10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(4):857-869. doi:10.7150/ijbs.30611 This issue Cite
Research Paper
MicroRNA-322 Regulates Self-renewal of Mouse Spermatogonial Stem Cells through Rassf8
1. Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders (Ministry of Education), Bio-X Institutes, Shanghai Jiao Tong University, Shanghai 200240, China
2. Key Laboratory of Fertility Preservation and Maintenance of Ministry of Education, Ningxia Medical University, Yinchuan 750004, China
3. Shanghai Key Laboratory of Reproductive Medicine, Shanghai 200025, China
4. State Key laboratory of Oncogenes and Related Genes, Shanghai Cancer Institute, Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200032, China
* These authors contribute equally to this work
Received 2018-10-11; Accepted 2019-1-11; Published 2019-3-1
Abstract
Spermatogonial stem cells (SSCs) are essential for spermatogenesis and male fertility. MicroRNAs (miRs) are key regulators of gene expression involved in self-renewal, differentiation, and apoptosis. However, the function and mechanisms of individual miR in regulating self-renewal and differentiation of SSCs remain unclear. Here, we report for the first time that miR-322 regulates self-renewal of SSCs. Functional assays revealed that miR-322 was essential for SSC self-renewal. Mechanistically, miR-322 promoted SSC self-renewal by targeting RASSF8 (ras association domain family 8). Moreover, the WNT/β-catenin signaling pathway was involved in the miR-322-mediated regulation. Furthermore, miR-322 overexpression increased GFRα1, ETV5 and PLZF expression but decreased STRA8, C-KIT and BCL6 expression. Our study provides not only a novel insight into molecular mechanisms regulating SSC self-renewal but also a basis for the diagnosis, treatment, and prevention of male infertility.
Keywords: miRNA-322, Rassf8, SSCs, slef-renewal, WNT/β-catenin signaling pathway
Introduction
Continual spermatogenesis is a stepping stone to male fertility, which mostly relies on spermatogonial stem cells (SSCs), accounting for only 0.02%-0.03% of the germ cell population[1]. SSCs settle on the basement membrane and then migrate toward the niche, a microenvironment that provides nutritional and structural support for SSC self-renewal[2, 3].
Spermatogenesis is a complex process and involves a variety of endocrine and paracrine signals to coordinate SSC self-renewal and differentiation[4, 5]. Numerous studies have revealed that SSC self-renewal is related to the intricate mechanisms of gene regulation, including the RNA-induced silencing complex (RISC) and microRNAs (miRs). Previous studies have demonstrated that miRs are vitally important for spermatogenesis[6, 7]. For example, deletion of RISC component Dicer in germ disrupts spermatogenesis and gives rise to infertility[8, 9].
MiRNAs are globally expressed in the murine testis, and numerous miRNAs play vital roles in spermatogenesis, especially SSC development[10]. For example, miR-34b/c and miR-449a/b/c are important for normal spermatogenesis and male fertility, and knockout of miR-34b/c and miR-449 leads to infertility because of severe spermatogenic disruptions[11]. MiR-100 is highly expressed in SSCs, and miR-100 promotes SSC proliferation via Stat3[10]. MiR-17-92 is a critical player in normal spermatogenesis of mice, and disruption of miR-17-92 suppresses sperm production because of the reduced number of SSCs[12]. MiR-10b regulates the self-renewal of SSCs by targeting Klf4[13]. There are approximately 1000 miRNAs encoded in the mouse genome. Therefore, more miRNAs may be involved in regulating the fate of SSCs[14].
Recent studies have shown that miR-322 promotes lipopolysaccharide-stimulated murine macrophage proliferation and negatively regulates the inflammatory response[15]. Another study reported that miR-322 protects against hypoxia-induced apoptosis in cardiomyocytes by targeting BDNF[16]. MiR-322 also plays a vital role in the proliferation of pulmonary arterial smooth muscle cells[17], vascular smooth muscle cells[18], and neural stem cells[19]. Nevertheless, there is little information about the function and target genes of miR-322 in controlling SSC fate determination.
In this study, we report that miR-322 is a novel intrinsic RNA molecule that regulates self-renewal and differentiation of SSCs via the WNT/β-catenin signaling by targeting Rassf8. Our findings revealed a novel mechanism regulating SSC fate determination and may have important implications in male infertility.
Methods
Animals
C57BL/6 mice purchased from the Shanghai SLAC Laboratory Animal Co. Ltd were used in this study. All mouse experiments were approved by the Institutional Animal Care and Use Committee of Shanghai. The procedures were conducted in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals.
Culture of mouse SSCs
The cells used in this study were an SSC line that we had established previously [13, 20]. The SSC culture system employed a mitotically inactivated STO (SIM mouse embryo-derived thioguanine and ouabainresistant feeder) feeder layer. STO cells were cultured in DMEM supplemented with fetal bovine serum (FBS, Life Technologies), 2 mM glutamine (Amresco), 100 µg/ml penicillin (Amresco), and 100 µg/ml streptomycin (Amresco) (STO culture medium). The culture medium for SSCs was alpha-minimum essential medium supplemented with 1 mM sodium pyruvate (Amresco), 10% fetal bovine serum (FBS, Life Technologies), 10 ng/ml mouse glial cell line‐derived neurotrophic factor (GDNF) (PeproTech), 1 mM non-essential amino acids (Invitrogen), 2 mM L-glutamine (Amresco), 0.1 mM β-mercaptoethanol (Biotech), 10 ng/ml leukemia inhibitory factor (Santa Cruz Biotechnology, CA, USA), 20 µg/ml transferrin (Sigma), 60 µM putrescine, 20 ng/ml mouse epidermal growth factor (PeproTech), 5 µg/ml insulin, 30 mg/l penicillin (Amresco), 75 mg/l streptomycin (Amresco) (SSC medium), and 10 ng/ml human basic fibroblast growth factor (PeproTech). The medium was changed every 2-3 days, and cells were subcultured at 1:2 or 1:3 ratios by enzymatic digestion every 5-7 days. The cells were maintained at 37°C in 5% CO2 [20].
Isolation of germ cells from postnatal mice
SSCs were isolated according to our previously described methods[20]. Briefly, testes were harvested from postnatal day 6 C57BL/6 mice and cut into small pieces. Collagenase (1 mg/ml) was added to the tissues, followed by incubation at 37°C with gentle agitation for about 20 minutes. Then, the tissues were placed in 0.2% trypsin and neutralized by adding 10% FBS. The suspension was centrifuged at 1000 rpm for 5 minutes, and the supernatant was aspirated. Cell pellets were resuspended and passed through a 40-µm nylon cell strainer. Cells expressing Thy1 were isolated by magnetic-activated cell sorting with magnetic microbeads conjugated to an anti-Thy1 antibody (BD Biosciences, USA). Pachytene spermatocytes(PS), round spermatids(RS) and sperms were isolated from 17-dpp, adult mice by the unit gravity sedimentation method according to others described one. For the isolation of PS and RS, briefly, testes of corresponding age mice were harveste, removed the albuginea and cut into small pieces. Then, using collagenase (1 mg/ml) and 0.25%trypsin to digest them in sequence. Dispersed cells were suspended in DMEM and then bottom-loaded into a settling basin followed by BSA solution of 2%-4% gradient in DMEM. After 3hours of sedimentation, the cell fractions were collected from the bottom of the settling basin at a rate of 10mL/min[21]. For the isolation of sperms, epididymides of adult mice were harvested in PBS solution, and removed the fat, then cut into small pieces. Then they were transferred into the centrifuge tube, put the tube at 4°C for 2 hours releasing sperms from epididymis. Then the suspension was centrifuged at 3500 rpm for 15 minutes to collect sperms.
