10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(5):1091-1103. doi:10.7150/ijbs.31533 This issue Cite
Research Paper
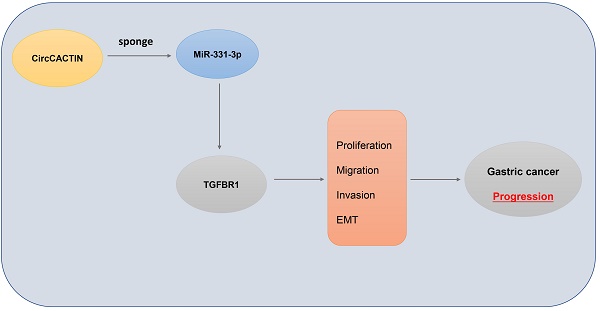
Circular RNA CircCACTIN Promotes Gastric Cancer Progression by Sponging MiR-331-3p and Regulating TGFBR1 Expression
1. Department of Tumor Biological Treatment, The Third Affiliated Hospital of Soochow University, Changzhou 213003, China.
2. Department of Radiation Oncology, The Third Affiliated Hospital of Soochow University, Changzhou 213003, China
3. Jiangsu Engineering Research Center for Tumor Immunotherapy, The Third Affiliated Hospital of Soochow University, Changzhou 213003, China
4. Department of Oncology, The Third Affiliated Hospital of Soochow University, Changzhou 213003, China.
Received 2018-11-15; Accepted 2019-2-27; Published 2019-4-22
Abstract
Background: Circular RNAs (circRNAs) are a new class of RNAs that play a significant role in regulating gene expression and biological function. However, the expression profile and function of circRNAs in gastric cancer (GC) remain mostly uncertain. In the present study, we researched the expression profile of circRNAs in human GC tissues and explored the role of circCACTIN (hsa_circ_0092303).
Methods: Circular RNA microarray assays were performed to detect circular RNA expression profiles of GC and circCACTIN was identified for further investigation. Quantitative real-time PCR was used to detect the expression of circCACTIN, miR-331-3p and TGFBR1 in GC specimens and cell lines. CircCACTIN was stably silenced and overexpressed in GC cells, and cell proliferation, migration, invasion, epithelial-mesenchymal transition (EMT), as well as tumorigenesis in nude mice were performed to assess the effect of circCACTIN on GC.
Results: CircCACTIN expression was obviously up-regulated in GC tissues and cell lines. Knockdown of circCACTIN inhibited GC cells proliferation, migration, invasion and EMT. Enforced-expression of circCACTIN promoted GC cells migration, invasion and EMT, but had no effect on GC cells proliferation. Moreover, in vivo experiments, circCACTIN up-regulation promoted GC tumor growth and EMT, and circCACTIN down-regulation inhibited GC tumor growth and EMT. Binding interactions were detected between circCACTIN and miR-331-3p, and between miR-331-3p and TGFBR1 by Dual-luciferase reporter assays. Mechanistically, we demonstrated that circCACTIN promoted gastric cancer progression by sponging miRNA-331-3p and regulating TGFBR1 mRNA expression.
Conclusion: The circCACTIN/miR-331-3p/TGFBR1 axis affected the proliferation, migration, invasion and EMT of GC through the mechanism of competing endogenous RNAs (ceRNA). Furthermore, our results identified circCACTIN as a novel oncogenic circRNA in GC.
Keywords: circCACTIN, gastric cancer, progression, miR-331-3p, TGFBR1
Background
Gastric cancer (GC) has become the sixth most common cancer worldwide and ranks as the third cause of cancer death[1]. Despite many advances in the diagnosis and treatment of GC, the prognosis of patients remains unfavourable, with a 5-year overall survival of less than 30% in most countries[2]. Therefore, to improve the diagnosis, prevention and treatment of GC, the discoveries of novel molecular mechanisms and therapeutic targets are of great importance.
Circular RNAs (circRNAs) are a new class of RNAs formed by a covalently closed loop with neither 5'-3' polarities nor polyadenylated tails[3, 4]. They are evolutionarily conserved in different species and often expressed tissue/developmental-stage specific [5]. With rapid advances in bioinformatics technologies and high-throughput sequencing, increasing circRNAs have been discovered. Plenty of evidence has indicated circRNAs play vital roles in several physiological and pathological processes, including cell proliferation, cell cycle progression, migration, invasion, metastasis and carcinogenesis[6]. Recent studies have confirmed that circRNAs participate in the occurrence and progress of many malignancies, such as glioma[7], hepatocellular carcinoma[8], pancreatic carcinoma[9], gastric cancer[10] and colon cancer[11], by serving as microRNA (miRNA) sponges and protecting the target genes from miRNA-mediated mRNA dysfunction. The functions of individual circRNA in GC remain largely unknown and need further investigation.
In our study, tissue microarrays were used to explore the differential expression profiles of circRNAs in GC. A large number of differentially expressed circRNAs were found between GC and matched para-carcinoma tissues. Here, we focus our attention on a circular RNA derived from CACTIN gene, termed circCACTIN. CircCACTIN is also regarded as hsa_circ_0092303 (http://www.circbase.org/cgi-bin/simplesearch.cgi), which is located at chr19: 3620270-3620690. CircCACTIN is remarkably up-regulated in GC tissues. Therefore we performed biological and molecular experiments in vitro and in vivo to examine the effect of circCACTIN on GC. Our research was the first to reveal the circCACTIN/ MiR-331-3p/Transforming growth factor-β receptor type 1 (TGFBR1) pathway involved in GC progress, which established a new molecular mechanism of GC development and indicated a potential therapeutic target for GC.
Materials and Methods
Clinical samples
Patients were collected from the department of gastrointestinal Surgery of the Third Affiliated Hospital of Soochow University (China) between October 19, 2017 and December 19, 2017. Gastric cancer tissues and their matched para-carcinoma tissues were collected from 2 gastroscopy patients and 30 surgical patients. All patients were pathologically diagnosed with gastric adenocarcinoma. All specimens collected were frozen in liquid nitrogen for future use. We used eight surgical specimens and two gastroscopic biopsy specimens to perform circRNA microarray analyses. Detailed clinical information for circRNA microarray detection is summarized in Table S1. Detailed clinical information for quantitative real-time PCR (qRT-PCR) is summarized in Table S2. GC patients were staged following the AJCC tumor-node-metastasis (TNM) staging system (8th ed.). This study was approved by the Human Research Ethics Committee of Soochow University.
Analyzing circRNA expression profile
Ten pairs of GC and para-carcinoma tissues including eight surgical specimens and two gastroscopic biopsy specimens, were selected for circRNA microarray detection. The patients enrolled did not receive any treatment before surgery. CircRNAs microarray detection (H1710082 AS-CR-005 Human Circular RNA Microarray v2) was performed and analyzed by KangChen Bio-tech Inc. (Shanghai, China), which detected 656 up-regulated and 761 down-regulated circRNAs.
