 2007;5:1263-75
14. O'Connor KW, Dejsuphong D, Park E, Nicolae CM, Kimmelman AC, D'Andrea AD. et al. PARI overexpression promotes genomic instability and pancreatic tumorigenesis. Cancer research. 2013;73:2529-39
15. Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H. et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer cell. 2011;20:427-42
16. Ye X, Zhang Y, Wang X, Li Y, Gao Y. Tumor-suppressive functions of long-chain acyl-CoA synthetase 4 in gastric cancer. IUBMB life. 2016;68:320-7
17. Wang H, Huang C. FOXM1 and its Oncogenic Signaling in Gastric Cancer. Recent patents on anti-cancer drug discovery. 2015;10:270-9
18. Dibb M, Han N, Choudhury J, Hayes S, Valentine H, West C. et al. FOXM1 and polo-like kinase 1 are co-ordinately overexpressed in patients with gastric adenocarcinomas. BMC research notes. 2015;8:676
19. Chen Y, Hao J, Jiang W, He T, Zhang X, Jiang T. et al. Identifying potential cancer driver genes by genomic data integration. Scientific reports. 2013;3:3538
20. Ge S, Li B, Li Y, Li Z, Liu Z, Chen Z. et al. Genomic alterations in advanced gastric cancer endoscopic biopsy samples using targeted next-generation sequencing. American journal of cancer research. 2017;7:1540-53
21. Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual review of genetics. 2010;44:113-39
22. Oda K, Tanikawa M, Sone K, Mori-Uchino M, Osuga Y, Fujii T. Recent advances in targeting DNA repair pathways for the treatment of ovarian cancer and their clinical relevance. International journal of clinical oncology. 2017;22:611-8
23. Young K, Starling N, Cunningham D. Targeting deficient DNA damage repair in gastric cancer. Expert opinion on pharmacotherapy. 2016;17:1757-66
24. Jung CK, Jung JH, Lee KY, Kang CS, Kim M, Ko YH. et al. Centrosome abnormalities in non-small cell lung cancer: correlations with DNA aneuploidy and expression of cell cycle regulatory proteins. Pathology, research and practice. 2007;203:839-47
25. van Dam S, Cordeiro R, Craig T, van Dam J, Wood SH, de Magalhaes JP. GeneFriends: an online co-expression analysis tool to identify novel gene targets for aging and complex diseases. BMC genomics. 2012;13:535
26. Lee JJ, Lee HJ, Son BH, Kim SB, Ahn JH, Ahn SD. et al. Expression of FOXM1 and related proteins in breast cancer molecular subtypes. International journal of experimental pathology. 2016;97:170-7
27. Li D, Wei P, Peng Z, Huang C, Tang H, Jia Z. et al. The critical role of dysregulated FOXM1-PLAUR signaling in human colon cancer progression and metastasis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:62-72
28. Li X, Qi W, Yao R, Tang D, Liang J. Overexpressed transcription factor FOXM1 is a potential diagnostic and adverse prognostic factor in postoperational gastric cancer patients. Clinical & translational oncology: official publication of the Federation of Spanish Oncology Societies and of the National Cancer Institute of Mexico. 2014;16:307-14
29. Sun Q, Dong M, Chen Y, Zhang J, Qiao J, Guo X. Prognostic significance of FoxM1 expression in non-small cell lung cancer. Journal of thoracic disease. 2016;8:1269-73
30. Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D. et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer research. 2009;69:3501-9
31. Francica P, Nisa L, Aebersold DM, Langer R, Bladt F, Blaukat A. et al. Depletion of FOXM1 via MET Targeting Underlies Establishment of a DNA Damage-Induced Senescence Program in Gastric Cancer. Clin Cancer Res. 2016;22(21):5322-36
Author contact
![]() Corresponding authors: Yandong Li, e-mail: yandongli2017edu.cn and Yong Gao, e-mail: gaoyoncom, 150 Ji-Mo Rd. Shanghai 200120, China.
Corresponding authors: Yandong Li, e-mail: yandongli2017edu.cn and Yong Gao, e-mail: gaoyoncom, 150 Ji-Mo Rd. Shanghai 200120, China.

 Global reach, higher impact
Global reach, higher impact