Lentiviral infection and selection of SSCs
For miR-322 lentiviral vectors, we purchased pGMLV-MI7 and pGMLV-MA2 lentiviral vectors and lentivirus packaging plasmids from Genomeditech Biotechnology Co., Ltd (Shanghai, China). The vector plasmid contains a puromycin selection site for selecting cells. We designed a miRNA inhibition sequence for miR-322 knockdown as follows. Primer (F): GATCCGACGGCGCTAGGATCATCAACTCCAAAACATGAATCTATTGCTGCTGCAAGTATTCTGGTCACAGAATACAACTCCAAAACATGAATCTATTGCTGCTGCAAGATGATCCTAGCGCCGTCTTTTTTG; Primer (R): AATTCAAAAAAGACGGCGCTAGGATCATCTTGCAGCAGCAATAGATTCATGTTTTGGAGTTGTATTCTGTGACCAGAATACTTGCAGCAGCAATAGATTCATGTTTTGGAGTTGATGATCCTAGCGCCGTCG. In addition, we designed a pri-miR sequence for miR-322 overexpression as follows. Primer (F): CCGCTCGAGCACCAAGACTTTGGAGCTGGC; Primer (R): CCGGGATCCTACTGTTCCCGCTGCTAGGGC).
For Rassf8 lentiviral vectors, we purchased pGMLV-SC5 RNAi and pGMLV-CMV-MCS-3*flag-EF1-ZsGreen1-T2A-Puro lentiviral vectors from Genomeditech Biotechnology. We designed a sequence against Rassf8 for knockdown Primer (F): GATCCGCCCAAGCTATAGGTCGAACTTTCAAGAGAAGTTCGACCTATAGCTTGGGCTTTTTTG; Primer (R):AATTCAAAAAAGCCCAAGCTATAGGTCGAACTTCTCTTGAAAGTTCGACCTATAGCTTGGGCG and pri-miR sequence for Rassf8 overexpression Primer (F): AACCGGTGCGGCCGCGCCACCATGGAACTTAAAGTGTGGG; Primer (R): ATGGTCT TTGTAGTCTACATAGATGCCTTCAGGATTAAAACCC).
Lentivirus particles were generated by cotransfection of inhibit or overexpression plasmids and lentivirus packaging plasmids into HEK293T cells using transgene reagent. Enhancing buffer was added to the medium after 12 h of transfection. Virus particles were harvested at 48 h after transfection and a standardized virus titer was obtained using HEK293T cells.
For lentivirus infection, 10000 SSCs were seeded on 48-well plate pre-coated with laminin and incubated with 1:1 mixture of culture medium and lentivirus concentrated solution (lentivirus titer: 1*109TU/ml), supplemented with 5µg/ml polybrene. After overnight infection, cells were re-plated onto puromycin-resistant STO feeder layers and cultured in SSC medium. After 12 hours of re-plating, we incubated the SSCs with 1:1 mixture of culture medium and lentivirus concentrated solution once more. After overnight infection, we changed fresh culture medium and cultured for 12 hours. We infected SSCs for the third time. After overnight infection, we changed fresh culture medium and cells were cultured at 37°C in 5% CO2. At the 6th day, cells were subcultured at 1:1 -1:2 ratio, and 100 ng/ml puromycin was added to the SSC culture medium to screen for puromycin-resistant SSCs. We will use puromycin to screen for 72 hours. The surviving SSCs colonies were passaged and analyzed by quantitative real-time polymerase chain reaction (qRT-PCR).
Reverse transcription PCR(RT-PCR) and qRT-PCR
Total RNA was extracted from SSCs using Trizol reagent, according to the manufacturer's protocol. Approximately 1000 ng RNA was used to synthesize cDNA(Complementary Deoxyribonucleic acid) using M-MLV reverse transcriptase in a 20 µl volume. PCR analysis was carried out with Taq DNA polymerase. qRT-PCR was conducted with SYBR Premix Ex Taq (Takara, Shanghai, China) in a 20 µl volume on a Applied Biosystems® 7500 Real-Time PCRSystem. The conditions were 95°C for 30 s, 40 cycles of 95°C for 5 s and 60°C for 34 s, followed by 95°C for 15 s, 60°C for 60 s, and then 95°C for 15 s. Quantitative analyses of miR-322 and Rassf8 employed U6 and Gapdh, respectively, as the internal references. The 2-ΔΔCt method was used to analyze data. The primers were as follows. For RT-PCR, miR-322: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCCAAAAC; U6: AACGCTTCACGAATTTGCGT. For PCR or qRT-PCR, Oct4-F: CCCGGAAGAGAAAGCGAACT;Oct4-R: GGAAAGGTGTCCCTGTAGCC; Plzf-F: TTCAGCCTCAAGCACCAGTT; Plzf-R: GGGCAGTATTCCGTGCAGAT; Gfrα1-F:GGAGGCCTTGAAGCAGAAGT; Gfrα1-R:AACGGGACTGCCCGGAATA; Etv5-F: CTGGGGAACGCTACGTCTAC; Etv5-R: CCAGGAGGTAAGCAGGGTTG; Mvh-F: GTGGAAATACTGGCAGAGCG; Mvh-R: CTGAAGCTGGGAGGCACATA; Gapdh-F: GGTTGTCTCCTGCGACTTCA; Gapdh-R: TAGGGCCTCTCTTGCTCAGT; U6-F: CTCGCTTCGGCAGCACA; U6-R: AACGCTTCACGAATTTGCGT; Rassf8-F: AAACGACGTGGAGATCGAGG; Rassf8-R: TGCCTTCACAGTCTGTCACC; β-catenin-F: AAGGAAGCTTCCAGACATGC; β- catenin-R: AGCTTGCTCTCTTGATTGCC; CyclinD1-F: CAAGGAGATTGGGGACAAC; CyclinD1-R: TTGCTTTGAGTCACACTGGT; Stra8-F:ACAACCTAAGGAAGGCAGTTTAC; Stra8-R: GACCTCCTCTAAGCTGTTGGG; C-kit-F: CTCCCCCAACAGTGTATTCAC; C-kit-R: TAGCCCGAAATCGCAAATCTT; miR-322-F: ACACTCCAGCTGGGCAGCAGCAATTCATGT; miR-322-R: TGGTGTCGTGGAGTCG; C-myc-F: CCACACATCAGACAACTACGCT; C-myc-R: GCATTTTCGGTTGTTGCTGATC; Bcl6-F: CCGGCACGCTAGTGATGTT; Bcl6-R: TGTCTTATGGGCTCTAAACTGCT.
Immunofluorescence staining
Cells cultured in 48-well plates were washed with 1× phosphate-buffered saline (PBS), fixed in 4% formaldehyde for 30 minutes at room temperature, and then washed three times with PBS for 5 minutes each wash. Then, the cells were incubated at 37°C for 10 minutes in blocking buffer (PBS containing 10% goat serum). Next, the cells were incubated overnight at 4°C with the primary rabbit anti-MVH antibody (1:200, Santa Cruz Biotechnology) or the primary rabbit anti-RASSF8 antibody(1:100, Abcam). After washing three times with PBS, the cells were incubated at 37°C for 30 minutes with a 1:150 dilution of tetramethylrhodamine isothiocyanate (TRITC)- conjugated secondary antibody (goat anti-rabbit IgG; ProteinTech). Then, the cells were incubated at 37°C for 10 minutes with 500 ng/mL 4′,6-diamidino-2- phenylindole (DAPI; Sigma). Images were acquired using a Leica digital camera under a fluorescence microscope (DM2500, DMI3000B; Leica).
For PLZF and GFRA1 staining, before incubation in blocking buffer, cells were permeabilized with 0.5% Triton X-100 for 30 minutes at room temperature, and then washed with PBS three times. The primary antibody was mouse-anti-PLZF (1:150, Santa Cruz Biotechnology) or anti-GFRA1 (1:100, ABclonal). The secondary antibody was a 1:150 dilution of goat anti-mouse IgG.