Cell culture
GES1, BGC-823, MGC-803 and SGC-7901 cell lines were purchased from Chinese Academy of Sciences, Shanghai Institutes for Biological Sciences. All cell lines were cultured in RPMI 1640 medium (Gibco, New York, USA) with 10% fetal bovine serum (FBS, Gibco, NY, USA). The cells were incubated at 37℃ in a humidified atmosphere containing 5% CO2.
RNA isolation and qRT-PCR
TRIzol reagent (Invitrogen, CA, USA) was used to extract total RNA according to the manufacturer's instructions. RNA concentration was measured by Beckman Coulter, and each paired RNA samples were adjusted to the uniform concentration. A One Step SYBR® PrimeScript™ RT-PCR Kit II (Takara, Kusatsu, Japan) was used to conduct qRT-PCR assays for circRNAs and TGFBR1. The amount of the target RNA was normalized to the expression of endogenous reference (GAPDH). A TaqMan MicroRNA Assays Kit (Applied Biosystems, Carlsbad, CA, USA) was used for miRNA-331-3p qRT-PCR. U6 was used as the internal control for miR-331-3p detection. The qRT-PCR reaction was performed on an ABI 7500 Real-Time PCR System (Applied Biosystems, CA, USA). Relative gene expression was shown as the fold change (2-ΔΔCT). All the primer sequences were listed in Table S3, and divergent primers were designed for circRNAs. Each qRT-PCR analysis was repeated three times.
Transient transfection and construction of stable cell lines
Small interference RNAs (siRNAs) specific to human circCACTIN and TGFBR1, miR-331-3p mimic, inhibitor and their negative controls were generated by Gene-Pharma (Shanghai, China). A fragment of 420bp of circCACTIN cDNA was cloned into PLCDH-cir vector (Ribobio, Guangzhou, China) to overexpress circCACTIN. For TGFBR1 overexpression, the 1509bp coding sequence of TGFBR1 (NM_004612) was cloned into pLVX-IRES-Puro (Promega, USA). The lentivirus in this study were constructed by Genelily (Shanghai, China). The above vectors were transfected by Lipofectamine® 3000 (Thermo fisher, China) according to the manufacturer's instructions. After transfection for 48h, the transfection efficiency of cells in each group was detected by qRT-PCR.
Cell proliferation assay
Cell Counting Kit (CCK8, Dojindo, Osaka, Japan) was used to perform the cell proliferation assay. BGC-823 and MGC-803 cells in different groups were seeded into 96-well plates with serum-free RPMI 1640 medium. Each group was replicated with four independent wells. Cell proliferation was observed at 24, 48 and 72h by measuring absorbance 450nm using a spectrophotometer. Before measurement, each well was added with 10μl CCK-8 reagent and then cells were cultured at 37℃ for another 3h.
Wound-healing assay
BGC-823 and MGC-803 cells in different groups were seeded into 6-well plates. The cell layers were scratched with 200ml pipette tips, and the cells were maintained in serum-free RPMI-1640 medium. The cells were photographed by an inverted microscope to record the wound width at 0 and 24h, and the wound width was measured using Image J software. Measuring was conducted 5 times in each group at random intervals along the wound length, and the results of three independent experiments were averaged and expressed as a percentage of the original width.
Cell invasion assay
BGC-823 and MGC-803 cells (5×104) in each group were resuspended in 200μl medium without serum and seeded into the upper chamber of each transwell (Corning, New York, USA) which was coated with Matrigel (BD Biosciences, New York, USA). 600μl medium containing 20% FBS was added to the bottom chamber as a chemoattractant. The cells were incubated at 37°C with 5% CO2 for 24h. Afterwards, cells were fixed with 4% paraformaldehyde for 30min and then stained with 0.1% crystal violet for 30min. Three randomly selected fields under an inverted microscope were used to count the number of invading cells.
Western blotting
RIPA lysis buffer (Beyotime, China) was used to extract total protein of each group according to the manufacturer's specification. Equivalent amounts of proteins from each group cells were subjected to SDS-PAGE electrophoresis and then transferred onto a polyvinylidene fluoride (PVDF) membrane (Sigma- Aldrich, USA). After being blocked in 5% fat-free milk for 1h at room temperature, membranes were incubated with specific primary antibodies overnight at 4°C with gentle shaking, and followed by detection with enhanced chemiluminescence system (SuperSignal West Femto trial kit, Pierce). Primary antibodies were as follows: TGFBR1 (1:3000; Abcam, MA, USA), E-cadherin (1:2000, Cell Signaling Technology, MA, USA), Vimentin (1:1000, Cell Signaling Technology, MA, USA), Snail1 (1:1000, Cell Signaling Technology, MA, USA). GAPDH (1:6000, Sigma, St. Louis, MO, USA) was used as loading control. The membranes were then incubated with HRP-labeled goat anti mouse/rabbit secondary antibodies (1:8000, Sigma Aldrich, St. Louis, MO, USA). After washes, Membranes were exposed to X-ray film (Kodak China Investment, China) to visualize the bands, then signals were detected using a chemiluminescence system (Bio-Rad, USA) and analyzed using Image Lab Software (Bio-Rad, CA, USA).
Dual-luciferase reporter assays
Circular RNA Interactome (https://circinteractome.nia.nih.gov) and TargetScan (http://www.targetscan.org) were used to predict potential binding sites. Dual-luciferase reporter assays were used to detect the binding interactions between circCACTIN and miR-331-3p, miR-331-3p and TGFBR1. BGC-823 cells were cultured in 12-well plates and co-transfected with PsiCHECK-2-Report vector (Promega, China) carrying either wild or mutated circCACTIN or TGFBR1 3'UTR sequences together with miR-331-3p mimics or mimics NC. Gene-Pharma (Shanghai, China) provided the circCACTIN and TGFBR1 3'UTR constructs. The miR-331-3p mimics were constructed by RiboBio (Guangzhou, China). Firefly and Renilla luciferase activities were measured 48 hours after transfection using the dual-luciferase reporter assay (Promega, Madison, WI, USA). The luciferase activity was calculated as the ratio of firefly luciferase intensity to renilla luciferase intensity.
In vivo tumor formation assay
Male athymic BALB/c nude mice, 4-6 weeks old, were purchased from Slac Laboratories (Shanghai, China). The animal care and experimental protocols were approved by the institutional guidelines of Animal Care and Use Committee of Soochow University. 1×107 BGC-823 cells stably transfected with si-circCACTIN and si-NC and MGC-803 cells stably transfected with Lv-circCACTIN and Lv-NC were individually resuspended in 200μL PBS medium and then subcutaneously injected into the flank of each nude mouse. The tumors were measured every 5 days and the tumor volume was calculated following the formula length × width2/2. The mice were killed on day 25 after inoculation.
