Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(6):1125-1138. doi:10.7150/ijbs.30543 This issue Cite
Research Paper
Synthetic-Evolution Reveals Narrow Paths to Regulation of the Saccharomyces cerevisiae Mitotic Kinesin-5 Cin8
1. Department of Chemistry and Ilse Katz Institute for Nanoscale Science and Technology, Ben-Gurion University of the Negev, PO Box 653, Beer-Sheva, 84105, Israel
2. Institute of Technology, University of Tartu, Estonia
3. Institute for Systems Genetics, New York University Langone Health, NY, USA.
Received 2018-10-10; Accepted 2019-2-4; Published 2019-5-2
Abstract
Cdk1 has been found to phosphorylate the majority of its substrates in disordered regions, but some substrates maintain precise phosphosite positions over billions of years. Here, we examined the phosphoregulation of the kinesin-5, Cin8, using synthetic Cdk1-sites. We first analyzed the three native Cdk1 sites within the catalytic motor domain. Any single site conferred regulation, but to different extents. Synthetic sites were then systematically generated by single amino-acid substitutions, starting from a phosphodeficient variant of Cin8. Out of 29 synthetic Cdk1 sites, 8 disrupted function; 19 were neutral, similar to the phospho-deficient variant; and only two gave rise to phosphorylation-dependent spindle phenotypes. Of these two, one was immediately adjacent to a native Cdk1 site. Only one novel site position resulted in phospho-regulation. This site was sampled elsewhere in evolution, but the synthetic version was inefficient in S. cerevisiae. This study shows that a single phosphorylation site can modulate complex spindle dynamics, but likely requires further evolution to optimally regulate the precise reaction cycle of a mitotic motor.
Keywords: phosphoregulation, Cin8, Cdk1, kinesin-5, anaphase B
Introduction
Mitosis is the process by which duplicated chromosomes are transmitted from mother to daughter cells. This essential process is mediated by the mitotic spindle, a microtubule (MT)-based structure that undergoes a distinct set of dynamic changes with precise temporal and spatial regulation. One of the major coordinators of the mitotic events is the conserved cyclin-dependent kinase 1 (Cdk1). Cdk1 depends upon binding of six cyclins for its activation. The levels of these cyclins oscillate during the cell cycle [1], thus helping to provide stage-specific functions for Cdk1.
Previous work identified a large class of “flexible” Cdk1 substrates that were enriched for phosphorylation sites throughout evolution, but where the precise site position was not strictly conserved [2]. These phosphorylation sites tended to occur in disordered regions of proteins. It was suggested that these phosphates might interact with modular phospho-binding motifs or disrupt a protein-protein interface, forms of regulation that are relatively tolerant of changes in site position [2, 3]. A well-characterized example of this type of flexible regulation is the mitogen-activated protein kinase scaffold protein Ste5 [4] which has gained and lost Cdk1 sites during evolution [3]. Another example of a Cdk1 target with a flexible phosphorylation cluster is the Sic1 protein, a Cdk1 inhibitor in Saccharomyces cerevisiae [5, 6]. For proper phosphoregulation of Sic1, a minimal set of priming phosphorylation sites and phospho-degrons is required, but the distances between the sites in the cluster can be flexible [7, 8]. A second, smaller set of substrates was found that maintained a precise position throughout long periods of evolution. These sites were often found in ancient metabolic enzymes, suggesting that they may have evolved early and adopted a highly efficient form of phosphoregulation that relies on exact conformational changes [2]. Although a recent study presented evidence for evolutionary sequence flexibility in short nuclear localization motifs, while maintaining robust function [9], there is very little experimental data directly testing the flexibility of phosphoregulation.
To address this issue, we experimentally examined the flexibility of phosphoregulation by Cdk1 using the S. cerevisiae mitotic kinesin-5 Cin8 as a model-protein. These conserved bipolar kinesin-5 motors perform essential roles in mitotic spindle dynamics by crosslinking and sliding apart antiparallel microtubules of the spindle (reviewed in [10-12]). Their function has been shown to be regulated by Cdk1 in various organisms [13-20]. The budding yeast S. cerevisiae encodes two kinesin-5 homologues, Cin8 and Kip1, that have partially overlapping functions in assembly and maintenance of the bipolar spindle structure [21-23], in focusing kinetochore clusters [20, 24, 25], in anaphase B spindle elongation [26, 27], and in stabilizing and organizing the middle-spindle midzone [28-32].
Kinesin motors undergo diverse phosphoregulation, including by Cdk1 (reviewed in [10, 11, 33]). In particular, the mitotic functions of both S. cerevisiae kinesins-5, Cin8 and Kip1, were shown to be regulated by Cdk1 [17, 18, 20]. For Cin8, this phosphoregulation occurs primarily at three Cdk1 sites, which are located in disordered loops 8 and 14 within the catalytic motor domain [18-20]. We therefore asked how easy it is to recapitulate this phosphoregulation by generating Cdk1 sites at novel positions within Cin8. To answer this question, we examined the phenotypes of 29 new possible Cdk1- sites that were systematically generated by a single amino-acid substitution starting from a phosphodeficient allele of Cin8. By combining a comprehensive genetic, cell biological and biochemical characterization of these mutants we found that out of 29 novel synthetic Cdk1 sites that we created, eight led to a loss of function, 19 resulted in a neutral spindle phenotype similar to the phosphodeficient variant, and only two gave rise to phosphorylation-dependent spindle localization phenotypes. This study shows that the gain of a single phosphorylation site can confer complex regulation. However, this regulation is likely to undergo subsequent optimization. Robust regulation of the complex dynamics of a mitotic kinesin-5 motor protein ultimately requires precise phosphoregulation that becomes evolutionarily constrained.
Results
Strategy for generation of synthetic Cdk1 phosphorylation sites in Cin8
Our previous work identified a large number of Cdk1 substrates. Most of these substrates were phosphorylated in disordered and loop regions, with the precise position of phosphorylation sites not being conserved in evolution. Only a small minority of substrates maintained precise phosphosite positions over long evolutionary trajectories [2]. Examination of the three native sites in Cin8 motor domain that conferred phosphoregulation [18, 20, 34] revealed that two of the three sites, at positions S277 and T285, were located in the disordered loop 8 within the Cin8 catalytic domain. These sites are likely to be evolutionarily young, as they are present in a large insert in loop 8 that is only present in strains from the Saccharomycetaceae clade (Fig. S1A). Nevertheless, in our previous studies, we found one of these sites, S277, to confer the most significant phosphoregulation out of the five Cdk1 sites in Cin8 [20]. On the other hand, the position of site S493, located in loop 14, is conserved among kinesin-5 homologs indicating that this site is evolutionarily constrained (Fig. S1B). This suggests that phosphoregulation of the S. cerevisiae kinesin-5 Cin8 is conferred by both evolutionary young and rigid Cdk1 sites. However, there has been almost no systematic evaluation of the flexibility or rigidity of phosphoregulation for any protein.