Meiotic spreads and SYCP3 staining
Meiotic spreads of cell samples were prepared as described previously[22]. For SYCP3 staining, the samples were incubated at 37°C for 10 minutes with blocking buffer (PBS containing 10% normal goat serum). Subsequently, cells were incubated overnight with a 1:200 dilution of a rabbit polyclonal anti-SYCP3 antibody (Abcam; ab150292) at 4°C. After washing in PBS, the samples were incubated with a 1:150 dilution of TRITC-conjugated secondary antibody (goat anti- rabbit IgG) at 37°C for 30 minutes. Then, cells were incubated at 37°C for 20 minutes with 500 ng/mL DAPI. The samples were mounted as described above. Images were obtained with a Leica DFC 550 digital camera under the DM2500 microscope.
CCK8 proliferation assay
Mouse SSCs were seeded at 2000 cells/well in 96-well plates and cultured for 3 days. Then, CVTK solution (Research Science, Shanghai, China) was diluted with cell culture medium at 1:10 ratio, then each well of the plate was added 110µL mixture. Be careful to avoid introducing bubbles to the wells, since the bubbles will interfere the O.D. reading. Then the plate was incubated for 2 hours at 37°C in 5% CO2. The incubation needs to avoid light. Absorbance was measured at 450 nm using a microplate reader.
EdU(5-ethynyl-2'- deoxyuridine) proliferation assay
A Cell-Light EdU Apollo567 in vitro imaging kit (RiboBio, Guangzhou, China) was used to analyze SSCs, according to the manufacturer's protocol. Cells were incubated with 50 µM EdU for 2 hours. Then, cells were fixed with 4% paraformaldehyde for 30 minutes at room temperature and then washed with 2 mg/ml glycine for 5 minutes on a shaker. Permeabilization was conducted by 0.5% Triton X-100. Then, 1× Apollo was added, followed by incubation for 30 minutes on a shaker. After washing 3 times with PBS containing 0.5% Triton X-100, 1× Hoechst 33342 was used to stain cell nuclei. Images were captured under the Leica fluorescence microscope.
3′-UTR luciferase reporter assays
psiCHECK-2 vectors including firefly and Renilla luciferase genes were purchased from Promega. Mouse Rassf8 3′-UTRs including the predicted binding site of miR-322 (named wt) or a site-directed gene mutated miR-322-binding site (named mt) were inserted downstream of the firefly luciferase gene of the psiCHECK2 vector. The wt or mt vector was cotransfected into SSCs with miR- 322-vectors or control vectors in 24-well plates. After 48 h, the cells were harvested and assayed by a Dual Luciferase Assay (Promega) in accordance with the manufacturer's protocol. Transfections were repeated at least three times in independent experiments.
Western blot analysis
Cells were lysed with RIPA buffer (Shanghai Yeasen Biotechnology Co., Ltd) containing a protease inhibitor cocktail. Proteins were separated on 12% SDS-PAGE gels and blotted on nylon membranes. Five percent non-fat powdered milk in Tris-buffered saline with Tween 20 (TBST) was used to block the membranes. Then, the membranes were incubated with primary antibodies (rabbit-anti- Rassf8, 1:5000, Abcam; mouse-anti-β-tubulin, 1:6000, Santa Cruz Biotechnology; rabbit-anti-CyclinD1, 1:12000, Abcam) at 4°C overnight. After incubation, the membranes were washed with TBST three times and incubated with a horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. The labeled proteins were visualized by enhanced chemiluminescence.
Cell cycle assay
For analysis of cell cycle, Cell cycle and Apoptosis Analysis Kit (Beyotime, Shanghai, China) was used. Cells were treated with 0.25%trypsin, washed twice in PBS, and then fixed in 70% precooled ethanol solution at 4°C overnight. The next day, the fixed cells were washed with PBS twice. The cells were resuspended with 535 µL propidium iodide(PI) solution(containing 500µL dye buffer, 25µL 20× PI stain and 10µL 50×RNase A) and incubated at 37°C for 30minutes under conditions avoid of light. Then the cell cycle was detected at a 488nm excitation wavelength, which was detected by Cytoflex (Beckman Coulter).
Statistical analysis
Results were shown as the mean ± SD of three independent experiments. Student's t-test was used to calculate differences between groups. Differences were considered significant at p < 0.05.
Results
Characterization of our SSC line
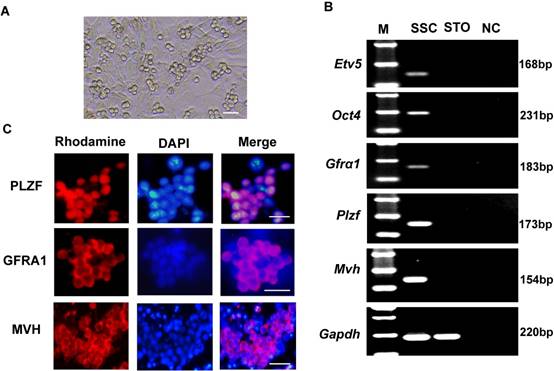
The cultured SSCs appeared as colonies with a grape-like shape (Figure 1A). To characterize these cells, we assessed marker gene expression of spermatogonial progenitor cells or germ cells in the mouse testis: Gfrα1 (GDNF family receptor alpha 1)[23], Plzf (also known as Zbtb16, zinc finger and BTB domain containing 16)[24], Etv5 (ets variant 5)[25], Oct4 (also known as Pou5f1, POU domain, class 5, transcription factor 1)[26], and Mvh (also known as Ddx4, DEAD (Asp-Glu-Ala-Asp) box polypeptide 4)[27]. RT-PCR results showed that the cells expressed Etv5, Oct4, Gfrα1, Plzf, and Mvh (Figure 1B). Immunofluorescence analysis also showed positivity for PLZF, GFRA1 and MVH proteins (Figure 1C).
MiR-322 is highly expressed in SSCs
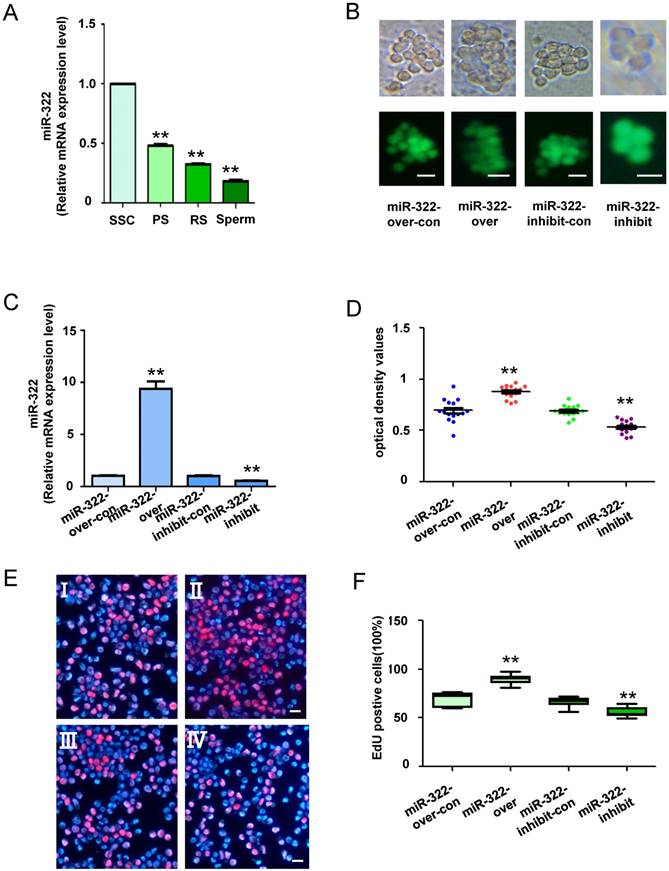
We previously reported expression of miR-322 in the testis of postnatal mice at various days, and its expression is gradually reduced by 6 days after birth[28]. These results indicate that miR-322 plays an important role in SSC development. To determine the expression level of miR-322 in mouse SSCs development, quantitative real-time PCR (qRT-PCR) analysis was conducted in various germ cell types, including the SSCs (isolation of Thy1+ cells from testes of postnatal day 6 C57BL/6 mice), PS, RS and sperms. We found that the expression of miR-322 was gradually decreased as the development of germ cells (Figure 2A). These results indicated high expression of miR-322 in SSCs.