Immunohistochemistry (IHC)
Xenograft tumor specimens from nude mice were fixed in 4% paraformaldehyde and subsequently embedded in paraffin. After dewaxing and hydration, antigen retrieval and blocking, the 5μm sections were incubated with specific primary antibodies overnight at 4°C. Primary antibodies were as follows: TGFBR1(1:200, Abcam, MA, USA), E-cadherin (1:300, Cell Signaling Technology, MA, USA), Vimentin (1:200, Cell Signaling Technology, MA, USA) and Snail1(1:200, Cell Signaling Technology, MA, USA). Then the sections were incubated with anti-mouse/ rabbit secondary antibody (Abcam, USA) for 30min at room temperature. Staining was performed using diaminobenzidine (DAB) kit (Sigma, USA). The images were obtained under a microscope (Olympus, Japan) with appropriate magnification.
Statistical analysis
SPSS v21.0 (SPSS Inc., USA) and GraphPad Prism v5.0 software (GraphPad Software, USA) were used for statistical analyses. Data difference between two groups was assessed using Student's t-test. Difference among multiple groups was determined by one-way analysis of variance (ANOVA) test. Data were presented as means ± standard error of the mean (SEM). P value < 0.05 was considered as statistically significant.
Results
Circular RNA expression profile in gastric cancer
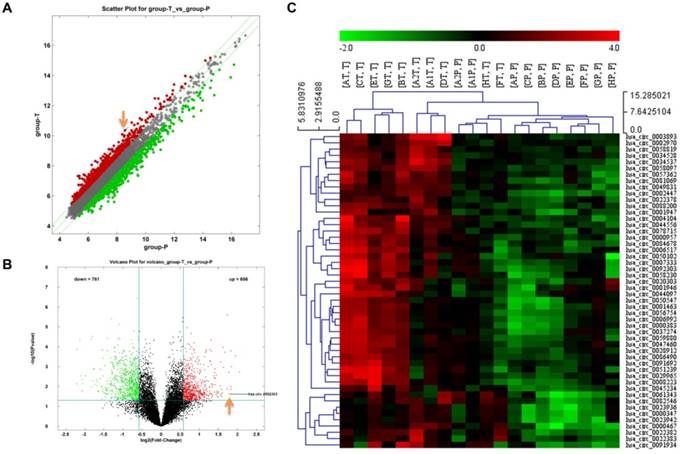
Differences of circRNA expression profiles between GC tissues and paired para-carcinoma tissues were detected by high-throughput human circRNA microarrays. Differentially expressed circRNAs between cancer tissues and para-carcinoma tissues were identified through Fold Change filtering (Fig. 1A) and Volcano Plot filtering (Fig. 1B). The top 50 high expressed circRNAs in GC extracted by hierarchical clustering are shown in Fig. 1C. The details of top 20 high expressed circRNAs in GC are now listed in Table 1.
The identification of circRNA expression profiles in gastric cancer. A Scatter plot of circRNAs expression in gastric cancer and para-carcinoma tissues. The circRNAs above the top green line and below the bottom green line are those up-regulated or down-regulated more than 1.5 folds. B Volcano plot visualizes the different expression in these two groups. The red dots represent more than 1.5 fold changes (log2 scaled) of circRNAs and with statistical significance (P < 0.05). C Hierarchical cluster analysis of the top 50 up-regulated circRNAs of gastric cancer in 10 pairs of matched tissues. T: gastric cancer tissues; P: para-carcinoma tissues.
Top 20 up-regulated circRNAs in gastric cancer.
| CircRNA | P-value | FC (abs) | regulation | chrom | circRNA_type | GeneSymbol | FC of qRT-PCR |
|---|---|---|---|---|---|---|---|
| hsa_circ_0003893 | 0.001908727 | 4.1497811 | up | chr15 | exonic | CEMIP | 3.45 |
| hsa_circ_0034537 | 0.000285534 | 3.869808 | up | chr15 | exonic | THBS1 | 1.98 |
| hsa_circ_0044556 | 0.001010661 | 3.5869766 | up | chr17 | exonic | COL1A1 | 2.94 |
| hsa_circ_0058230 | 0.013816053 | 3.5639838 | up | chr2 | exonic | PNKD | 2.84 |
| hsa_circ_0004104 | 0.010071999 | 3.5493467 | up | chr5 | exonic | SPARC | 1.58 |
| hsa_circ_0058097 | 0.000161903 | 3.5450916 | up | chr2 | exonic | FN1 | 1.90 |
| hsa_circ_0092303 | 0.024196064 | 3.4657038 | up | chr19 | intronic | CACTIN | 2.67 |
| hsa_circ_0078715 | 0.00195079 | 3.4266263 | up | chr6 | exonic | THBS2 | 1.82 |
| hsa_circ_0002447 | 2.55292E-06 | 3.3414571 | up | chr2 | exonic | COL6A3 | - |
| hsa_circ_0008223 | 0.013097578 | 3.3061326 | up | chr16 | exonic | XPO6 | - |
| hsa_circ_0022382 | 0.000190467 | 3.189447 | up | chr11 | exonic | FADS2 | - |
| hsa_circ_0081069 | 0.00108704 | 3.1113818 | up | chr7 | exonic | COL1A2 | - |
| hsa_circ_0000467 | 0.000107203 | 3.0778227 | up | chr13 | exonic | SKA3 | - |
| hsa_circ_0007333 | 0.021394251 | 3.0338617 | up | chr2 | exonic | PAX3 | - |
| hsa_circ_0051239 | 0.031807734 | 3.0103916 | up | chr19 | exonic | ATP5SL | - |
| hsa_circ_0006517 | 0.013508829 | 2.9909962 | up | chr3 | exonic | LPP | - |
| hsa_circ_0029965 | 0.023720536 | 2.9900042 | up | chr13 | exonic | PDS5B | - |
| hsa_circ_0001947 | 0.000584194 | 2.9564012 | up | chrX | exonic | AFF2 | - |
| hsa_circ_0000383 | 0.02708055 | 2.9419336 | up | chr12 | antisense | KRAS | - |
| hsa_circ_0001463 | 0.009668386 | 2.922215 | up | chr5 | intronic | BRD9 | - |
FC: fold change.
