10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2018; 14(8):983-991. doi:10.7150/ijbs.23817 This issue Cite
Research Paper
Improving Prediction of Self-interacting Proteins Using Stacked Sparse Auto-Encoder with PSSM profiles
1. University of Chinese Academy of Sciences, Beijing 100049, China;
2. Xinjiang Technical Institute of Physics and Chemistry, Chinese Academy of Science, Urumqi 830011, China;
3. Institute of Machine Learning and Systems Biology, School of Electronics and Information Engineering, Tongji University, Caoan Road 4800, Shanghai 201804, China;
4. College of Computer Science and Technology, and Key Laboratory of Symbolic Computation and Knowledge Engineering of Ministry of Education, Jilin University, Changchun, Jilin 130012, China.
*These authors contributed equally to this work.
Abstract
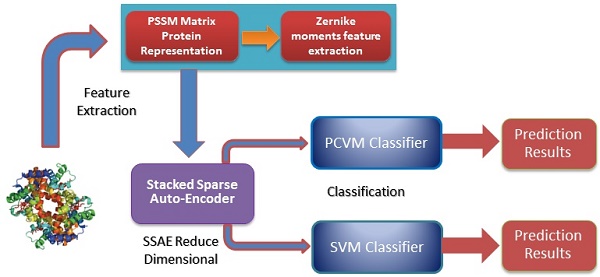
Self-interacting proteins (SIPs) play a significant role in the execution of most important molecular processes in cells, such as signal transduction, gene expression regulation, immune response and enzyme activation. Although the traditional experimental methods can be used to generate SIPs data, it is very expensive and time-consuming based only on biological technique. Therefore, it is important and urgent to develop an efficient computational method for SIPs detection. In this study, we present a novel SIPs identification method based on machine learning technology by combing the Zernike Moments (ZMs) descriptor on Position Specific Scoring Matrix (PSSM) with Probabilistic Classification Vector Machines (PCVM) and Stacked Sparse Auto-Encoder (SSAE). More specifically, an efficient feature extraction technique called ZMs is firstly utilized to generate feature vectors on Position Specific Scoring Matrix (PSSM); Then, Deep neural network is employed for reducing the feature dimensions and noise; Finally, the Probabilistic Classification Vector Machine is used to execute the classification. The prediction performance of the proposed method is evaluated on S.erevisiae and Human SIPs datasets via cross-validation. The experimental results indicate that the proposed method can achieve good accuracies of 92.55% and 97.47%, respectively. To further evaluate the advantage of our scheme for SIPs prediction, we also compared the PCVM classifier with the Support Vector Machine (SVM) and other existing techniques on the same data sets. Comparison results reveal that the proposed strategy is outperforms other methods and could be a used tool for identifying SIPs.
Keywords: Deep learning, Zernike Moments, Probabilistic Classification Vector Machines

 Global reach, higher impact
Global reach, higher impact