Characteristics of cultured SSCs. (A) Representative morphology of cultured SSCs. (B) Reverse transcription PCR detection of Gfrα1, Plzf, Etv5, Oct4, and Mvh mRNA expression in SSCs. M, 100 bp DNA markers. Gapdh served as a loading control. NC: negative control. (C) Immunofluorescence analysis of SSCs with antibodies against PLZF, GFRA1 and MVH. Scale bar: 20 µm
MiR-322 regulates SSC self-renewal
To investigate the biological function of miR-322 in SSCs, miR-322 overexpression, miR-322 overexpression control, miR-322 inhibition, and miR-322 inhibition control lentiviral vectors were used to infect SSCs (Figure 2B). qRT-PCR analysis showed that miR-322 expression was significantly increased in SSCs infected with miR-322 overexpression compared with cells infected with the miR-322 overexpression control (Figure 2C). Conversely, miR-322 expression was significantly decreased in SSCs infected with the miR-322 inhibition lentiviral vector compared with SSCs infected with the miR-322 inhibition control (Figure 2C). In addition, CCK8 assays demonstrated that the optical density values of miR-322- overexpressing cells were significantly higher than those of control cells after 3 days of culture, whereas the values of miR-322-inhibited cells were decreased significantly (Figure 2D). Furthermore, we used an EdU incorporation assay to assess the proliferation of mouse SSCs infected by miR-322 overexpression or inhibition lentiviral vectors. EdU-positive cells were significantly increased among SSCs infected with the miR-322 overexpression vector compared with the miR-322 overexpression control (Figure 2E and 2F). In contrast, infection with the miR-322 inhibition lentivirus vector led to a significant decrease in EdU- positive cells among SSCs compared with the miR-322 inhibition control (Figure 2E and 2F). Taken together, these results suggested that miR-322 enhances the proliferation of SSCs in vitro.
MiR-322 regulates expression of stemness and differentiated markers
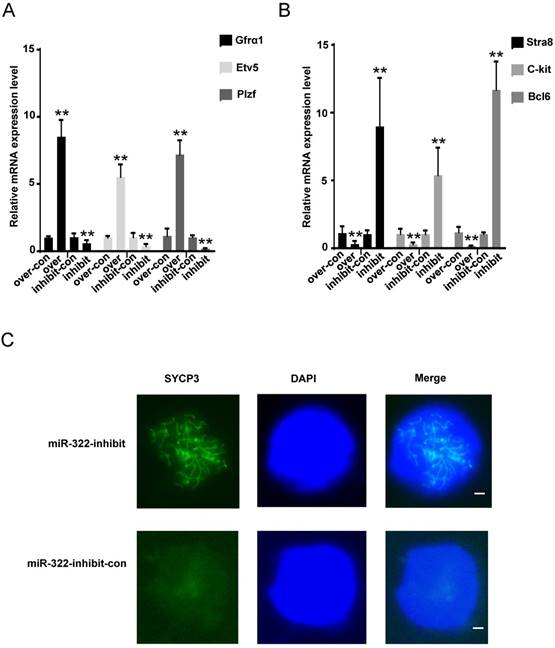
We further explored changes in molecular markers. Gfrα1, Etv5 and Plzf are known to be expressed in undifferentiated spermatogonia [23-25], and Stra8, C-kit and Bcl6 are differentiation markers[29-31]. We found that the mRNA levels of Gfrα1, Etv5 and Plzf were significantly elevated (Figure 3A), but those of Stra8, C-kit and Bcl6 were significantly decreased in SSCs infected with the miR-322 overexpression lentivirus compared with the overexpression control (Figure 3B). In contrast, the expression levels of Gfrα1, Etv5 and Plzf were significantly decreased (Figure 3A), but those of Stra8, C-kit and Bcl6 were increased in SSCs infected with the miR-322 inhibition lentivirus compared with the inhibition control (Figure 3B). Immunofluorescence analysis detected the expression of SYCP3 in SSCs infected with the miR-322 inhibition lentivirus, suggesting that some cells had entered meiosis (Figure 3C). These results revealed that miR-322 plays a role in regulation of SSC self-renewal and differentiation.
MiR-322 expression in various types of cells, and the biological function of miR-322 in SSCs. (A) MiR-322 mRNA levels in SSCs, PS, RS and sperms were measured by qRT-PCR. To compare miR-322 expression among cell types, miR-322 expression in SSCs was set as 1. **P< 0.01 compared with SSC group. (B) Fluorescence and bright field image for SSCs infected with lentivirus. (C) qRT-PCR examined the expression level of miR-322 in SSCs infected with miR-322 overexpression lentivirus control (miR-322-over-con), and miR-322 overexpression lentivirus (miR-322-over), miR-322 inhibition lentivirus control (miR-322-inhibit-con), or miR-322 inhibition lentivirus (miR-322-inhibit). (D, E) CCK-8 assays (D) and EdU incorporation assays (E) were conducted using cells infected with the miR-322 overexpression lentivirus control (miR-322-over-con), miR-322 overexpression lentivirus (miR-322-over), miR-322 inhibition lentivirus control (miR-322-inhibit-con), or miR-322 inhibition lentivirus (miR-322-inhibit). Ⅰ: miR-322-over-con, Ⅱ: miR-322-over, Ⅲ: miR-322-inhibit-con, Ⅳ: miR-322-inhibit. (F) Quantification of EdU assay results. Data are presented as the mean ± SD of three independent experiments. **P< 0.01 compared with the control group. Scale bars: 10 µm
Effects of miR-322 on stemness and differentiation markers of SSCs. (A, B) SSCs were infected with the miR-322 overexpression lentivirus control (over-con), miR-322 overexpression lentivirus (over), miR-322 inhibition lentivirus control (inhibit-con), or miR-322 inhibition lentivirus (inhibit). Expression of Gfrα1, Etv5, Plzf, Stra, C-kit and Bcl6 was measured by qRT-PCR. Data are presented as the mean ± SD of three independent experiments. **P< 0.01 compared with the control group. (C) Meiotic spread assays of SYCP3 expression. SSCs infected with miR-322 inhibition lentivirus control (miR-322-inhibit-con), or miR-322 inhibition lentivirus (miR-322-inhibit) were detected. Scale bar: 1 µm
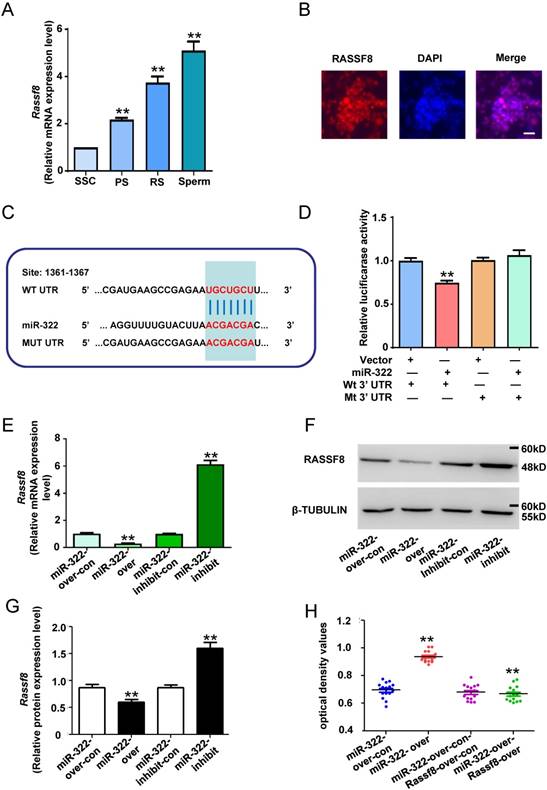
Rassf8 is a direct target of miR-322 in SSCs
To explore the mechanism of miR-322 promoting self-renewal of SSCs, we applied TargetScan to predict the possible target genes. The result showed that Rassf8 is a target gene of miR-322, and miR-322 can bind to the Rassf8 mRNA 3′-untranslated region (UTR). In addition, Rassf8 is a member of the RAS association domain family (RASSF) involved in a myriad of biological processes including cell death, proliferation, and microtubule stability[32]. RASSF8 inhibits cell growth and regulates the Wnt signaling pathway[33], while the Wnt pathway is involved in various cellular processes such as proliferation, differentiation, apoptosis, fate determination, and migration[34]. Furthermore, little is known about the function of Rassf8 in regulating the self-renewal and differentiation of SSCs. For determining the expression level of Rassf8 in mouse SSCs development, qRT- PCR analysis was performed. The expression level of Rassf8 was significantly increased as the development of germ cells (Figure 4A). And immunofluorescence analysis showed positivity for RASSF8 in SSCs (Figure 4B). Next, we conducted qRT-PCR analysis to determine the expression levels of miR-322 and Rassf8.
