10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(8):971-982. doi:10.7150/ijbs.23350 This issue Cite
Research Paper
Predicting Potential Drugs for Breast Cancer based on miRNA and Tissue Specificity
School of Computer Science and Technology, Xidian University, Xi'an, 710071, P.R. China.
Abstract
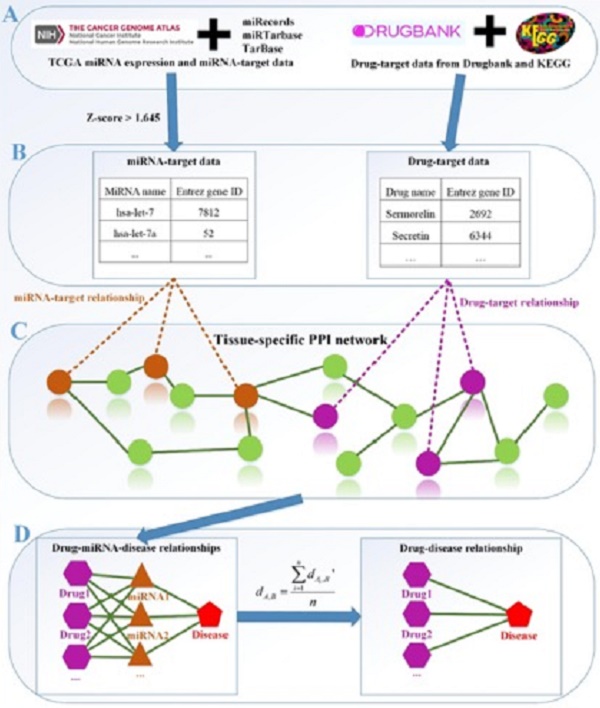
Network-based computational method, with the emphasis on biomolecular interactions and biological data integration, has succeeded in drug development and created new directions, such as drug repositioning and drug combination. Drug repositioning, that is finding new uses for existing drugs to treat more patients, offers time, cost and efficiency benefits in drug development, especially when in silico techniques are used. MicroRNAs (miRNAs) play important roles in multiple biological processes and have attracted much scientific attention recently. Moreover, cumulative studies demonstrate that the mature miRNAs as well as their precursors can be targeted by small molecular drugs. At the same time, human diseases result from the disordered interplay of tissue- and cell lineage-specific processes. However, few computational researches predict drug-disease potential relationships based on miRNA data and tissue specificity. Therefore, based on miRNA data and the tissue specificity of diseases, we propose a new method named as miTS to predict the potential treatments for diseases. Firstly, based on miRNAs data, target genes and information of FDA (Food and Drug Administration) approved drugs, we evaluate the relationships between miRNAs and drugs in the tissue-specific PPI (protein-protein) network. Then, we construct a tripartite network: drug-miRNA-disease Finally, we obtain the potential drug-disease associations based on the tripartite network. In this paper, we take breast cancer as case study and focus on the top-30 predicted drugs. 25 of them (83.3%) are found having known connections with breast cancer in CTD (Comparative Toxicogenomics Database) benchmark and the other 5 drugs are potential drugs for breast cancer. We further evaluate the 5 newly predicted drugs from clinical records, literature mining, KEGG pathways enrichment analysis and overlapping genes between enriched pathways. For each of the 5 new drugs, strongly supported evidences can be found in three or more aspects. In particular, Regorafenib (DB08896) has 15 overlapping KEGG pathways with breast cancer and their p-values are all very small. In addition, whether in the literature curation or clinical validation, Regorafenib has a strong correlation with breast cancer. All the facts show that Regorafenib is likely to be a truly effective drug, worthy of our further study. It further follows that our method miTS is effective and practical for predicting new drug indications, which will provide potential values for treatments of complex diseases.
Keywords: drug repositioning, miRNAs, tissue specificity, module distance

 Global reach, higher impact
Global reach, higher impact