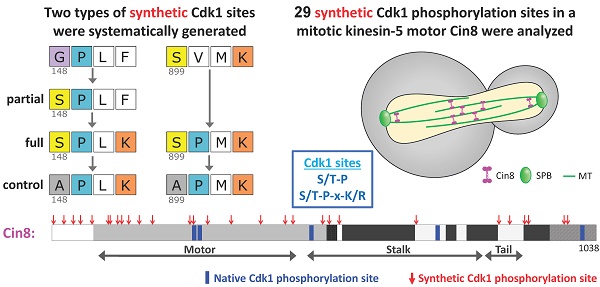
To examine the flexibility of phosphoregulation of Cin8 by Cdk1 we introduced novel consensus Cdk1 sites in the Cin8 ORF in a systematic manner. As a basis for mutagenesis, we used a phosphodeficient variant of Cin8 (Cin8-5A) carrying mutations of the phosphoacceptor serines and a threonine to alanine within the five native Cdk1 sites of Cin8, at positions S277, T285, S493, S736, and S1010. New Cdk1 sites were systematically created by single amino-acid replacement in most locations such that replacement resulted in a novel partial or full Cdk1 consensus site (Fig. 1). We comprehensively created mutations around every proline as this amino acid is the major specificity determinant for Cdk1. For every proline, a serine was introduced at position -1 to generate a Cdk1 phosphorylation consensus sequence (S-P or S-P-x-K/R). If lysine or arginine were already present at the +3 position relative to the new serine, this mutation generated a full site. Otherwise, a partial (or minimal) site was created. In these cases, a second synthetic allele was generated with a lysine at position +3 to the serine, to produce the full Cdk1 phosphorylation consensus [S-P-x-K] (Fig. 1B class 1). In addition, we created Cdk1 sites by substitution of a proline only in cases where full Cdk1 phosphorylation consensus sites were created by this single change (i.e., [S-x-x-K] to [S-P-x-K]; Fig. 1B class 2). We limited the proline substitutions in this fashion because proline substitutions are often poorly tolerated due to the structural constraints of this amino acid. To avoid any bias, we generated the new sites based on the sequence only, without excluding structured and conserved regions. Using this approach, we reintroduced three native Cdk1 sites at the motor domain of Cin8, termed Cin8 native phosphorylation variants (Cin8nat), and additionally generated 29 novel sites termed Cin8 synthetic- phosphorylation variants (Cin8syn) (Figs. 1A, C and D). For variants that exhibited spindle localization phenotypes (see below), we created control mutations in which the serine within the consensus sequence was changed to alanine, which lacks a phosphoacceptor hydroxyl group (Fig. 1B) (see Materials and Methods).
Single Cdk1 sites at the native positions confer regulation
Re-generation of the native Cdk1 sites by the above described approach allowed us to examine the functionality of these sites as a sole source of Cdk1 phosphoregulation. We previously demonstrated that each of the native Cdk1 phosphorylation sites in the motor domain of Cin8, S277, T285, and S493, are independently phosphorylated by Cdk1 in vitro and that mutations to alanine of each of these sites cause different phenotypes during anaphase [20]. Here, the native sites were examined as partial (suffixed by “p”) and full (suffixed by “f”) Cdk1 consensus sequences since they were generated as variants of class 1 (Fig. 1B). The native variants were first examined by general viability assays, then by two dynamic spindle-localization features: translocation to the midzone at mid-anaphase and detachment from the spindle at late anaphase. Finally, the functionality of the variants was evaluated by analysis of anaphase spindle elongation dynamics [20].
We first conducted a yeast viability assay in which the Cin8nat variants were expressed from a centromeric (CEN) plasmid as a sole source of kinesin-5 function in cells deleted for the chromosomal copies of CIN8 and KIP1. We found that cells expressing native partial and full Cin8 phosphorylation variants (Cin8nat) and their controls, in which the serine at the Cdk1 site was mutated to alanine, are viable at all temperatures (Fig. 2A). Therefore, these single phosphorylation site alleles are sufficient for the essential functions of Cin8.
Next, we examined the localization of Cin8 variants, tagged with 3GFP, during anaphase B spindle elongation (Fig. 2B). These experiments were performed in cells deleted for CIN8, with the homologous Kip1 remaining functional. We performed line-scan intensity analysis of Cin8 distribution along the spindle at different anaphase spindle lengths [20]. We found that the phosphodeficient mutant Cin8-5A is mainly concentrated at the SPBs (Fig. 2B and 2C, black arrows), while wt Cin8 exhibited a lower concentration at the SPBs and higher levels along the spindle (Fig. 2C gray arrow), indicating that Cin8 translocates from the SPB's to the midzone in a phosphorylation dependent manner.
We next investigated the relative importance of each of the native Cdk1 sites in Cin8. We examined the spindle distribution of a Cin8 variant lacking phosphorylation sites in the stalk and tail, but with all sites in the motor domain intact (Cin8-2A). Cells expressing this mutant as a sole source of kinesin-5 were fully functional in terms of viability and did not show significantly different spindle localization dynamics from wt Cin8 (Fig. 2A and B). We concluded that the two C-terminal sites do not play a major role in Cin8 spindle distribution but rather, the three sites in the motor domain confer this regulation. Quantification of line-scans indicated that, of these sites, the native partial and full Cin8nat277 variants exhibited spindle distributions similar to wt Cin8. The Cin8nat493 variant had an intermediate phenotype between that of the wt Cin8 and Cin8-5A, and the Cin8nat285 variants were similar to the phospho- deficient Cin8-5A variant (Fig. 2C). These results were consistent with our previous report [20]. Therefore, we concluded that a single Cdk1 in the motor domain can confer near wild-type phosphoregulation of Cin8.
To characterize the detachment of the native Cin8 phosphorylation variants from the spindle at late anaphase (Fig. 2B, yellow arrows), we analyzed Cin8 fluorescence signal perpendicular to the spindle (Fig. 2D) [20]. We found that the detachment of wt Cin8 from the spindle in late stages of anaphase B was abolished by the phosphodeficient mutant Cin8-5A, but not by the Cin8-2A mutant phosphodeficient in the stalk and tail only (Figs. 2E), indicating that phosphorylation by Cdk1 in the catalytic domain of Cin8 is required for this detachment [18, 20]. Consistent with previous reports we also found that the S277 site contributes most strongly to Cin8 detachment from the spindle (Fig. 2E and S3), indicating that although the S277 site is located in a recent large insert in loop 8, and is therefore evolutionarily young, it is the most important phosphosite for Cin8 regulation.
A strategy to generate synthetic Cdk1 sites. (A) Schematic representation of Cin8. Indicated by blue rectangles: native Cdk1; red arrows: synthetic phosphorylation sites; CC: coiled coil. (B) Mutagenesis strategy to create the synthetic phosphorylation sites. Class 1 synthetic sites are more structurally conservative; amino acids prior to existing prolines were mutated to serine to create minimal Cdk1 consensus sites. For synthetic sites that did not result in full Cdk1 phosphorylation consensus, we also generated a full consensus site by additional mutation of the position +3 relative to the potential phosphoacceptor serine to lysine. Finally, control mutants were generated by mutating the serine at the phosphoacceptor position to alanine; see Materials and Methods. In class 2 synthetic sites, the amino-acid proline was introduced at +1 relative to an existing serine or threonine with a lysine or arginine at the +3 position, resulting in a full consensus Cdk1 site. As in class 1, alanine control mutants were generated for cases that conferred localization phenotypes. (C) Sequence of Cin8 motor domain with secondary structure annotated according to [43]. Key domains: ATPase p-loop in purple, Switch I in orange, Switch II in yellow, MT biding domain (helix 4, loop 12 and helix 5) in pink. Native and Synthetic Cdk1 phosphorylation sites are indicated by blue and red, respectively. (D) Model of the Cin8 motor domain predicted by Swiss-model based on human kinesin-5 Eg5 (PDB ID: 3HQD). Cdk1 sites are represented by spheres, blue for native and red for synthetic sites. Key domains are color coded as in 1C.
