10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(8):892-900. doi:10.7150/ijbs.24548 This issue Cite
Research Paper
Identifying differentially expressed genes from cross-site integrated data based on relative expression orderings
1. Fujian Key Laboratory of Medical Bioinformatics, Key Laboratory of Ministry of Education for Gastrointestinal Cancer, Fujian Medical University, Fuzhou, 350122, China
2. Fujian Key Laboratory of Tumor Microbiology, Fujian Medical University, Fuzhou 350122, China
3. Department of Systems Biology, College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150086, China
Abstract
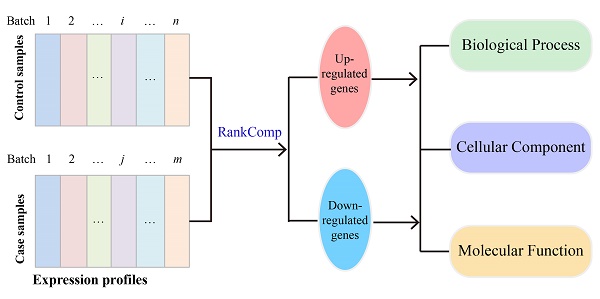
It is a basic task in high-throughput gene expression profiling studies to identify differentially expressed genes (DEGs) between two phenotypes. But the weakly differential expression signals between two phenotypes are hardly detectable with limited sample sizes. To solve this problem, many researchers tried to combine multiple independent datasets using meta-analysis or batch effect adjustment algorithms. However, these algorithms may distort true biological differences between two phenotypes and introduce unacceptable high false rates, as demonstrated in this study. These problems pose critical obstacles for analyzing the transcriptional data in The Cancer Genome Atlas where there are many small-scale batches of data. Previously, we developed RankComp to detect DEGs for individual disease samples through exploiting the incongruous relative expression orderings between two phenotypes and further improved it here to identify DEGs using multiple independent datasets. We demonstrated the improved RankComp can directly analyze integrated cross-site data to detect DEGs between two phenotypes without the need of batch effect adjustments. Its usage was illustrated in detecting weak differential expression signals of breast cancer drug-response data using combined datasets from multiple experiments.
Keywords: Differentially expressed genes, Relative expression orderings, Batch effect, Drug response

 Global reach, higher impact
Global reach, higher impact