We used the dual luciferase reporter system to confirm whether miR-322 directly downregulated the expression of Rassf8. We subcloned 3′-UTRs of Rassf8 mRNA, including the predicted miR-322-binding sequence (wild-type) or a mutated sequence (mutant type), into luciferase reporter plasmids (Figure 4C). The relative luciferase activity of the miR-322-vactor group co-transfected with the Rassf8 3′-UTR wild-type was significantly decrease compared with the control group, whereas the Rassf8 3′-UTR mutant group had an insignificant difference compared with the control group (Figure 4D). The results suggested binding between miR-322 and the Rassf8 3′UTR. Subsequently, we found that overexpression of miR-322 significantly reduced Rassf8 expression at mRNA and protein levels compared with control cells (Figure 4E-4G), whereas inhibition of miR-322 significantly increased the expression of Rassf8 at mRNA and protein levels compared with control cells (Figure 4E-4G).
To confirm the interaction between miR-322 and Rassf8, we used a Rassf8 overexpression lentivirus to infect SSCs that had been infected with the miR-322 overexpression lentivirus. CCK8 assays demonstrated that the optical density values of miR-322 and Rassf8-overexpressing cells was significantly decreased compared with those of miR-322-overexpressing cells (Figure 4H). These results suggested that miR-322 and Rassf8 interact.
Collectively, these results indicated that miR-322 directly regulates Rassf8 expression at transcription and translation levels by targeting its 3′-UTR.
MiR-322 directly targets Rassf8. (A) Rassf8 mRNA levels in SSCs, PS, RS and sperms were measured by qRT-PCR. To compare Rassf8 expression among cell types, Rassf8 expression in SSCs was set as 1. **P< 0.01 compared with SSC group. (B) Immunofluorescence analysis of SSCs with antibodies against RASSF8. Scale bar: 20 µm. (C) Target region of the Rassf8 3′-UTR for miR-322 and the mutant type of Rassf8 3′-UTR. (D) Effects of miR-322 on the activity of firefly luciferase reporters containing either wild-type (Wt) or mutant type (Mt) 3′-UTRs were assessed by luciferase reporter gene assays. (E, F, G) Effects of miR-322 on Rassf8 expression levels were examined by qRT-PCR (E) and western blot analyses (F, G). Data are presented as the mean ± SD of three different independent experiments. **P< 0.01 compared with the control group. (H) CCK8 assays detected cell proliferation of SSCs infected with miR-322-over-con, miR-322-over, miR-322-over-con-Rassf8-over-con, or miR-322-over-Rassf8-over. Scale bar: 20 µm
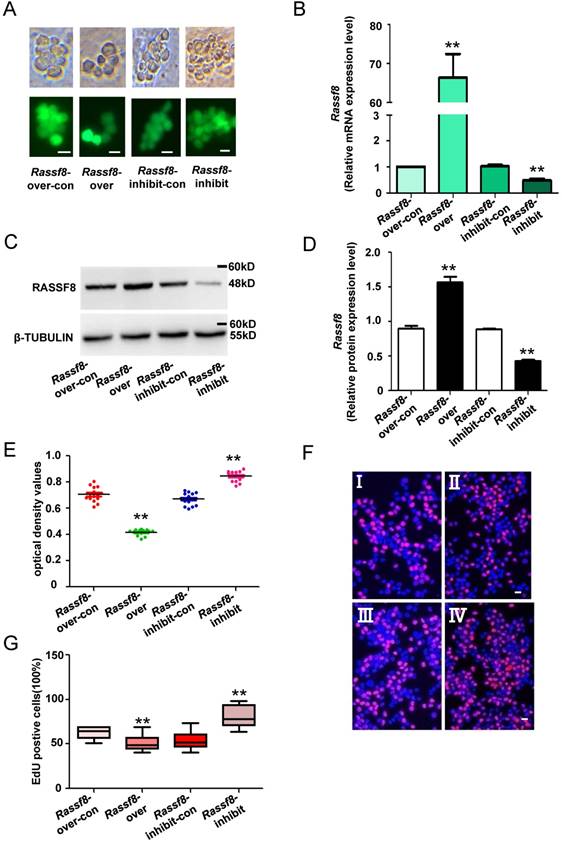
Knockdown of Rassf8 enhances the proliferation of SSCs in vitro
We next explored the function of Rassf8 in regulating SSC proliferation. Rassf8 inhibition and Rassf8 overexpression lentivirus were used to infect SSCs and regulate endogenous Rassf8 expression. Both qRT-PCR and western blot analyses revealed that the mRNA and protein levels of Rassf8 were significantly increased in SSCs after Rassf8 overexpression lentivirus infection compared with cells infected with the Rassf8 overexpression lentivirus control (Figure 5A-5D). Conversely, Rassf8 expression was significantly decreased in SSCs infected with the Rassf8 inhibition lentivirus compared with SSCs infected with the Rassf8 inhibition lentivirus control (Figure 5A-5D). In addition, CCK8 assays demonstrated that the optical density values of Rassf8 inhibition lentivirus-infected cells were significantly higher than those of control cells at 3 days (Figure 5E), whereas values of Rassf8 overexpression lentivirus-infected cells were significantly lower than those of controls (Figure 5E). Furthermore, EdU assays revealed that EdU-positive cells were significantly increased among SSCs infected with the Rassf8 inhibition lentivirus compared with the Rassf8 inhibition lentivirus control (Figure 5F and 5G). In contrast, infection by the Rassf8 overexpression lentivirus led to a significant decrease in EdU-positive cells among SSCs compared with infection by the Rassf8 overexpression lentivirus control (Figure 5F and 5G). These results suggested that Rassf8 inhibits the proliferation of SSCs in vitro.