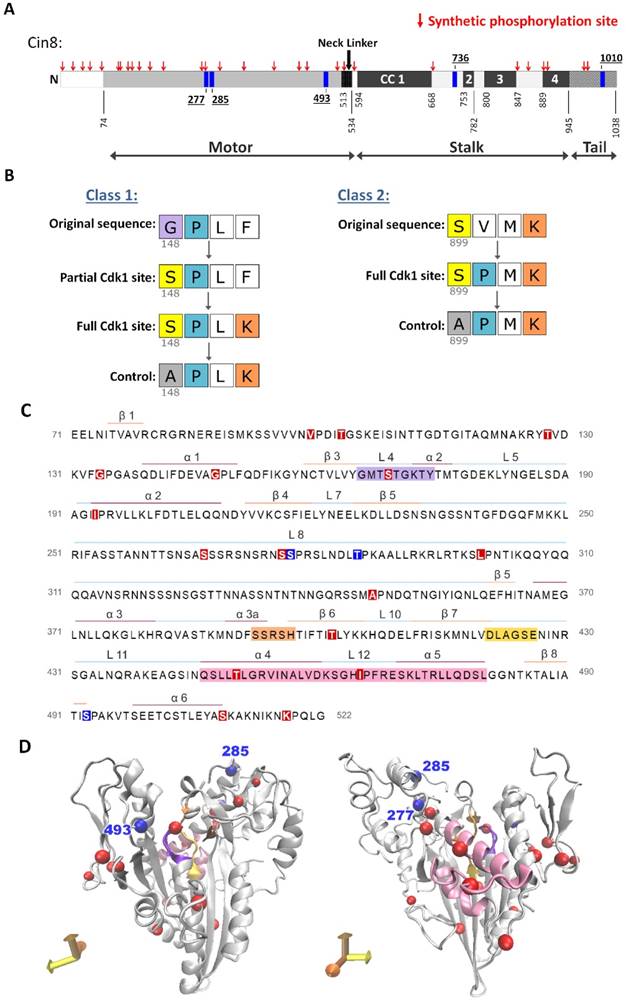
A single native phosphorylation site is sufficient for viability and each site regulates the distribution of kinesin-5 differently. (A) Viability assay of a shuffle yeast strain deleted for its chromosomal copies of CIN8 and KIP1 and containing a recessive chromosomal cycloheximide resistance gene (cin8Δ;kip1Δ;cyhr) on plates containing 7.5 µg/ml cycloheximide. Cells contain centromeric (CEN) plasmids expressing the Cin8 phospho-variants with one native Cdk1 site as a sole source of Cdk1 phospho-regulation (indicated on top, e.g., Cin8nat277 is a cin8Δ; kip1Δ strain with a plasmid containing Cin8 with only the CDK1 consensus site at position 277 intact). Cells were plated in serial dilution, grown at 26°C and 35°C indicated at the left; *grown at 37°C [18, 52]. (B) Localization of Cin8 native phospho-variants (Cin8nat) to anaphase spindles. Anaphase spindles are divided into length categories, indicated on the left. Representative 2D projections of a 3D stack of fluorescence images of cells expressing 3GFP-tagged Cin8 phospho-variants from a CEN plasmid are shown. Micrographs are oriented with mother cells at the bottom. Scale bar: 5 µm; Yellow arrows: Cin8 detachment from the SPBs; white arrows: Cin8 accumulation at the SPBs; and white arrowheads: Cin8 localization at the midzone. (C) Fluorescence intensity along the spindle measured starting at the mother pole and divided to 100 segments of equal length. The signal was normalized to the integral of spindle-bound Cin8-3GFP signal. The signal for each variant was compared to that of Cin8-5A (orange), indicated on top. For Cin8nat277 and Cin8nat285 the color coding for the partial and full Cdk1 phosphorylation consensus variants are indicated. Spindle length-ranges are indicated on the right. Black arrows: Cin8 accumulation at the SPBs; light gray arrow: elevated distribution of Cin8 at the midzone region; red arrows: reduced Cin8 distribution at the SPBs compared to Cin8-5A. At each spindle length, the average intensity ratio in arbitrary units ± s.e.m. of 15-20 cells is presented. Significance was determined for Cin8-2A, Cin8nat277f, Cin8nat285f and Cin8nat493f compared to wt Cin8 (gray asterisks) and Cin8-5A (orange asterisks). Significance of T-test is indicated as *p < 0.05, **p < 0.005, ***p < 0.0005. (D) Method to quantify Cin8 detachment from the spindle. A 41-pixel line was drawn perpendicular to the spindle at the mother SPB. The Y-axis represents the intensity measured along the yellow line. The corresponding cell is shown at the top right; scale bar: 2 µm. The brightest 5 pixels were assigned as Cin8 attached to the center of the SPB and are indicated on the graph as a dashed gray area. The 12 distal pixels on the far ends of the line were averaged and subtracted as background (dark gray dashed area). The fluorescence intensity of the 6 pixels immediately flanking the SPB was assigned Cin8 detached from the spindle (green area). (E) Average intensity ratio of the Cin8 detachment at spindle length of 6-7 µm, as explained in C. Average ± s.e.m of 18-20 cells is shown. α (orange): p-value compared to Cin8-5A, β (grey): p-value compared to wt Cin8. *p < 0.05, **p < 0.001. (A)-(E): p - partial phosphorylation consensus (S-P), f - full phosphorylation consensus (S-P-x-K), fc - full control phosphorylation consensus (A-P-x-K).
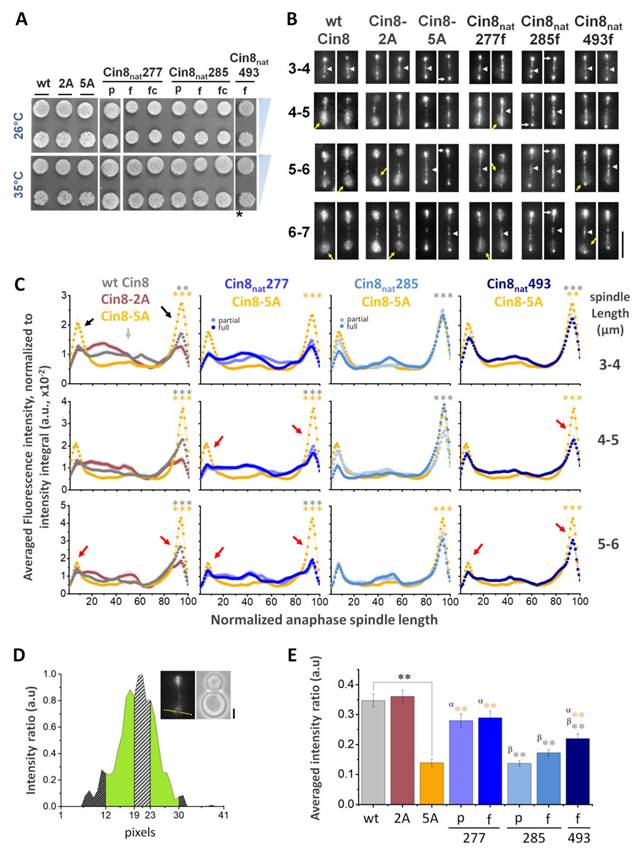
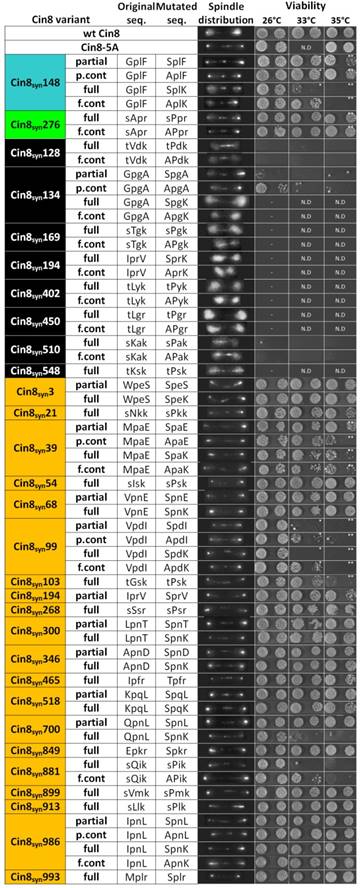
Phenotypes of all the Cin8syn mutants examined in this study. On the left, the names of the mutants, including the position of phosphorylation of the synthetic Cdk1 consensus, are indicated. The mutants are color coded: Cin8syn148 (cyan) and Cin8syn276 (green) which exhibit phosphorylation dependent detachment from the spindle; mutants exhibiting no detachments from the spindle (orange), therefore classified as neutral and mutants that are non-viable at 26°C (black). In the following column, each type mutation is indicated: partial [S/T-P] and full [S/T-P-x-K/R] Cdk1 sequences; and full and partial control mutations (f.cont and p.cont, respectively). In the next two columns, the original and mutated sequences are indicated; capital letters represent position of mutations. Next, representative images of the phosphovariant distribution during anaphase cin8Δ strains (containing the endogenous KIP1 gene to allow analysis of non-functional cin8 alleles) and expressing the phosphovariants, are indicated at the spindle distribution column. On the right, viability of a shuffle cin8Δ;kip1Δ;cyhr strains expressing Cin8syn mutants as a sole source for kinesin-5 function, at 26°C, 33°C, and 35°C are shown. For each strain, two colonies at reducing concentration of yeast cells are shown. *the temperature was 34°C, **the temperature was 37°C. N.D. - not determined.