CircRNA selection and circCACTIN expression is up-regulated in GC tissues and cell lines
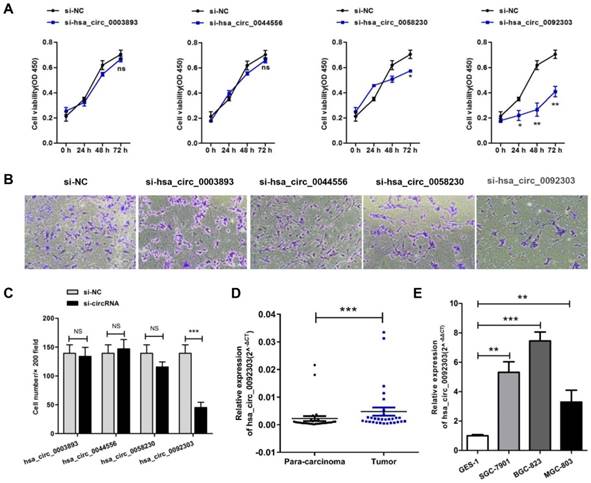
The first eight highly expressed circRNAs were verified by qRT-PCR. The results confirmed the expressions of hsa_circ_0003893, hsa_circ_0044556, hsa_circ_0058230, hsa_circ_0092303 in GC tissues were more than doubled their expression in para-carcinoma tissues, as shown in Table 1 and Fig S1. Therefore, hsa_circ_0003893, hsa_circ_0044556, hsa_circ_0058230, hsa_circ_0092303 were chosen to explore the function of circRNAs in GC. We constructed four siRNAs to silence the four circRNAs individually in BGC-823 cells. Their interferential efficiencies were shown in Fig. S2. CCK8 assays revealed only hsa_circ_0058230 and hsa_circ_0092303 had effects on GC cells proliferation, and hsa_circ_0092303 was the most influential (Fig. 2A). We also conducted transwell invasion assays and found only hsa_circ_0092303 had effect on GC cells invasion (Fig. 2B, C). Therefore circCACTIN (hsa_circ_0092303) marked with orange arrow in Fig. 1A and Fig. 1B was chosen as the research emphasis in subsequent studies. To rule out the effect of linear RNA, qRT-PCR was used to detect the CACTIN mRNA expression in circCACTIN knockdown BGC-823 cells, and no significant expression change was observed (Fig. S3). So far there was no report about CACTIN in GC detected in PubMed, Web of Science or Embase. Therefore, we analyzed the data from TCGA database. Kaplan-Meier survival curves showed that there was no statistically significant difference in survival between the low CACTIN expression group and the high CACTIN expression group in GC (N = 397, P = 0.088, HR=1.45, 95% CI=0.91-2.19) (Fig. S4). The expression of circCACTIN was further assessed with paired GC and para-carcinoma tissues from 30 patients by qRT-PCR. Compared with para-carcinoma tissues, circCACTIN was remarkably increased in GC tissues (Fig. 2D). Furthermore, the expression levels of circCACTIN in three GC cell lines (SGC-7901, BGC-823 and MGC-803) were dramatically higher than those in normal human gastric epithelial cell line GES-1, and circCACTIN expression was the highest in BGC-823 cells and lowest in MGC-803 cells (Fig. 2E). Thus BGC-823 and MGC-803 cell lines were used for subsequent functional analyses. Our data indicated that circCACTIN expression was increased in GC tissues and cell lines.
CircCACTIN promotes proliferation, invasion, and migration of GC cells in vitro
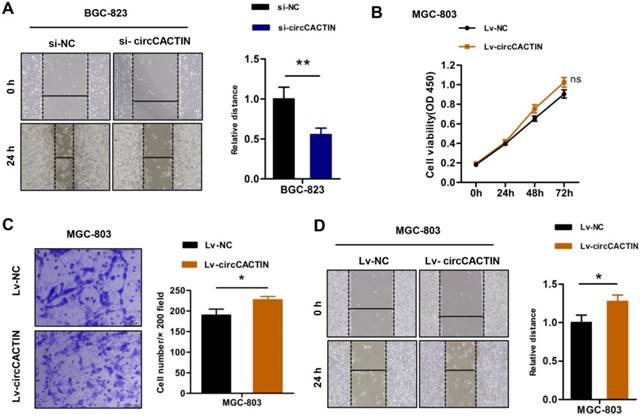
The previous results showed that down-regulation of circCACTIN decreased BGC-823 cells proliferation and invasion. Wound-healing assays indicated down-regulation of circCACTIN decreased BGC-823 cells migration when compared with the control group (Fig. 3A). Besides down-regulation of circCACTIN in BGC-823 cells we also constructed a retroviral stable expression of circCACTIN in MGC-803 cells. The expression level of circCACTIN in MGC-803 cells was validated by qRT-PCR (Fig. S5). However, up-regulation of circCACTIN had little impact on MGC-803 cells proliferation, as shown by the results of the CCK8 assays (Fig. 3B). Transwell assays and Wound-healing assays suggested up-regulation of circCACTIN significantly promoted MGC-803 cells invasion and migration (Fig. 3C, D). These results indicated that circCACTIN played a key role in promotion of proliferation, invasion and migration of GC cells in vitro.
CircRNA selection and CircCACTIN expression is up-regulated in gastric cancer tissues and cell lines. A CCK8 assays were performed in BGC-823 cells transfected individually with si-hsa_circ_0003893, si-hsa_circ_0044556, si-hsa_circ_0058230, si-hsa_circ_0092303. B, C Transwell assays were used to measure the invasion ability of BGC-823 cells transfected individually with si-hsa_circ_0003893, si-hsa_circ_0044556, si-hsa_circ_0058230, si-hsa_circ_0092303. D Relative expression levels of circCACTIN in gastric cancer tissues (T) and para-carcinoma tissues (P) (n = 30) detected by qRT-PCR. Results are presented as the fold change (2-ΔCT) (Wilcoxon matched-pairs signed rank test). E Relative expression levels of circCACTIN in three gastric cancer cell lines and the immortalized gastric normal mucosal cell line GES-1 detected by qRT-PCR. Experiments were repeated 3 times and data were presented as means ± s.d. ns (not significant), *P < 0.05, **P < 0.01, ***P < 0.001.
Binding interaction exists between circCACTIN and miR-331-3p
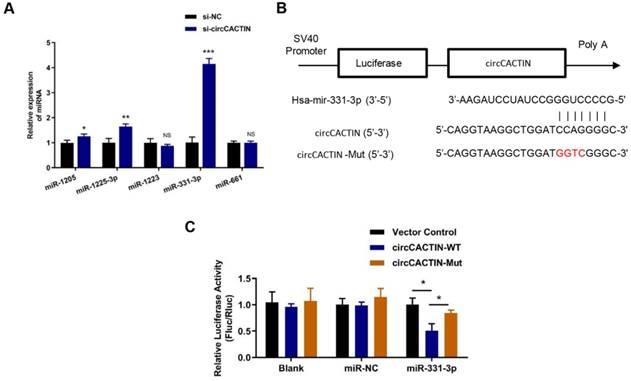
17 miRNAs were predicted as the potential targets of circCACTIN by the circular RNA Interactome Database. Firstly, 5 miRNAs with scores above 90 including hsa-miR-1205, hsa-miR-1225-3p, hsa-miR-1233, hsa-miR-331-3p, hsa-miR-661 were selected. QRT-PCR was used to detect the expression changes of these miRNAs in the circCACTIN down-regulation BGC-823 cells. The results indicated that hsa-miR-331-3p expression was most significantly affected by circCACTIN (Fig. 4A). Dual-luciferase reporter assays were performed to verify if there was a binding site between circCACTIN and miR-331-3p. The alignments of potential binding site and mutant type of circCACTIN were constructed through bioinformatics analysis (Fig. 4B). The firefly luciferase reporter activity was significantly decreased in the group of co-transfection of hsa-miR-331-3p mimics and circCACTIN WT in BGC-823 cells, and the circCACTIN mutant group showed no notable change of luciferase reporter activity (Fig. 4C). This finding confirmed that binding interaction existed between circCACTIN and miR-331-3p.