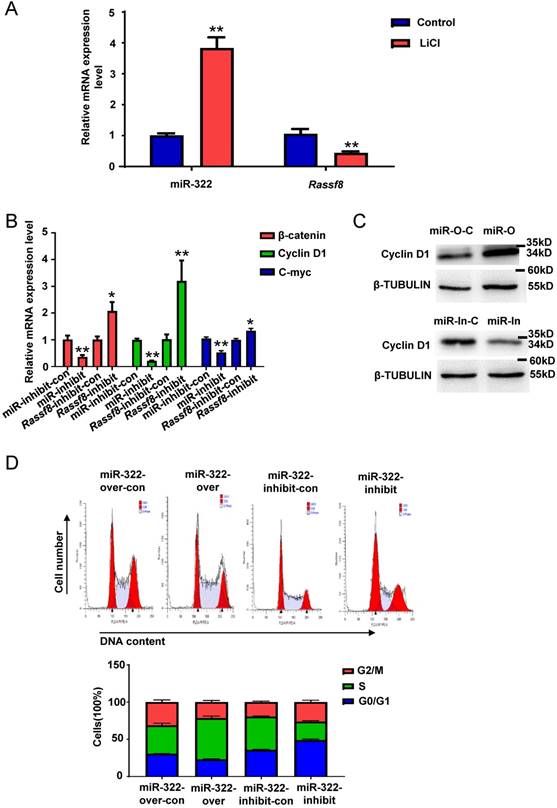
MiR-322 regulates SSC self-renewal via WNT/β-catenin signaling
WNT/β-catenin signaling plays a vital role in self-renewal and differentiation of SSCs[35]. To explore whether miR-322 is related to the WNT/β-catenin pathway in mouse SSCs, we treated SSCs with lithium chloride (LiCl) that inhibits β-catenin degradation. qRT-PCR analysis showed that miR-322 expression was increased significantly and Rassf8 expression was decreased significantly in SSCs by LiCl treatment (Figure 6A). We also found that the mRNA levels of β-catenin, Cyclin D1 and C-myc, which are downstream of the WNT/β catenin pathway, were significantly decreased after infection with the miR-322 inhibition lentivirus compared with cells infected with the miR-322 inhibition control (Figure 6B). However, the mRNA levels of β-catenin, Cyclin D1 and C-myc were significantly increased by infection with the Rassf8 inhibition lentivirus (Figure 6B). We found that the protein level of Cyclin D1 was significantly increased infected with miR-322 overexpression lentivirus vectors compared with control group (Figure 6C), whereas the protein level of Cyclin D1 was significantly decreased infected with miR-322 inhibition lentivirus vectors compared with control group (Figure 6C). While Cyclin D1 is crucial for the G1/S transition of cell cycle[36]. When Cyclin D1 is declined, it will induce G0/G1 arrest[36]. And our cell cycle analysis showed that miR-322 overexpression lentivirus increased cell number of S phase compared to control cells (Figure 6D), whereas miR-322 inhibition lentivirus increased cell number of G0/G1 phase and decrease cell number of S phase and G2/M phase compared to control group (Figure 6D). Collectively, these results suggested that the WNT/ β-catenin pathway is involved in miR-322-mediated regulation of SSC self-renewal.
Discussion
Adult male germ cells include SSCs that serve as a reservoir to guarantee the continuation of spermatogenesis. SSCs are unipotent for commitment to the germ cell lineage within the testis, but SSCs have the capacity to become pluripotent under certain conditions in vitro[37]. Recently, increasing studies have indicated that SSC unipotency is reversible, and SSCs can acquire pluripotency to differentiate into all cell lineages of the three germ layers under the certain condition [38]. Therefore, SSCs can be used in autologous organ regeneration therapies for human diseases and have no immune rejection[39]. The regulation of SSCs requires both intrinsic factors and extrinsic signals, and studies have suggested that intrinsic factors play a leading role in the regulation of SSC renewal[38]. Therefore, it is imperative to uncover how intrinsic factors regulate the renewal or differentiation of SSCs.
MiRNA plays crucial roles in the regulation of cellular proliferation[40], apoptosis[41], and differentiation [42, 43]. Previous studies attested that microRNAs might play a vital role in spermatogenesis of mammals[44-48]. For example, miRNA-202 is highly expressed in mouse SSCs and negatively regulated by GDNF and retinoic acid. MiRNA-202 maintains SSCs by inhibiting cell cycle regulators and RAN-binding proteins[49]. MiRNA microarray and fluorescence in situ hybridization revealed that miRNA-20 and miRNA-106a are expressed preferentially in SSCs, which promote proliferation and DNA synthesis of SSCs by targeting STAT3 and Ccnd1[39]. MiRNA-21 is preferentially expressed in SSCs and regulates the self-renewal of mouse SSCs by targeting ETV5 which is downstream of GDNF signaling and essential for SSC self-renewal[38]. MiRNA-34c is testis-specific and highly expressed in the testis of sexually mature mice, which promotes meiosis by interacting with Nanos2, leading to upregulation of Stra8 in mouse SSCs[50]. We demonstrated that miR-322 is mainly expressed in Thy1+ cells that are highly expressed in the testis of mice at postnatal day 6. It is known that the amount of SSCs reaches its peak at postnatal day 6. These findings indicate that miR-322 strongly affects the regulation of SSCs. Indeed, CCK8 and EDU assays revealed that miR-322 promoted the proliferation of SSCs. Moreover, the mRNA expression of Gfrα1, Etv5 and Plzf was increased significantly, while that of Stra8, C-kit and Bcl6 was decreased significantly by miR-322 overexpression. Thus, we concluded that miR-322 promotes the self-renewal of SSCs and inhibits their differentiation.
Effects of Rassf8 on SSCs. (A) Fluorescence and bright field image for SSCs infected with lentivirus. (B) mRNA expression levels of Rassf8 in cells infected with the Rassf8 overexpression lentivirus control (Rassf8-over-con), Rassf8 overexpression lentivirus (Rassf8-over), Rassf8 inhibition lentivirus control (Rassf8-inhibit-con), Rassf8 inhibition lentivirus (Rassf8-inhibit) were examined by qRT-PCR. (C, D) western blot analyses detected the protein level of Rassf8 in cells infected with the Rassf8 overexpression lentivirus control (Rassf8-over-con), Rassf8 overexpression lentivirus (Rassf8-over), Rassf8 inhibition lentivirus control (Rassf8-inhibit-con), Rassf8 inhibition lentivirus (Rassf8-inhibit). (E, F) CCK-8 assays (E) and EdU incorporation assays (F) were conducted using SSCs infected with the Rassf8 overexpression lentivirus control (Rassf8-over-con), Rassf8 overexpression lentivirus (Rassf8-over), Rassf8 inhibition lentivirus control (Rassf8-inhibit-con), or Rassf8 inhibition lentivirus (Rassf8-inhibit). Ⅰ: Rassf8-over-con, Ⅱ: Rassf8-over, Ⅲ: Rassf8-inhibit-con, Ⅳ: Rassf8-inhibit. (G) Quantification of EdU assay results. Data are presented as the mean ± SD of three independent experiments. **P< 0.01 compared with the control group. Scale bars: 10 µm
MiR-322 activates WNT/β-catenin signaling in SSCs. (A) qRT-PCR analysis of the gene expression levels of miR-322 and Rassf8 in SSCs treated with or without LiCl (20 mmol/L). (B) qRT-PCR analysis of β-catenin, Cyclin D1 and C-myc mRNA expression levels in SSCs infected with the miR-322 inhibition lentivirus control (miR-322-inhibit-con), miR-322 inhibition lentivirus (miR-322-inhibit), Rassf8 inhibition lentivirus control (Rassf8-inhibit-con), or Rassf8 inhibition lentivirus (Rassf8-inhibit). (C) Western blot analysis was conducted to detect the protein level of Cyclin D1 in SSCs infected with miR-322 overexpression control lentivirus vector (miR-O-C), miR-322 overexpression lentivirus vectors (miR-O), miR-322 inhibition control lentivirus (miR-In-C) or miR-322 inhibition lentivirus (miR-In). (D) Cell cycle analysis of SSCs infected with miR-322 overexpression lentivirus control (miR-322-over-con), miR-322 overexpression lentivirus (miR-322-over), miR-322 inhibition lentivirus control (miR-322-inhibit-con), or miR-322 inhibition lentivirus (miR-322-inhibit). Data are presented as the mean ± SD of three independent experiments. *P< 0.05, **P< 0.01 compared with the control group
Ras association domain family 8 is ubiquitously expressed in the murine embryo and normal human adult tissues such as the kidney, brain, liver, heart, and lung[51]. Rassf8 is found both in the nucleus and cell membrane[33]. Rassf8 is related to the cell cycle, accelerates apoptosis, and restrains migration and invasion[52]. Overexpression of Rassf8 induces G0/G1 arrest, apoptosis, and downregulation of Cyclin D1 downstream of the WNT/β-catenin pathway. In our study, bioinformatics analysis and luciferase reporter assays revealed that the RASSF8 3′-UTR has a specific miR-322-binding sequence. Therefore, we used a Rassf8 overexpression lentivirus to infect miR-322- overexpressing cells, and CCK8 assays demonstrated the proliferation was suppressed. Functional analysis showed that the Rassf8 inhibition lentivirus promoted SSC self-renewal, which is consistent with previous results. These findings suggest that miR-322 directly targets Rassf8.