Categorization of the synthetic Cdk1 sites
The results presented so far demonstrate that a single Cdk1 site is sufficient to confer some regulation of Cin8 (Fig. 2). We next asked whether sites at new positions could also result in regulation. That is, is regulation flexible or constrained, and are there unexplored paths to regulation? To address this question, we created synthetic phospho-variants of Cin8 (Cin8syn). We postulated several hypothetical outcomes. We expected some mutations to result in loss of function due to perturbation of Cin8 structure. Other mutants may result in no structural perturbation but also confer no observable phosphoregulation; these would be neutral mutations. Finally, there may be mutants that result in regulation of Cin8 function, either by recapitulating the original phosphoregulation or by creating novel forms of regulation.
To distinguish between these possibilities, the 29 Cin8syn variants described above were each assessed for their ability to support viability as a sole source for kinesin-5 function in cells deleted for the endogenous kinesins-5, CIN8 and KIP1 (Fig. 3). These variants were also tagged with 3GFP and expressed in a strain deleted for CIN8 but that maintains wild type KIP1 (to enable analysis of non-functional CIN8syn alleles) to determine their localization to anaphase spindles by live-cell imaging (Fig. 3). Out of the 29 Cin8syn mutants that were generated, eight (28%) were found to carry deleterious mutations that resulted in cell death. These mutants exhibited mostly diffusive detachment of Cin8 from the spindle that was not abolished by the alanine control variants and thus were not due to phosphorylation (Fig. 3). Of these eight variants, five carried proline insertion mutations (Fig. 1B Class 2). The largest class of mutants exhibited no detachment from the spindle, therefore were categorized as “neutral”. We found 19 such mutants (~65%) that had no discernible effect on localization, i.e., Cin8-5A-like phenotype of lack of detachment from the mitotic spindle during anaphase (Fig. 3) (see Materials and Methods). These sites may not be accessible for Cdk1 phosphorylation, phosphatases may keep steady-state phosphorylation levels low, or it could be that phosphorylation at these sites does not influence Cin8 detachment from the spindle. Viability of four of the 19 neutral variants was reduced at elevated temperatures (Fig. 3 and Materials and Methods). The reason for this temperature sensitivity is difficult to determine. The most likely explanation is that the protein fold is slightly destabilized. However, it is also possible that spurious phosphorylation at these positions accounts for some of this destabilization. Finally, two novel Cdk1 positions (7%) resulted in a phosphorylation- dependent spindle-localization phenotype, i.e., dynamic regulation of Cin8 localization was to some degree restored and non-phosphorylatable alanine control variants abolished or corrected part of the phenotypes induced by the novel Cdk1 sites S148 and S276 (Figs. 3, 4 and S2). The last were further investigated by quantitative dynamic localization analysis (Figs. 4 and S3), in vitro kinase assays, and spindle elongation measurements (Figs. 4E, 5, and S5).
Two synthetic Cdk1 sites confer differential phosphoregulation
The novel sites at positions S148 and S276 exhibited differential effects on the dynamic detachment of Cin8 from the spindle (Figs. 3, 4A, B, S2 and S3). The partial Cin8syn148p variant and its control Cin8syn148pc showed no detachment, similar to the control Cin8-5A (Figs. 3, 4A, B and S2). However, the full Cin8syn148f variant exhibited high levels of detachment from the spindle at anaphase, comparable to wt Cin8. This novel regulation was lost in the non- phosphorylatable alanine control allele Cin8syn148fc (Figs. 3, 4A, B and S2), indicating that detachment from the spindle of the full Cin8syn148f is dependent on phosphorylation at this site. The full Cin8syn276f variant also exhibited detachment from the spindle but to a lesser extent. This detachment was also abolished by the control Cin8syn276fc mutation (Figs. 3, 4A, B and S2), indicating that it is also phosphorylation dependent.
Synthetic Cin8 variants can confer novel phosphoregulation. (A) Spindle localization of 3GFP-tagged Cin8syn phospho-variants at various spindle lengths (indicated on the left). Mother cell is oriented to the bottom of micrographs. Yellow arrows: Cin8 detachment from the spindle; white arrows: Cin8 concentration at the SPBs. Scale bar: 5 µm. (B) Cin8 detachment from 6-7 µm spindles, measured as in Figs. 2D and E. Average ± s.e.m. of 18-20 cells. p: partial Cdk1 consensus sequence [S/T-P], pc: control of partial consensus sequence [A-P], f: full Cdk1 consensus sequence [S/T-P-x-K/R], fc: control of full consensus sequence [A-P-x-K/R]. α: p-value of t-test compared to Cin8-5A. *p < 0.05, **p < 0.01. (C) Viability of shuffle cin8Δ;kip1Δ;cyhr strains expressing Cin8syn mutants. Cells were plated in serial dilution and grown on YPD plates with 7.5 µg/ml cyclohexamide at 26°C and 33°C. (D) Average fluorescence intensity along the spindle of Cin8syn mutants, indicated on top, compared to the fluorescence distribution of the corresponding non-phosphorylatable alanine control mutant and of Cin8-5A as in Fig. 2C. Color coding of the mutants and their control is indicated. Black arrows: Cin8 accumulation at the SPBs; light gray arrow: elevated distribution of Cin8 at the midzone; red arrows: reduced distribution at the SPBs compared to Cin8-5A. At each spindle length, the average intensity ratio in arbitrary units ± s.e.m. of 15-20 cells is presented. Significance was determined for Cin8syn148p, Cin8syn148f and Cin8syn276f compared to wt Cin8 (gray asterisks), Cin8-5A (orange asterisks), the respective control variant on the same plot (light cyan, cyan and green for Cin8syn148p, Cin8syn148f and Cin8syn276f respectively) and between Cin8syn148p and Cin8syn148f (black asterisks). *p < 0.05, **p < 0.005, ***p < 0.0005. (E) Spindle elongation rates of the first-fast and the second-slow phases of the full native and synthetic variants. Averages ± s.e.m. of 9-12 cells are shown. Phospho-variants are indicated on the bottom, 5A: Cin8-5A. α (orange): p-value compared to Cin8-5A. *p < 0.05, **p < 0.01.
Cin8syn276f, which contains a full Cdk1 phosphorylation consensus sequence in position S276 and is of Class 2 (Fig. 1B), was viable at all temperatures, similar to the wt Cin8. In contrast, the mutant bearing a full Cdk1 consensus sequence at position S148 (Cin8syn148f) was temperature sensitive at temperatures higher than 33°C (Figs. 3 and 4C). The control variant Cin8syn148fc partially rescued this phenotype at 33°C but not at 35oC (Fig. 3 and 4C), indicating that loss of fitness at 33oC results from combination of mutation of phenylalanine to a lysine (F to K) at position 151 and a serine or gain of phosphate at position 148 (Fig. 4C), while the mutation of phenylalanine to a lysine alone, with no possibility of phosphorylation, is sufficient to destabilize the protein at 35oC (Fig. 3). It is, in principle possible, that the reduced viability at higher temperatures is due to a neomorphic mutations at position 148 that causes aberrant regulation that makes the cells temperature sensitive. However, we believe that the simplest explanation is that these mutations destabilize Cin8. Indeed, we observed that the expression levels of the full Cin8syn148f and its control Cin8syn148fc variants are somewhat reduced at 33.5oC (Fig S4), supporting the notion that the reduced viability at elevated temperatures of Cin8syn148f is at least in part due to its reduced stability.