CircCACTIN functions as an efficient sponge of miR-331-3p in GC
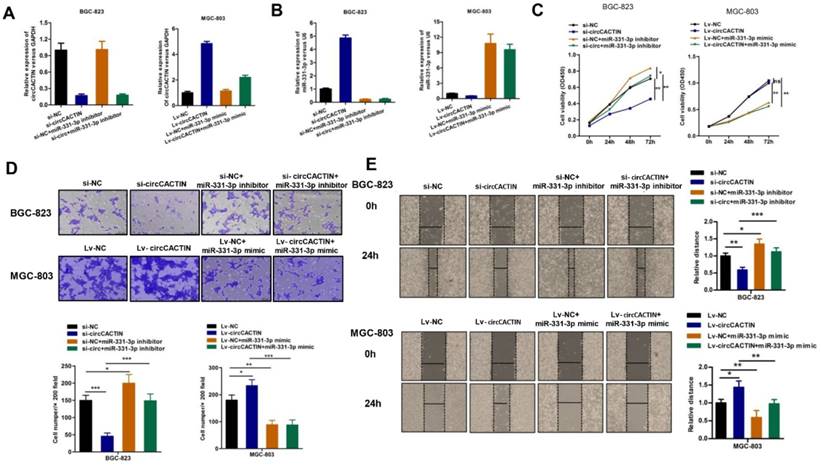
We further explored whether circCACTIN functioned as an oncogene by serving as a sponge of miR-331-3p. Firstly, BGC-823 cells were transfected individually with si-NC, si-circCACTIN, si-NC+miR-331-3p inhibitor, si-circCACTIN+miR-331-3p inhibitor; MGC-803 cells were transfected individually with Lv-NC, Lv-circCACTIN, Lv-NC+miR-331-3p mimic, and Lv-circCACTIN+miR-331-3p mimic. QRT-PCR was used to determine the transfection efficiency (Fig. 5A, B), and it was revealed miR-331-3p expression was inversely related to circCACTIN in si-circCACTIN transfected BGC-823 cells and LV-circCACTIN stable transfected MGC-803 cells (Fig. 5B). Subsequently, CCK-8 assays, transwell assays and wound-healing assays were performed. The results indicated that transfection of miR-331-3p inhibitor promoted BGC-823 cells proliferation, invasion and migration; transfection of miR-331-3p mimic suppressed MGC-803 cells proliferation, invasion and migration. Moreover, miR-331-3p reversed the changes of cells proliferation, invasion, and migration induced by variation of circCACTIN in GC (Fig. 5C-E).
circCACTIN promotes proliferation, invasion, and migration of GC cells in vitro. A Wound-healing showed down-regulated circCACTIN inhibited BGC-823 cells migration. B CCK8 assays were performed in MGC-803 cells transfected with Lv-NC and Lv-circCACTIN individually. C Transwell assays were performed to measure the invasion ability of MGC-803 cells transfected with Lv-NC and Lv-circCACTIN individually. D Up-regulated circCACTIN promoted MGC-803 cells migration. Experiments were repeated 3 times and data were presented as means ± s.d. ns (not significant), *P < 0.05, **P < 0.01.
The interaction between circCACTIN and miR-331-3p. A. QTR-PCR was performed to detect the expression of hsa-miR-1205, hsa-miR-1225-3p, hsa-miR-1233, hsa-miR-331-3p and hsa-miR-661 in circCACTIN down-regulation BGC-823 cells. B. Putative miR-331-3p binding site in the 3′-UTR of circCACTIN is shown. The sequence in the circCACTIN 3′-UTR at the complementary site of miR-331-3p was mutated. C. Compared to mutant group, the luciferase activity of circCACTIN wild type group had decreased. The relative luciferase activities were normalized with Vector activity. Experiments were repeated 3 times and data were presented as means ± s.d. ns (not significant), *P < 0.05, **P < 0.01, ***P < 0.001.
Binding interaction exists between miR-331-3p and TGFBR1
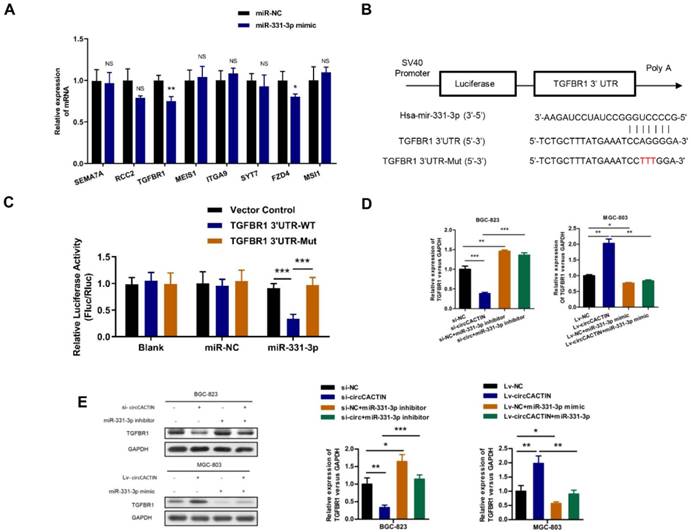
For the effects of circCACTIN on GC cell invasion and migration, we selected 8 targets (SEMA7A, RCC2, TGFBR1, MEIS1, ITGA9, SYT7, FZD4, MSI1) related to tumor invasion and migration from hundreds of miR-331-3p targets predicted by Targetscan, and found TGFBR1 expression was most significantly affected by miR-331-3p (Fig.6A). Dual-luciferase reporter assays were performed to verify if there was a binding site between miR-331-3p and TGFBR1. The alignments of potential binding site and mutant type of TGFBR1 were constructed through bioinformatics analysis (Fig. 6B). Co-transfection of hsa-miR-331-3p mimics and TGFBR1 WT cells were observed with significantly reduction of firefly luciferase reporter activity and the TGFBR1 mutant group showed no notable change of luciferase reporter activity (Fig. 6C). This finding confirmed that binding interaction existed between miR-331-3p and TGFBR1 mRNA. In addition, we also proved both TGFBR1 mRNA and protein expression were regulated by circCACTIN through miR-331-3p (Fig. 6D, E).