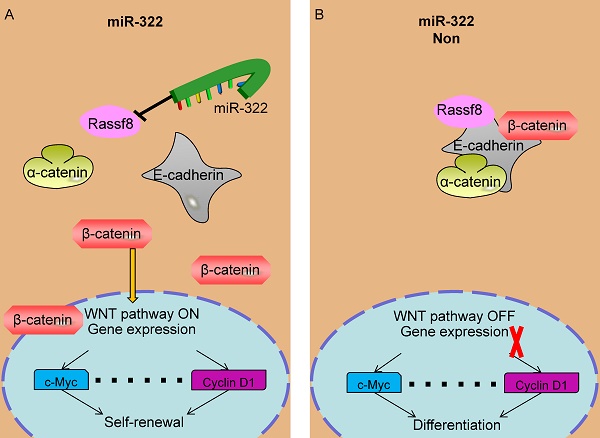
A recent study reported that Rassf8 colocalizes with the adherens junction component β-catenin, binds to E-cadherin, and then regulates the Wnt/ β-catenin pathway[33]. The Wnt/β-catenin pathway is a critical player in the regulation of mouse and human spermatogonia[35]. Therefore, miR-322 may regulate the self-renewal and differentiation of SSCs via Rassf8, which was confirmed by our study. When miR-322 is overexpressed, Rassf8 expression is decreased significantly, leading to insufficient Rassf8 to bind to E-cadherin and dissociation of the E-cadherin- β-catenin-α-catenin complex. Then, adherens junctions destabilize and E-cadherin is lost from the cell membrane. β-Catenin relocalizes to the nucleus, and this change in location is a hallmark of activation of canonical Wnt signaling[53]. In the nucleus, β-catenin facilitates activation of target genes such as uPAR, P53, MMP7, c-Myc, and Cyclin D1. Overexpression of miR-322 induced the self-renewal of SSCs. When miR-322 was inhibited, Rassf8 expression was evaluated. Therefore, Rassf8 bound to E-cadherin, and the E-cadherin complex localized to the membrane. Thus, β-catenin could not enter the nucleus and initiate gene expression, leading to inactivation of Wnt/β-catenin signaling. Ultimately, inhibition of miR-322 gave rise to the differentiation of SSCs.
Our study is the first to reveal the regulatory role of miR-322 in SSC development in vitro, suggesting that miR-322 is essential for fate determination of SSCs by targeting Rassf8. This study provides novel mechanisms that regulate SSC fate and may have vital implications for understanding SSC development.
Abbreviations
BSA: Bovine serum albumin; cDNA: Complementary DNA; DNA: Deoxyribonucleic acid; EDU: 5-ethynyl-2'- deoxyuridine; FBS: Fetal bovine serum; GDNF: Glial cell line- derived neurotrophic factor factor; RNA: Ribonucleic acid; MACS: Magnetic activated cell sorting; MMC: Mitomycin C; mRNA: Messenger RNA; MVH: Mouse Vasa Homolog; PFA: Paraformaldehyde; PLZF: Promyelocytic leukemia zinc finger; GFRA1: GDNF family receptor alpha 1; ETV5: Ets variant 5; OCT4(POU5F1): POU domain, class 5, transcription factor 1; RT: Reverse transcript; SSC: Spermatogonial stem cell; PBS: Phosphate buffer solution; EGFP: Enhanced green fluorescent protein; PCR: Polymerase chain reaction; STO: SIM mouse embryo-derived thioguanine and ouabainresistant feeder.
Acknowledgements
This study was supported by National Basic Research Program of China (2018YFC1003501) and the National Nature Science Foundation of China (81720108017).
Authorship
Yinjuan Wang, and Xiaoyong Li conducted the major experiments, data analysis and wrote the manuscript; Xiaoweng Gong provided miR-322 relevant lentivirus vectors; Ji Wu initiated and supervised the entire project. All authors read and approved the final manuscript.
Ethics Approval
All mouse experiments were approved by the Institutional Animal Care and Use Committee of Shanghai. The procedures were conducted in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tagelenbosch RAJ, Rooij DGD. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F 1 hybrid mouse. Mutation Research. 1993;290:193-200
2. Kostereva N, Hofmann MC. Regulation of the spermatogonial stem cell niche. Reproduction in Domestic Animals. 2010;43:386-92
3. Kanatsu-Shinohara M, Shinohara T. Spermatogonial stem cell self-renewal and development. Annual Review of Cell & Developmental Biology. 2013;29:163-87
4. Brinster RL. Male germline stem cells: from mice to men. Science. 2007;316:404-5
5. Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annual Review of Cell & Developmental Biology. 2008;24:263-86
6. González-González E, López-Casas PP, Mazo JD. Gene silencing by RNAi in mouse Sertoli cells. Reproductive Biology & Endocrinology. 2008;6:29 -
7. Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W. et al. The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2647-52
8. Hayashi K, Lopes SMCDS, Kaneda M, Tang F, Hajkova P, Lao K. et al. MicroRNA Biogenesis Is Required for Mouse Primordial Germ Cell Development and Spermatogenesis. Plos One. 2008;3:e1738
9. Papaioannou MD, Nef DS. microRNAs in the Testis: Building Up Male Fertility. Journal of Andrology. 2010;31:26-33
10. Huang YL, Huang GY, Lv J, Pan LN, Luo X, Shen J. miR-100 promotes the proliferation of spermatogonial stem cells via regulating Stat3. Molecular Reproduction & Development. 2017;84:693
11. Yuan S, Tang C, Zhang Y, Wu J, Bao J, Zheng H. et al. mir-34b/c and mir-449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. 2015; 4: 212-23.