Quantitative examination of anaphase spindle distribution [20] revealed that the synthetic variants regulate the translocation of Cin8 from the SPBs to the midzone, as indicated by reduced intensity at the SPBs, compared to the phosphodeficient Cin8-5A variant (Fig. 4D). The Cin8syn276f variant exhibits significantly reduced distribution to the SPBs compared to the control variant (Cin8syn276fc) at all spindle lengths (Fig. 4D). However, the full Cin8syn148f variant only exhibited significantly lower distribution to the SPBs at short spindles compared to the control Cin8syn148fc variant (Fig. 4D 3-4 µm, red arrows). Finally, the translocation from the SPBs to the spindle of the full Cin8syn276f variant was less pronounced compared to the full variants at the S148 site (Fig, 4D 3-4 µm). Taken together, our data indicate that the synthetic Cdk1 sites regulate the localization of Cin8 on anaphase spindles, with S276 mainly affecting detachment from the spindle (Figs. 3, 4A, B and S3), and the S148 site affecting both Cin8 translocation to the midzone at mid anaphase and detachment from the spindle at late anaphase (Figs. 3, 4A, B and S3).
We next examined the control of spindle dynamics by Cin8 phospho-variants. Spindle length was measured as a function of time, and rate was determined for the first (fast) and second (slow) phases of spindle elongation [20]. Fig. 4E shows a comparison of spindle elongation rates during the two phases between the full Cin8 variants, while Fig. S5 presents the full comparison including the partial and control counterparts of these sites. We found that the full variants at the native sites have no effect on spindle elongation during either elongation phase compared to wt Cin8 and Cin8-5A. On the other hand, the synthetic variants at positions S148 and S276 did alter anaphase spindle dynamics. The full synthetic variant at position S148 significantly reduced the rate of elongation during the early fast elongation compared to the full native variants Cin8nat277 and Cin8nat285 (Fig. 4E up). This effect was reversed by the control mutation Cin8syn148fc (Fig. S5). On the other hand, there was no difference in spindle elongation rate between the partial Cin8syn148p and its control Cin8syn148pc counterpart (Fig. S5). These results indicate that the reduction of rate of spindle elongation induced by the full Cdk1 site at position 148 is phosphorylation dependent. The full synthetic variant at position S276 significantly reduced the elongation rate during the slower second phase compared to the full native sites Cin8nat277 and Cin8nat285 (Fig. 4E bottom), with no effect on the fast phase. This suggests that whereas single native sites bearing full Cdk1 phosphorylation sites have little to no effect on spindle elongation rate, the synthetic sites do affect Cin8 function during spindle elongation. Interestingly, although the synthetic site S276 is in very high proximity to the native site S277, its regulation of spindle elongation is very different (Fig. 4E bottom). These results indicate that the synthetic phospho-variants affect spindle elongation in a unique manner, different from that of the native variants, likely affecting spindle localization and/or motor activity by mechanisms distinct from those of wt Cin8.
To examine whether the newly created Cdk1 sites are accessible for phosphorylation by Clb2/Cdk1, we performed in vitro phosphorylation assays using a purified Cin8 motor domain as a substrate [18, 20]. Our data indicate that all functional variants of Cin8, excluding the negative controls undergo in vitro phosphorylation. The three native variants, S277, S285, and S493, undergo substantial phosphorylation in vitro. Mutation of the S277 and S285 sites, which are minimal sites in wild-type Cin8, to full Cdk1 phosphorylation consensuses resulted in an ~20-fold increase in phosphorylation rates (Fig. 5 right). Interestingly, these higher in vitro phosphorylation levels did not correlate with significant differences in Cin8 localization patterns in vivo (Fig. 2). Cin8 syn148f exhibited a lower but still significant increase in phosphorylation levels, and this full-site conferred significant regulation in vivo (Fig. 4). We also included the Cin8syn168f variant in this assay, which exhibits serine/threonine independent diffusive Cin8 localization and is also non-viable at 26°C (Fig. 3). This mutant exhibited no phosphorylation in vitro (Fig. 5 left), indicating that phenotypes exhibited by this mutant are a result of a deleterious mutation that disrupts Cin8 structure and prevents Cin8 from performing its essential roles. We also tested the Cin8syn268f mutant, which has a neutral phenotype and is close to the native Cdk1 phosphorylation at position S277 in the disordered loop 8. This mutant underwent substantial phosphorylation in vitro, ~3-fold higher than wt Cin8 (Fig. 5 right); however, no phosphoregulation of Cin8 localization was evident with this variant (Fig. 3). Similarly, the Cin8syn99 variants, which exhibited temperature sensitive behavior exhibited a high degree of phosphorylation, but did not cause detachment from the spindle (Figs. 3 and 5 left). Taken together, these results indicate that a high degree of phosphorylation is insufficient to confer regulation. Rather, regulatory phosphorylation only functions within a limited set of positions.
Discussion
In this study we examined the plasticity of phosphoregulation of the Cin8 mitotic kinesin-5 motor by Cdk1 in S. cerevisiae cells. We employed an extensive synthetic mutation experiment and characterized the functionality of Cin8 harboring both native and synthetic sites by quantification of four in vivo parameters: ability to support viability, translocation to the midzone in mid anaphase, release from the spindle in late anaphase, and rate of anaphase spindle elongation. We revealed the complexity of regulation at the native Cdk1 sites and found two synthetic sites that conferred novel phosphoregulation.
Possible mechanisms of phosphoregulation of Cin8 by native and synthetic Cdk1 sites
Most Cdk1 substrates contain multiple phosphorylation sites [2, 35]. To initiate this study, we first investigated to what extent single sites could confer regulation. We found that each of the three native Cdk1 sites in the catalytic domain of Cin8 were functional alone. The three phosphorylation sites together could give more robust regulation, or perhaps fine-tune the activity of Cin8. Indeed, not all sites were equally efficient and, consistent with our previous work [20], there was evidence of cooperative regulation of Cin8. The mechanisms by which phosphorylation of kinesins leads to regulation are a matter of ongoing research.
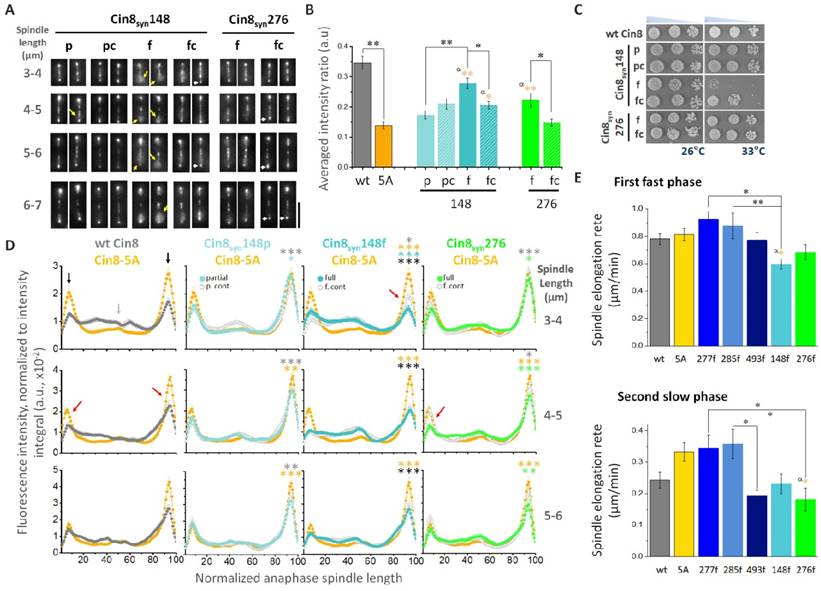
Synthetic sites are phosphorylated by Cdk1 in vitro. In vitro Clb2/Cdk1 kinase assay of the Cin8 motor domain. Top - Coomassie brilliant blue staining and 32P autoradiograms of SDS-PAGE analysis of phosphorylation reactions. Clb2/Cdk1 is the kinase, substrate is bacterially expressed, purified motor domain of Cin8 alleles. The phosphorylation reaction mixture without Cin8 served as negative control (no Cin8). Bottom: quantification of phosphorylation rates normalized to wild-type. Cdk1 consensus sequences are indicated as “p” for partial and “f” for full.