TGFBR1 is overexpressed in GC tissues and regulated by circCACTIN
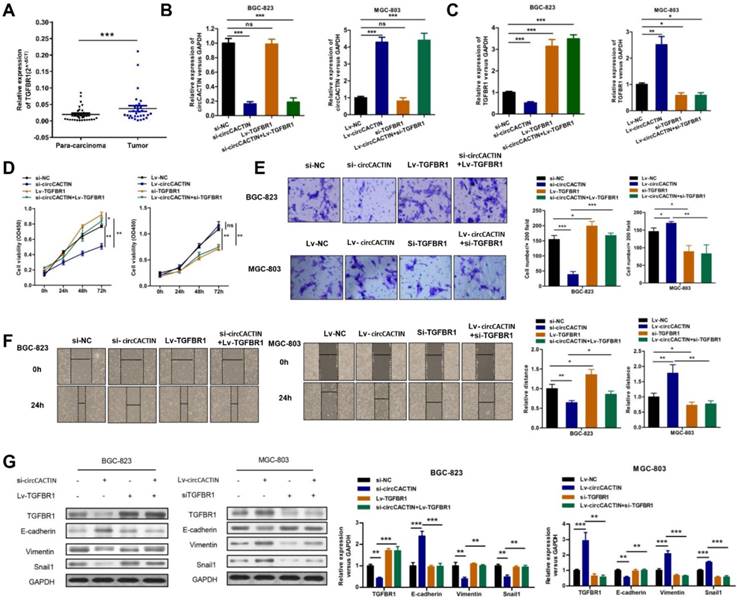
Given the previous results, we considered circCACTIN/MiR-331-3p/TGFBR1 pathway might be involved in gastric cancer progress. TGFBR1 mRNA expression detected in paired GC and para-carcinoma tissues from 30 patients showed TGFBR1 was significantly overexpressed in GC tissues (Fig. 7A). BGC-823 cells were transfected individually with si-NC, si-circCACTIN, Lv-TGFBR1, si-circCACTIN+Lv-TGFBR1; MGC-803 cells were transfected individually with Lv-NC, Lv-circCACTIN, si-TGFBR1, and Lv-circCACTIN+si-TGFBR1. QRT-PCR was used to determine the expression levels of circCACTIN and TGFBR1 mRNA for each group (Fig. 7B, C). Obviously, TGFBR1 expression was positively related to circCACTIN in si-circCACTIN group BGC-823 cells and LV-circCACTIN group MGC-803 cells (Fig. 7C). As for the cell function experiments, TGFBR1 was proved to promote cell viability, invasion, and migration in GC, which was consistent with the function of circCACTIN; an over-expressed TGFBR1 could rescue biological changes induced by knocking down of circCACTIN, and silencing of TGFBR1 could rescue biological changes caused by overexpression of circCACTIN (Fig. 7D-F). For TGFBR1 in Transforming growth factor-β (TGF-β) signaling pathway and its close relationship to cell epithelial-mesenchymal transition (EMT), western blotting was used to detect TGFBR1 protein and EMT marker proteins. The results showed that the expressions of TGFBR1, Vimentin, Snail1 were remarkably down-regulated and E-cadherin was significantly up-regulated in si-circCACTIN group BGC-823 cells; TGFBR1, Vimentin, Snail1 were remarkably up-regulated and E-cadherin was significantly down-regulated in Lv-circCACTIN group MGC-803 cells; TGFBR1 could rescue these protein expression changes induced by interference of circCACTIN (Fig. 7G). Above all, TGFBR1 played a vital role in GC cells proliferation, invasion, migration, and EMT, which was positively regulated by circCACTIN.
MiR-331-3p reverses the effects of circCACTIN on gastric cancer cells. A The expression level of circCACTIN was analyzed by qRT-PCR in each group. B QRT-PCR was used to determine the expression of miR-331-3p in different groups. C CCK8 assays were performed to determine the proliferation of BGC-823 cells and MGC-803 cells in different groups. D The cell invasion capability was detected by transwell assay. E Migration capability was measured by Wound-healing assay. ns (not significant), *P < 0.05, **P < 0.01, ***P < 0.001.
The interaction between miR-331-3p and TGFBR1. A. QRT-PCR was performed to detect the expression of SEMA7A, RCC2, TGFBR1, MEIS1, ITGA9, SYT7, FZD4 and MSI1 in miR-331-3p up-regulation MGC-803 cells. B Putative miR-331-3p binding site in the 3′-UTR of TGFBR1 mRNA is shown. The sequence in the TGFBR1 3′-UTR at the complementary site of miR-331-3p was mutated. C Compared to mutant group, the luciferase activity of TGFBR1 wild type group had decreased. The relative luciferase activities were normalized with Vector activity. D QRT-PCR was used to determine the expression of TGFBR1 mRNA. E Western blotting was used to detected the TGFBR1 protein expression. Experiments were repeated 3 times and data were presented as means ± s.d. ns (not significant), *P < 0.05, **P < 0.01, ***P < 0.001.
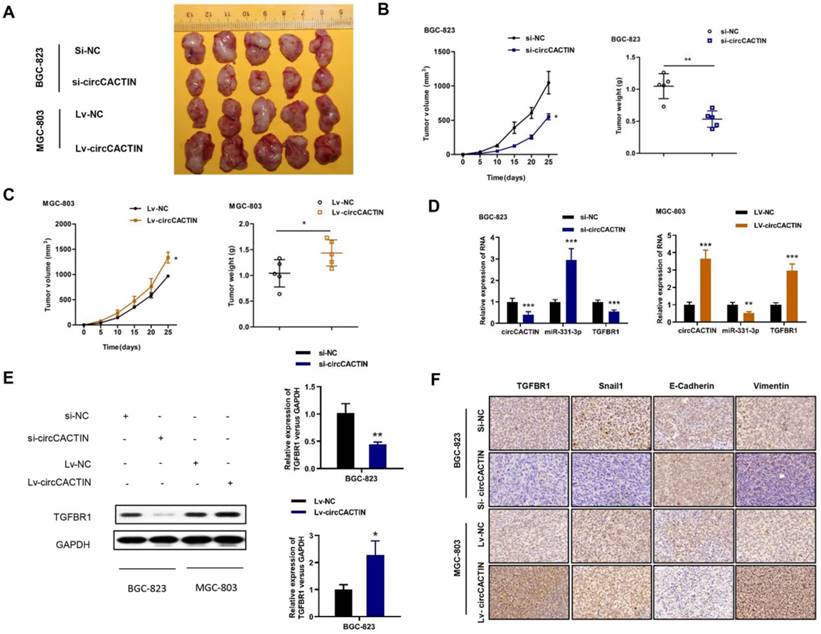
CircCACTIN promotes GC tumor growth in vivo
We then explored the effect of circCACTIN on tumor progression in vivo. Stably down-regulated circCACTIN BGC-823 cells and up-regulated circCACTIN MGC-803 cells and their corresponding control cells were subcutaneously injected into nude mice. The mice were killed on day 25 after inoculation and then xenograft tumors were extracted (Fig. 8A). Decreased tumor sizes and weights were observed in the circCACTIN down-regulation group compared to those in the si-NC group (Fig. 8B). Additionally, increased tumor sizes and weights were observed in circCACTIN up-regulation group compared to those in the Lv-NC group (Fig. 8C). The results of qRT-PCR showed miR-331-3p expression was inversely related to circCACTIN in xenograft tumors. The expression of TGFBR1 mRNA and protein was decreased in circCACTIN down-regulation group, and increased in circCACTIN up-regulation group (Fig. 8D, E). IHC staining showed silencing of circCACTIN down-regulated the expression of TGFBR1, Vimentin, and Snail1, and up-regulated the expression of E-cadherin (Fig. 7F). While over-expressed circCACTIN had the opposite results (Fig. 8F). Above all, these results demonstrated that circCACTIN efficiently promoted GC tumor formation and EMT in vivo.