12. Xie R, Lin X, Du T, Xu K, Shen H, Wei F. et al. Targeted Disruption of miR-17-92 Impairs Mouse Spermatogenesis by Activating mTOR Signaling Pathway. Medicine. 2016;95:e2713
13. Li J, Liu X, Hu X, Tian GG, Ma W, Pei X. et al. MicroRNA-10b regulates the renewal of spermatogonial stem cells through Kruppel-like factor 4. Cell Biochemistry & Function. 2017;35:184-91
14. He Z, Jiang J, Kokkinaki M, Tang L, Zeng W, Gallicano I. et al. MiRNA-20 and MiRNA-106a Regulate Spermatogonial Stem Cell Renewal at the Post-transcriptional Level via Targeting STAT3 and Ccnd1. Stem Cells. 2013;31:2205-17
15. Zhang K, Song F, Lu X, Chen W, Huang C, Li L. et al. MicroRNA-322 inhibits inflammatory cytokine expression and promotes cell proliferation in LPS-stimulated murine macrophages by targeting NF-κB1 (p50). Bioscience Reports. 2016:37
16. Yang L, Song S, Hang L. MicroRNA-322 protects hypoxia-induced apoptosis in cardiomyocytes via BDNF gene. American Journal of Translational Research. 2016;8:2812
17. Zeng Y, Liu H, Kang K, Wang Z, Hui G, Zhang X. et al. Hypoxia inducible factor-1 mediates expression of miR-322: potential role in proliferation and migration of pulmonary arterial smooth muscle cells. Scientific Reports. 2015;5:12098
18. Merlet E, Atassi F, Motiani RK, Mougenot N, Jacquet A, Nadaud S. et al. miR-424/322 regulates vascular smooth muscle cell phenotype and neointimal formation in the rat. Cardiovascular Research. 2013;98:458-68
19. Gu H, Yu J, Dong D, Zhou Q, Wang JY, Yang P. The miR-322-TRAF3 circuit mediates the pro-apoptotic effect of high glucose on neural stem cells. Toxicological Sciences. 2015;144:186-96
20. Yuan Z, Hou R, Wu J. Generation of mice by transplantation of an adult spermatogonial cell line after cryopreservation. Cell Proliferation. 2010;42:123-31
21. Gan H, Lin X, Zhang Z, Zhang W, Liao S, Wang L. et al. piRNA profiling during specific stages of mouse spermatogenesis. RNA (New York, NY). 2011;17:1191-203
22. Peters AHFM, Plug AW, Vugt MJV, Boer PD. SHORT COMMUNICATIONS A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Research An International Journal on the Molecular Supramolecular & Evolutionary Aspects of Chromosome Biology. 1997;5:66
23. He Z, Jiang J, Hofmann MC, Dym M. Gfra1 Silencing in Mouse Spermatogonial Stem Cells Results in Their Differentiation Via the Inactivation of RET Tyrosine Kinase1. Biology of Reproduction. 2007;77:723-33
24. Buaas FW, Kirsh AL, Sharma M, Mclean DJ, Morris JL, Griswold MD. et al. Plzf is required in adult male germ cells for stem cell self-renewal. Nature Genetics. 2004;36:647-52
25. Simon L, Ekman GC, Tyagi G, Hess RA, Murphy KM, Cooke PS. Common and distinct factors regulate expression of mRNA for ETV5 and GDNF, Sertoli cell proteins essential for spermatogonial stem cell maintenance. Experimental Cell Research. 2007;313:3090-9
26. Dann CT, Alvarado AL, Molyneux LA, Denard BS, Garbers DL, Porteus MH. Spermatogonial stem cell self-renewal requires OCT4, a factor downregulated during retinoic acid-induced differentiation. Stem Cells. 2010;26:2928-37
27. Kobayashi T, Kajiurakobayashi H, Nagahama Y. Differential expression of vasa homologue gene in the germ cells during oogenesis and spermatogenesis in a teleost fish, tilapia, Oreochromis niloticus. Mechanisms of Development. 2000;99:139-42
28. Xing MX, Yong LX, Ji WU, Institutes BX. Expressions of a group of miRNAs during testis development and regulation effect of miR-125a on development of spermatogonial stem cells. Journal of Shanghai Jiaotong University. 2015;35:625-30
29. Shinohara T, Avarbock MR, Brinster RL. β1- and α6-integrin are surface markers on mouse spermatogonial stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5504-9
30. Lu XY, Yang B, Xu SF, Zou T. [STRA8 as a specific expression marker in postnatal male germ cells]. Zhonghua Nan Ke Xue. 2010;16:161-5
31. Liu X, Lu H, Chen T, Nallaparaju KC, Yan X, Tanaka S. et al. Genome-wide Analysis Identifies Bcl6-Controlled Regulatory Networks during T Follicular Helper Cell Differentiation. Cell reports. 2016;14:1735-47
32. Sherwood V, Recino A, Jeffries A, Ward A, Chalmers AD. The N-terminal RASSF family: a new group of Ras-association-domain-containing proteins, with emerging links to cancer formation. Biochemical Journal. 2010;425:303-11
33. Lock FE, Underhillday N, Dunwell T, Matallanas D, Cooper W, Hesson L. et al. The RASSF8 candidate tumor suppressor inhibits cell growth and regulates the Wnt and NF-|[kappa]|B signaling pathways. Oncogene. 2010;29:4307
34. Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781-810
35. Golestaneh N, Beauchamp E, Fallen S, Kokkinaki M, Uren A, Dym M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction (Cambridge, England). 2009;138:151-62
36. Hedberg Y, Ljungberg B, Roos G, Landberg G. Expression of cyclin D1, D3, E, and p27 in human renal cell carcinoma analysed by tissue microarray. British journal of cancer. 2003;88:1417-23
37. Payne CJ, Braun RE. Human Adult Testis-Derived Pluripotent Stem Cells: Revealing Plasticity from the Germline. Cell Stem Cell. 2008;3:471-2
38. Z N, SM G, S R, X W, JW T, MR A. et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12740
39. He Z, Jiang J, Kokkinaki M, Tang L, Zeng W, Gallicano I. et al. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells. 2013;31:2205-17
40. Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25-36
41. Ambros V. MicroRNA Pathways in Flies and Worms: Growth, Death, Fat, Stress, and Timing. Cell. 2003;113:673-6
42. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science. 2004;303:83-6
43. Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing 'stemness'. Nature. 2008;452:225-9
44. BjorRk JK, Sandqvist A, Elsing AN, Kotaja N, Sistonen L. miR-18, a member of Oncomir-1, targets heat shock transcription factor 2 in spermatogenesis. Development. 2010;137:3177-84
45. Luo L, Ye L, Liu G, Shao G, Zheng R, Ren Z. et al. Microarray-based approach identifies differentially expressed microRNAs in porcine sexually immature and mature testes. Plos One. 2010;5:e11744
46. Mciver SC, Stanger SJ, Santarelli DM, Roman SD, Nixon B, Mclaughlin EA. A Unique Combination of Male Germ Cell miRNAs Coordinates Gonocyte Differentiation. Plos One. 2012;7:e35553
47. Ro S, Song R, Park C, Zheng H, Sanders KM, Yan W. Cloning and expression profiling of small RNAs expressed in the mouse ovary. Rna-a Publication of the Rna Society. 2007;13:2366
48. Tong MH, Mitchell DA, Mcgowan SD, Evanoff R, Griswold MD. Two miRNA Clusters, Mir-17-92 (Mirc1) and Mir-106b-25 (Mirc3), Are Involved in the Regulation of Spermatogonial Differentiation in Mice1. Biology of Reproduction. 2012;86:72
49. Chen J, Cai T, Zheng C, Lin X, Wang G, Liao S. et al. MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins. Nucleic Acids Research. 2017;45:4142
50. Yu M, Mu H, Niu Z, Chu Z, Zhu H, Hua J. miR-34c enhances mouse spermatogonial stem cells differentiation by targeting Nanos2. Journal of Cellular Biochemistry. 2013;115:232-42
51. Wang L, Liu W, Zhang YP, Huang XR. The miR-224 promotes non-small cell lung cancer cell proliferation by directly targeting RASSF8. European Review for Medical & Pharmacological Sciences. 2017;21:3223
52. Chen Y, Bian L, Zhang Y. MiR-505 mediates methotrexate resistance in colorectal cancer by targeting RASSF8. Journal of Pharmacy & Pharmacology. 2018
53. Brown AM. Canonical Wnt signaling: high-throughput RNAi widens the path. Genome Biology. 2005;6:1-5
Author contact
![]() Corresponding author: Ji Wu, Bio-X Institutes, Shanghai Jiao Tong University, No. 800. Dongchuan Road, Minhang District, Shanghai, 200240, China. Phone: 86-21-34207263; Fax: 86-21-34204051; E-mail: jiwuedu.cn
Corresponding author: Ji Wu, Bio-X Institutes, Shanghai Jiao Tong University, No. 800. Dongchuan Road, Minhang District, Shanghai, 200240, China. Phone: 86-21-34207263; Fax: 86-21-34204051; E-mail: jiwuedu.cn

 Global reach, higher impact
Global reach, higher impact