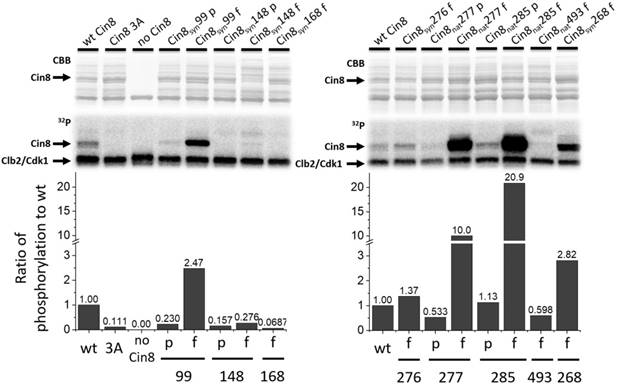
Position and location of the synthetic phosphorylation sites. (A) Model of Cin8 motor domain in complex with αβ-tubulin predicted by Swiss-model based on pseudo-atomic model of MT-bound S. pombe kinesin-5 motor domain in the AMPPNP state (PDB ID: 5M5I). Key-role domains are color-coded; ATPase p-loop in purple (loop 4 with first 3 residues of helix2), MT binding domain and switch II “MTDB+SWII” (loop 11, helix 4, loop 12 and helix 5) in pink loop-5 in orange [43, 53, 54]. Native sites are colored in blue, synthetic phospho-variants at positions 276 and 148 are colored in green and cyan, respectively. (B) Multiple sequence alignment of Cin8 with other kinesin-5 proteins as in Fig S1. Alignment was performed by Unipro UGENE alignment tool using MUSCLE algorithm; degree of identity in the sequence is indicated by blue highlight according to 50% threshold, Cdk1 phosphorylation consensuses are highlighted in yellow. Secondary structure is annotated on top according to [43]. The different clades are indicated on the left. Saccharomycetaceae clade (orange), Candida clade (green), Eurotiomycetaes clade (dark pink), Dothideomycetes clade (gray) and Schizosaccharomyces clade (blue). Species are listed in Fig. S1.
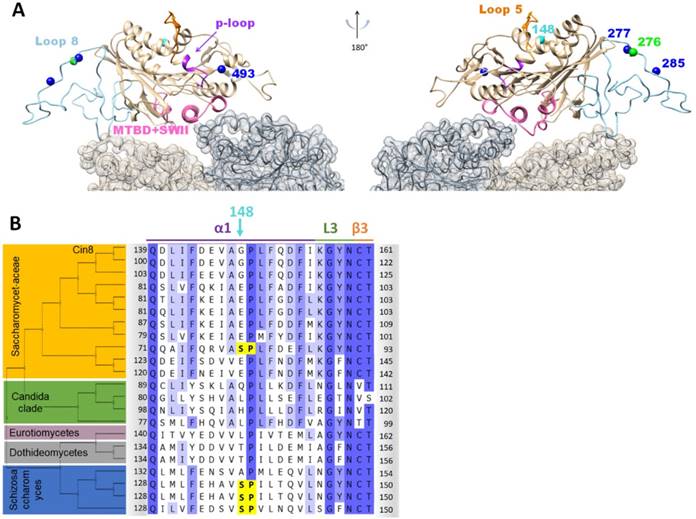
A number of kinesin-related proteins have been shown to be regulated by phosphorylation at residues in their catalytic domain. In these cases, phosphorylation affected the motile properties of kinesin and their interaction with MTs [18, 19, 36-39]. The native S493 site is located in loop 14 and is conserved among the kinesin-5 homologs (Fig. S1B). This site is located in the vicinity of the Cin8 ATP-binding pocket (Fig. 6A), raising the possibility that phosphorylation may affect ATPase activity. The two other native sites, S277 and T285, are located in loop 8 of Cin8, which contains a large insertion not present in the paralogous motor Kip1, or metazoan kinesin-5 motors. Interestingly, only members of the Saccharomycetaceae clade contain a large insert in loop 8, suggesting a recently evolved mode of kinesin-5 regulation (Fig. S1A). In the MT-bound state, loop 8 is located near the MT lattice [10, 40, 41]. Loop 8 was shown to regulate the directionality of Cin8 [37] and the non-canonical binding of Cin8 to MTs [42]. One of the synthetic phospho-variants, S276, is located in loop 8, one amino-acid apart from the native S277 site (Figs. 6A and S1A). The phenotypes of this site resemble the phenotypes of the wt Cin8 and of the native Cin8nat277 variant (Figs. 2, 3, 4, and S3), although they differ in spindle elongation dynamics (Figs. 4E and S5), indicating that the mechanisms of regulation of the S276 and S277 sites are similar, but not identical even though the position of these two sites only differs by a single amino acid. Two additional synthetic sites, Cin8syn286 and Cin8syn300 were introduced into loop 8, and one of these was efficiently phosphorylated in vitro (Fig. 5), but these sites conferred no regulation (Fig. 3). It is possible that phosphorylation within loop 8, at the S277nat and S276syn positions, changes the conformation of loop 8, altering its interaction with the MT lattice but this conformational change requires a precise coordination of the phosphate, explaining why only a subset of phosphorylation events in the loop confer regulation.
The second functional synthetic site, Cin8syn148 is located in α-helix 1 [43] in the vicinity of loop 5 (Fig. 6A) within the kinesin motor domain [44, 45]. Although the function of loop 5 is unknown, its proximity to the conserved nucleotide-binding P-loop element and the fact that several kinesin-5 inhibitors bind to this loop [46, 47] indicate that loop 5 is important for kinesin-5 function. Thus, phosphorylation of the S148 site may affect the function of kinesin-5 motors by affecting the conformation of loop 5. Sequence alignment reveals that the S148 site is present in some kinesin-5 homologs (Figs. 6B and S6), indicating that this site has been employed for kinesin-5 regulation elsewhere in evolution. Moreover, in some species, both S148 and the conserved S493 sites are present (Fig. S6). Interestingly, most of the kinesin-5 motors from the Saccharomycetaceae clade, which are characterized by the large young insert in loop 8 (Fig. S1), lost the Cdk1 phosphorylation consensus at position 148 (Fig. 6). The one strain from the Saccharomycetaceae clade that contains a Cdk1 consensus at position 148 (Fig. 6), lacks the large insert in loop 8 (Fig. S1). We speculate that the Cdk1 sites in loop 8 and at position 148 might interfere which each other. Our finding that the full variant at position S148 leads to destabilization and temperature sensitivity of Cin8 (Figs. 3 and 4C), may suggest an incompatibility with the large loop 8 insert.
Cell cycle networks are flexible, Cin8 regulation is precise
To date there has been very little experimental data systematically testing the flexibility of phosphoregulation. Cell cycle networks have been extensively rewired over long evolutionary time-scales. For example, while the general cell-cycle network topologies are conserved from yeast to humans, many of the molecular components have been replaced [48]. Thus, it is clear that there is considerable flexibility in cell cycle regulation. This general feature is likely related to the fact that >90% of Cdk1-dependent phosphorylation events occur in disordered regions of proteins [2]. Nevertheless, there is clearly constraint on some phosphorylation sites, indicating that the position is important [49]. As a number of kinesin-related proteins have been shown to be regulated by phosphorylation at different regions of the molecule we chose to investigate the flexibility of phosphoregulation using a mitotic kinesin-5 Cin8 as a model.
We found that two out of 29 Cin8syn mutations resulted in phosphorylation-dependent detachments. One of these sites, Cin8syn276, was immediately adjacent to a native site, Cin8nat277. These two alleles differed slightly in their distribution along the spindle (Figs. 2C and 4D), timing of their detachment from the spindle (Fig. S3), and in rate of anaphase spindle elongation (Figs. 4E and S5). The other site Cin8syn148, was completely novel in S. cerevisiae but had been sampled in other organisms during evolution (Figs. 6B and S6). This site also conferred regulation but at lower efficiency than the native sites. Thus, single synthetic sites conferred Cin8 regulation, and represent potential evolutionary paths to new regulation, but none of the new synthetic sites precisely recapitulated the phenotypes of the native sites.