Discussion
CircRNAs have gained increasing attention in recent years due to rapid advances in high-throughput sequencing and bioinformatics technologies. CircRNAs can act as miRNA sponges or decoys, protein sponges or decoys, protein scaffolding, Splicing and transcription to participate in a variety of biological processes[12]. Moreover, circRNAs can also serve as translatable RNAs[13, 14]. Growing evidence shows circRNAs are involved in the occurrence and development of various diseases[15-18], especially in cancers[19, 20]. CircRNAs were considered to act as miRNA sponges to compete with mRNAs, reversing the inhibitory effect of miRNA downstream genes, thus increasing the expression and functions of target genes.
In this study, we firstly performed a circRNA microarray detection and detected 656 up-regulated and 761 down-regulated circRNAs in GC. Then four up-regulated circRNAs (hsa_circ_0003893, hsa_circ_0044556, hsa_circ_0058230, hsa_circ_0092303) were silenced individually to explore the function of circRNAs in GC, and the results suggested circCACTIN played a role in BGC-823 cells proliferation and invasion. Thus circCACTIN was established as the research target. Elevated expression of circCACTIN was further confirmed by qRT-PCR both in GC tumor tissues and cell lines. Overexpressed circCACTIN promoted cell invasion and migration, but have little effect on cell proliferation in MGC-803 cells. Dual-luciferase reporter assays indicated that miR-331-3p targeted both circCACTIN and TGFBR1, which provided powerful evidence that circCACTIN functioned as a sponge of miR-331-3p to modulate TGFBR1 expression. It was assumed that the circCACTIN/miR-331-3p/TGFBR1 pathway played a critical role in GC progression. MiR-331-3p was reported to play a suppressive role in gastric cancer by targeting E2F1 and HER2 [21, 22]. In si-circCACTIN+miR-331-3p inhibitor group, miR-331-3p inhibitor did not show an increase in circCACTIN expression, while the miR-331-3p mimic decreased circCACTIN expression in Lv-circCACTIN+miR-331-3p mimic group. The reasons could be that to ensure the efficiency of circCACTIN silencing, the siRNA was sufficient during the experiment. The expression of miR-331-3p was at a relative low level in GC cells. Therefore, additional miR-331-3p inhibitors might be not enough to impact the effect of circCACTIN siRNA. However, the miR-331-3p mimics were capable to decrease the overexpressed circCACTIN. Cytological function experiments revealed that circCACTIN and miR-331-3p had reverse effects on cell proliferation, invasion and migration in GC. MiR-331-3p inhibitors could rescue biological changes resulted by silencing of circCACTIN, and miR-331-3p mimics could rescue biological changes caused by overexpression of circCACTIN.
TGFBR1 is overexpressed in GC tissues and regulated by circCACTIN. A Relative expression levels of TGFBR1 in gastric cancer tissues (T) and para-carcinoma tissues (P) (n = 30) were detected by qRT-PCR. Results are presented as the fold change (2-ΔCT) (Wilcoxon matched-pairs signed rank test). B, C QRT-PCR was used to determine the expression of circCACTIN and TGFBR1 in BGC-823 cells and MGC-803 cells in different groups. D CCK8 assays were performed to determine the proliferation of BGC-823 cells and MGC-803 cells in different groups. E The cell invasion capability was detected by transwell assay. F Migration capability was measured by Wound-healing assay. G Western blotting was used to determine the expression levels of TGFBR1, Vimentin, E-cadherin and Snail1. GAPDH was used as a loading control. Experiments were repeated 3 times and data were presented as means ± s.d. ns (not significant), *P < 0.05, **P < 0.01, ***P < 0.001.
CircCACTIN significantly affects tumor growth and EMT in vivo. A Xenograft tumor were extracted from nude mice. B Down-regulation of circCACTIN decreased the ability of tumorigenesis of BGC-823 cells, which reflected in the tumor weight and tumor volume. C Up-regulation of circCACTIN increased tumor weight and tumor volume in nude mice. D qRT-PCR was performed to measure the expression of circCACTIN, miR-331-3p and TGFBR1 in xenograft tumors. E Western blotting was used to determine the expression of TGFBR1 protein in xenograft tumors. F TGFBR1, E-cadherin, Vimentin and Snail1 expression was assessed by immunohistochemical method to determine the effect of circCACTIN on gastric cancer cells EMT function. *P < 0.05, **P < 0.01, ***P < 0.001.
TGFBR1 is a receptor for TGF-β ligands, and TGF-β signaling pathway is typically associated with induction and maintenance of EMT[23]. EMT is a cellular reprogramming process in which epithelial cells visibly alter their shape, acquire a mesenchymal phenotype and exhibit increased motility, and this cellular change is essential for tumor metastasis[24, 25]. When TGF-β ligands are present, they first bind to TGF-β receptor type 2 (TGFBR2), and TGFBR2 is phosphorylated and then activates TGFBR1[26]. TGFBR1 consequently activates Smad signaling and forms Smad complex translocating into the nucleus to regulate gene transcription. Following the activation of TGF-β signaling pathway, cells undergo malignant transformation, EMT, metastatic conversion, angiogenesis and cancer progression[27]. TGFBR1 is overexpressed in multiple malignancies, and dysregulation of TGFBR1 leads to tumorigenesis by disrupting cellular processes[28-30]. TGFBR1 up-regulation promoting EMT was detected in NSCLC cells[31]. However, the role of TGFBR1 in GC is unclear. In this study, we showed that silencing of TGFBR1 inhibited cell proliferation, migration, invasion and EMT in GC cells. Moreover, circCACTIN and TGFBR1 had consistent effects on cell phenotypes. An overexpressed TGFBR1 could rescue biological changes induced by silencing of circCACTIN, and a silencing of TGFBR1 could rescue biological changes induced by overexpression of circCACTIN. Today, some clinical trials of TGFBR1 inhibitor in cancers have been performed. Galunisertib (LY2157299), an TGFBR1 inhibitor, was reported with a function of anti-tumor in glioma[32, 33], hepatocellular carcinoma[34], pancreatic cancer [35] and Ovarian Cancer[36], but the results were not entirely satisfactory. Therefore, circCACTIN might be served as a new target for cancer therapy. For further confirmation and exploration of the function of circCACTIN, experiments in vivo were carried out. Down-regulation of circCACTIN inhibited GC tumor formation and EMT, and up-regulation of circCACTIN promoted GC tumor formation and EMT in vivo. The inconsistency in tumor growth between in vitro and in vivo experiments when circCACTIN was overexpressed might be attributed to other factors related to circCACTIN in the tumor microenvironment. Further research needs to be performed to explore the problem.
In this study, we confirmed that circCACTIN and TGFBR1 were overexpressed in GC tissues. CircCACTIN/miR-331-3p/TGFBR1 axis might exist in GC. CircCACTIN promoted GC progression by up-regulating TGFBR1 expression via sponging miR-331-3p.