In summary, the data presented here indicate that the immediate evolutionary paths to phosphoregulation of the kineisn-5 Cin8 are narrow. We sampled most of the possible single amino-acid substitutions that create a new Cdk1 consensus phosphoacceptor site, and two of these were functional. These single mutations created novel but suboptimal regulation. Kinesin-5 motors function by binding and moving along MTs via a precise catalytic cycle, with directionality, motor activity, and spindle binding all subject to phosphoregulation. Thus, while it is striking that single novel sites can confer regulation, precise control must ultimately evolve to allow mitotic kinesin motors to efficiently orchestrate the complex dynamics of the mitotic spindle.
Materials and Methods
Generation of synthetic Cdk1 phospho-variants and their categorization
The synthetic Cdk1 sites of Cin8 were generated on the basis of the phosphodeficient variant of Cin8 in which serines or threonines in all five native Cdk1 sites (S277, T285, S493, S736, and S1010) were mutated to alanine. Sites for novel synthetic Cdk1 phospho- variants were chosen in a systematic manner, with each original proline in the sequence of Cin8 being targeted as a potential Cdk1 phosphorylation site. Out of 20 prolines in the sequence, five are occupied by the native sites and the remaining fifteen prolines were tested for potential Cdk1 phosphoregulation sites. In these cases, to create a Cdk1 site, a serine was added replacing an amino-acid at position -1 to the proline, resulting in a partial ([S/T]-P) or full ([S/T]-P-x-[K/R]) Cdk1 phosphorylation site (Fig. 1B class 1). In the case where a partial Cdk1 phosphorylation site is created, a sequential mutation introduced a lysine at position +3 to the serine to result in a full Cdk1 phosphorylation consensus ([S/T]-P-x-[K/R]) was generated (Fig. 1B class 1). In addition, if a [S/T]-x-x-[K/R] sequence is present in a non-coiled coil region of Cin8, a proline is introduced at position +1 to the serine/threonine, resulting in a full Cdk1 phosphorylation consensus [S/T]-P-x-[K/R] (Fig. 1B class 2). The addition of serine at position -1 to the proline and the addition of lysine at position +3 to the serine were preferred over threonine and arginine, respectively, since it results in a stronger phosphorylation consensus [50, 51]. By this strategy, three of the five native sites, at positions 277, 285, and 493, were converted to [S-P] sites although position 285 originally contained a threonine followed by a proline [T-P]. The two native sites outside of the motor domain, at positions 736 and 1010, were not sampled since they were previously shown to have no phosphoregulation properties [18]. Two sites of class 2, at positions 712 and 271, were not sampled due to technical reasons. Overall, 29 new Cdk1 sites were generated. The novel sites were examined by live cell imaging for possible anaphase spindle-detachment phenotypes [20]. Mutants that were viable at 26°C and exhibited no detachment of Cin8 from the spindle were categorized as Cin8-5A-like mutants, to indicate they are not phosphoregulated compared to wt Cin8 (orange in Fig. 3). Typically, Cin8-5A-like mutants were viable at all temperatures with four exceptions: Cin8syn39, Cin8syn99, Cin8syn103, and Cin8syn700. Since these variants did not detach from the spindles, they were classified as Cin8-5A-like variants and not examined further by quantitative analyses.
Mutants that exhibited detachment from the spindle were targeted as possible Cdk1 phosphoregulation sites and additional control mutants were generated at these positions to determine if the observed phenotypes were phosphorylation-dependent. These control mutants contained a phosphodeficient alanine replacing the serine or threonine within the Cdk1 consensus sequence. In addition, the variants were examined by yeast viability assay to assess their ability to function in cells as a sole source of kinesin-5 activity (see below). Eight of the variants that exhibited detachment from the spindles at anaphase were not viable at 26oC as full Cdk1 sites (colored in black in Fig. 3). One site in this category, at position 134, was temperature sensitive as a partial control consensus (Cin8syn134pc). Since the partial and full variants at this position (Cin8syn134p and Cin8syn134f respectively) are non-viable, this site was not examined further by quantitative analysis. Another exception is at position 194, in which the partial variant Cin8syn194p was viable at 26oC and 33oC but temperature sensitive at 35oC, with no detachments from the spindle at 26oC. However, its full counterpart was non-viable at 26°C (Fig. 3).
Yeast viability assay
Viability of cells expressing the Cin8nat and Cin8syn variants as the sole source of kinesin-5 function was examined as previously described [18, 20, 52]. Yeast strains used for this assay were deleted for their chromosomal copies of CIN8 and KIP1 and contained an endogenic recessive cycloheximide resistance gene (cin8Δ;kip1Δ;cyhr). These cells were supplemented with a shuffle plasmid (pMA1208) encoding for wt Cin8 and a wt cycloheximide sensitivity gene. After transformation with a plasmid encoding the Cin8 variant of interest, the initial pMA1208 plasmid was shuffled out by growth on YPD medium containing 7.5 µg/mL cycloheximide for 3 to 4 days, at 26oC and elevated temperatures, in a serial dilution (1:1, 1:10, 1:102, 1:103, 1:104) starting with 0.2 O.Dλ=600nm (approximately 2.55×106 cells/ml).
Live cell imaging
The S. cerevisiae strains and plasmids used in this study are described in SI Tables S1, S2, and S3. Live cell imaging was performed as previously described [18, 28, 30], in cells deleted for CIN8, expressing functional copy of the homologous Kip1. Images were acquired using a Zeiss Axiovert 200M-based microscope setup equipped with a cooled CCD Andor Neo sCMOS camera. Images of Z stacks of eleven planes were obtained in three channels with 0.5 μm separation. Time-lapse images were obtained using a Zeiss Axiovert 200M-based Nipkow spinning-disc confocal microscope (UltraView ESR, Perkin Elmer, UK) with an EMCCD camera. Z stacks of 32-36 slices with 0.2 μm separation were acquired at one-minute intervals for 70 minutes. Data analysis was performed using MetaMorph (MDS Analytical Technologies) and open source ImageJ software. Spindle elongation rates were determined as previously described [18]. Statistical analysis of spindle elongation rates was done using post-ANOVA all-pairwise comparison performed with Tukey procedure using OriginLab software.
Fluorescence intensity distribution of Cin8 along the spindle
Line scan analysis along the spindle was employed to quantify the distribution of 3GFP-tagged Cin8 variants, as previously described [20]. The fluorescence intensity profile was determined along a line tracing the spindle from mother to bud (for 10 to 20 cells in each spindle length category). The background signal was calculated by averaging the intensity outside the nucleus and that value was subtracted from the fluorescence intensity measured at each point. Finally, intensity was interpolated and divided into 100 segments of equal length using Origin software (OriginLab). Normalization of the Cin8-3GFP fluorescent signals was performed by dividing the intensity at each point by the total Cin8-3GFP fluorescence intensity at each spindle (Figs. 2C and 4D). To calculate the relative averaged intensity at the bud SPB in Figs. 2C and 4D, the intensity of the normalized interpolated fluorescence signal was averaged over three maximal points at the SPB, for 10-20 cells. Statistical significances between these averages for different variants were determined by Student's t test.