Conclusion
In summary, this is the first study to explore the function of circCACTIN in GC and the interaction among circCACTIN, miRNA-331-3p and TGFBR1. Through the circCACTIN/miR-331-3p/TGFBR1 axis, circCACTIN performed specific regulatory roles in the progression of GC. CircCACTIN could be a novel biomarker and therapeutic target for GC patients.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This project is supported by the National Key R&D Program (2018YFC1313400), the National Science and Technology supporting Program (2015BAI12B12), the Joint Research Fund for Overseas Chinese, Hong Kong and Macao Scholars (31729001), the National Natural Science Foundation of China (31570877, 31570908), the Key R&D Project of Science and Technology Department of Jiangsu Province (BE2018645).
Authors' contributions
CPW and JTJ conceived of the study; LZ, XS and XC performed all experiments; QW and XZ collected tissue samples and the clinical data; LZ and XS analyzed and interpreted the data; LZ and XS drafted the manuscript. CPW and JTJ revised the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The clinical protocol was approved by the Human Research Ethics Committee of Soochow University and all patients signed a written informed consent form before specimen collection. The animal care and experimental protocols were approved by the institutional guidelines of Animal Care and Use Committee of Soochow University.
Consent for publication
Consent to publish has been obtained from all authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians. 2018
2. Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS. et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977-1010
3. Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555-65
4. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-8
5. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS genetics. 2013;9:e1003777
6. Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA biology. 2017;14:514-21
7. Li GF, Li L, Yao ZQ, Zhuang SJ. Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochemical and biophysical research communications. 2018;499:765-71
8. Han D, Li J, Wang H, Su X, Hou J, Gu Y. et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151-64
9. Huang WJ, Wang Y, Liu S, Yang J, Guo SX, Wang L. et al. Silencing circular RNA hsa_circ_0000977 suppresses pancreatic ductal adenocarcinoma progression by stimulating miR-874-3p and inhibiting PLK1 expression. Cancer letters. 2018;422:70-80
10. Chen J, Li Y, Zheng Q, Bao C, He J, Chen B. et al. Circular RNA profile identifies circPVT1 as a proliferative factor and prognostic marker in gastric cancer. Cancer letters. 2017;388:208-19
11. Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC genomics. 2017;18:680
12. Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Molecular cell. 2018;71:428-42
13. Begum S, Yiu A, Stebbing J, Castellano L. Novel tumour suppressive protein encoded by circular RNA, circ-SHPRH, in glioblastomas. Oncogene. 2018;37:4055-7
14. Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O. et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Molecular cell. 2017;66:22-37 e9
15. Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF. et al. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Molecular cell. 2017;67:214-27 e7
16. Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017:357
17. Zhou Z, Jiang R, Yang X, Guo H, Fang S, Zhang Y. et al. circRNA Mediates Silica-Induced Macrophage Activation Via HECTD1/ZC3H12A-Dependent Ubiquitination. Theranostics. 2018;8:575-92
18. Cheng X, Zhang L, Zhang K, Zhang G, Hu Y, Sun X. et al. Circular RNA VMA21 protects against intervertebral disc degeneration through targeting miR-200c and X linked inhibitor-of-apoptosis protein. Annals of the rheumatic diseases. 2018;77:770-9
19. Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D. et al. circEPSTI1 as a Prognostic Marker and Mediator of Triple-Negative Breast Cancer Progression. Theranostics. 2018;8:4003-15
20. Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS. et al. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer research. 2017;77:2339-50
21. Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD. et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Molecular cancer. 2014;13:92
22. Guo X, Guo L, Ji J, Zhang J, Zhang J, Chen X. et al. miRNA-331-3p directly targets E2F1 and induces growth arrest in human gastric cancer. Biochemical and biophysical research communications. 2010;398:1-6
23. Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764-74
24. De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nature reviews Cancer. 2013;13:97-110
25. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275-92
26. Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685-700
27. Inman GJ. Switching TGFbeta from a tumor suppressor to a tumor promoter. Current opinion in genetics & development. 2011;21:93-9
28. Gu D, Li S, Du M, Tang C, Chu H, Tong N. et al. A genetic variant located in the miR-532-5p-binding site of TGFBR1 is associated with the colorectal cancer risk. Journal of gastroenterology. 2018
29. Zhou B, Guo W, Sun C, Zhang B, Zheng F. Linc00462 promotes pancreatic cancer invasiveness through the miR-665/TGFBR1-TGFBR2/SMAD2/3 pathway. Cell death & disease. 2018;9:706
30. Hu Q, Hisamatsu T, Haemmerle M, Cho MS, Pradeep S, Rupaimoole R. et al. Role of Platelet-Derived Tgfbeta1 in the Progression of Ovarian Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:5611-21
31. Yang Z, He J, Gao P, Niu Y, Zhang J, Wang L. et al. miR-769-5p suppressed cell proliferation, migration and invasion by targeting TGFBR1 in non-small cell lung carcinoma. Oncotarget. 2017;8:113558-70
32. Rodon J, Carducci MA, Sepulveda-Sanchez JM, Azaro A, Calvo E, Seoane J. et al. First-in-human dose study of the novel transforming growth factor-beta receptor I kinase inhibitor LY2157299 monohydrate in patients with advanced cancer and glioma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2015;21:553-60
33. Brandes AA, Carpentier AF, Kesari S, Sepulveda-Sanchez JM, Wheeler HR, Chinot O. et al. A Phase II randomized study of galunisertib monotherapy or galunisertib plus lomustine compared with lomustine monotherapy in patients with recurrent glioblastoma. Neuro-oncology. 2016;18:1146-56
34. Cao Y, Agarwal R, Dituri F, Lupo L, Trerotoli P, Mancarella S. et al. NGS-based transcriptome profiling reveals biomarkers for companion diagnostics of the TGF-beta receptor blocker galunisertib in HCC. Cell death & disease. 2017;8:e2634
35. Ikeda M, Takahashi H, Kondo S, Lahn MMF, Ogasawara K, Benhadji KA. et al. Phase 1b study of galunisertib in combination with gemcitabine in Japanese patients with metastatic or locally advanced pancreatic cancer. Cancer chemotherapy and pharmacology. 2017;79:1169-77
36. Zhang Q, Hou X, Evans BJ, VanBlaricom JL, Weroha SJ, Cliby WA. LY2157299 Monohydrate, a TGF-betaR1 Inhibitor, Suppresses Tumor Growth and Ascites Development in Ovarian Cancer. Cancers. 2018:10
Author contact
![]() Corresponding authors: Changping Wu, Address: Department of Oncology, The Third Affiliated Hospital of Soochow University 185 Juqian Street, Changzhou, China, 213003. Email: wcpjjtcom and Jingting Jiang, Address: Department of Tumor Biological Treatment, The Third Affiliated Hospital of Soochow University, 185 Juqian Street, Changzhou, China, 213003. Email: jiangjingtingedu.cn.
Corresponding authors: Changping Wu, Address: Department of Oncology, The Third Affiliated Hospital of Soochow University 185 Juqian Street, Changzhou, China, 213003. Email: wcpjjtcom and Jingting Jiang, Address: Department of Tumor Biological Treatment, The Third Affiliated Hospital of Soochow University, 185 Juqian Street, Changzhou, China, 213003. Email: jiangjingtingedu.cn.

 Global reach, higher impact
Global reach, higher impact