Cin8 detachment from the spindle determined from fluorescence intensity perpendicular to the spindle
Line scan analysis perpendicular to the spindle was performed to quantify the detachment of Cin8 variants from the spindle during anaphase as previously described [20]. First, the Cin8-3GFP fluorescence profile was measured along a line of 41 pixels (5.24 µm) perpendicular to the spindle near the SPB (Figs. 2D). Twelve outer pixels on each side were assigned as background (Fig. 2D, dashed dark grey). The intensity of 5 pixels in the center of the line was assigned as the intensity near the SPB (Fig. 2D, dashed light grey). The intensities of pixels 12-18 and 23-29 were considered as the intensity resulting from nucleus-localized Cin8-3GFP, due to its detachment from the spindle (Fig. 2D, green). The intensity of Cin8-3GFP detached from the spindle (pixels 12-18 and 23-29) was averaged for each cell and normalized to the maximum intensity of the SPB. Finally, the normalized average Cin8-3GFP intensity perpendicular to the spindles of 6-7 µm, was calculated in all cells expressing the same variant (10-20 cells). Statistical significances between the averages of normalized Cin8 intensity perpendicular to spindles of different variants (Figs. 2E, 4B and S3) were determined by Student's t test.
In vitro phosphorylation assay
In vitro phosphorylation assays were performed as previously described [18, 20]. In brief, bacterially expressed Cin8 (590)-TEV-EGFP-6His variants were purified using standard nickel affinity chromatography, eluted with 300mM imidazole that was subsequently removed using Zeba Spin Desalting Columns 40K (Thermo Scientific). For a phosphorylation assay, equal concentrations of Cin8 variants were mixed with TAP-purified Clb2-Cdk1- Cks1 complex in kinase assay mixture [50mM HEPES, pH 7.4, 150mM NaCl, 5mM MgCl2, 8% glycerol, 0.2 mg/ml BSA, 500nM Cks1, and 500μM ATP (with added γ-32P-ATP (PerkinElmer)]. Reactions were stopped after 10 and 20 min with SDS-PAGE sample buffer and proteins were separated by SDS-PAGE. Gels were stained with Coomassie Brilliant Blue (CBB) R-250 (Sigma) and incorporation of 32P into the proteins was visualized by autoradiography.
Abbreviations
MT: microtubules; Cdk1: cyclin-dependent kinase 1; SBP: spindle pole body; wt: wild type; GFP: green fluorescent protein; nat: native; syn: synthetic; p: partial; f: full; c: control; pc: partial site, alanine control; fc: full site, alanine control; CBB: coomassie brilliant blue; ATP: adenosine 5' triphosphate; SDS: sodium dodecyl sulfate; PAGE: polyacrylamide gel electrophoresis; PDB: protein data bank; SEM: standard error of the mean; YPD: yeast peptone dextrose.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
We thank Kumar-Singh Sudhir, Pandey Himanshu, Siegler Nurit, and Popov Mary from Ben-Gurion University in the Negev, Beer-Sheva, Israel, for critical reading of this manuscript.
This work was supported in part by the Israel Science Foundation (ISF) (grants 165/13 and 386/18) awarded to L.G.; The United States - Israel Binational Science Foundation grant (BSF-2015851), awarded to L.G.; the William Bowes Foundation and Vilcek Foundation grants awarded to L.J.H.; The ERC Consolidator Grant Nr 649124, Phosphoprocessors, awarded to M.L.; and Estonian Science Agency grant Nr. IUT2-21 and PRG550, awarded to M.L.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Morgan DO, Roberts JM. Oscillation sensation. Nature. 2002;418:495-6
2. Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682-6
3. Landry CR, Freschi L, Zarin T, Moses AM. Turnover of protein phosphorylation evolving under stabilizing selection. Front Genet. 2014;5:245
4. Choi KY, Satterberg B, Lyons DM, Elion EA. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499-512
5. Mendenhall MD, Jones CA, Reed SI. Dual regulation of the yeast CDC28-p40 protein kinase complex: cell cycle, pheromone, and nutrient limitation effects. Cell. 1987;50:927-35
6. Schwob E, Bohm T, Mendenhall MD, Nasmyth K. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell. 1994;79:233-44
7. Koivomagi M, Ord M, Iofik A, Valk E, Venta R, Faustova I. et al. Multisite phosphorylation networks as signal processors for Cdk1. Nat Struct Mol Biol. 2013;20:1415-24
8. Koivomagi M, Valk E, Venta R, Iofik A, Lepiku M, Balog ER. et al. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature. 2011
9. V K, M O, M B. Evidence for Stabilizing Selection Driving Mutational Turnover of Short Motifs in. G3. 2018;8:3803-12
10. Goulet A, Moores C. New insights into the mechanism of force generation by kinesin-5 molecular motors. Int Rev Cell Mol Biol. 2013;304:419-66
11. Waitzman JS, Rice SE. Mechanism and regulation of kinesin-5, an essential motor for the mitotic spindle. Biol Cell. 2014;106:1-12
12. Singh SK, Pandey H, Al-Bassam J, Gheber L. Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles. Cell Mol Life Sci. 2018
13. Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159-69
14. Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD. et al. The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol. 1999;144:125-38
15. Crasta K. et al. Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins. 2006.
16. Cahu J, Olichon A, Hentrich C, Schek H, Drinjakovic J, Zhang C. et al. Phosphorylation by Cdk1 increases the binding of Eg5 to microtubules in vitro and in Xenopus egg extract spindles. PLoS ONE. 2008;3:e3936
17. Chee MK, Haase SB. B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet. 2010;6:e1000935
18. Avunie-Masala R, Movshovich N, Nissenkorn Y, Gerson-Gurwitz A, Fridman V, Koivomagi M. et al. Phospho-regulation of kinesin-5 during anaphase spindle elongation. J Cell Sci. 2011;124:873-8
19. Shapira O, Gheber L. Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain. Sci Rep. 2016;6:25597
20. Goldstein A, Siegler N, Goldman D, Judah H, Valk E, Koivomagi M. et al. Three Cdk1 sites in the kinesin-5 Cin8 catalytic domain coordinate motor. Cell Mol Life Sci. 2017;28:017-2523
21. Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109-20
22. Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95-108
23. Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451-8
24. Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. JCB. 2006;172:861-74
25. Gardner MK, Bouck DC, Paliulis LV, Meehl JB, O'Toole ET, Haase J. et al. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894-906
26. Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617-24
27. Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687-94
28. Gerson-Gurwitz A, Movshovich N, Avunie R, Fridman V, Moyal K, Katz B. et al. Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cell Mol Life Sci. 2009;66:301-13
29. Fridman V, Gerson-Gurwitz A, Movshovich N, Kupiec M, Gheber L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep. 2009;10:387-93
30. Movshovich N, Fridman V, Gerson-Gurwitz A, Shumacher I, Gertsberg I, Fich A. et al. Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. J Cell Sci. 2008;121:2529-39
31. Ibarlucea-Benitez I, Ferro LS, Drubin DG, Barnes G. Kinesins relocalize the chromosomal passenger complex to the midzone for spindle disassembly. J Cell Biol. 2018
32. Fridman V, Gerson-Gurwitz A, Shapira O, Movshovich N, Lakamper S, Schmidt CF. et al. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. J Cell Sci. 2013;126:4147-59
33. Gibbs KL, Greensmith L, Schiavo G. Regulation of Axonal Transport by Protein Kinases. Trends Biochem Sci. 2015;40:597-610
34. Shapira O, Goldstein A, Al-Bassam J, Gheber L. A potential physiological role for bi-directional motility and motor clustering of mitotic kinesin-5 Cin8 in yeast mitosis. J Cell Sci. 2017;130:725-34
35. Moses AM, Heriche JK, Durbin R. Clustering of phosphorylation site recognition motifs can be exploited to predict the targets of cyclin-dependent kinase. Genome Biol. 2007;8:R23
36. Garcia K, Stumpff J, Duncan T, Su TT. Tyrosines in the kinesin-5 head domain are necessary for phosphorylation by Wee1 and for mitotic spindle integrity. Curr Biol. 2009;19:1670-6
37. Gerson-Gurwitz A, Thiede C, Movshovich N, Fridman V, Podolskaya M, Danieli T. et al. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. Embo J. 2011;30:4942-54
38. Mennella V, Tan DY, Buster DW, Asenjo AB, Rath U, Ma A. et al. Motor domain phosphorylation and regulation of the Drosophila kinesin 13, KLP10A. J Cell Biol. 2009;186:481-90
39. Padzik A, Deshpande P, Hollos P, Franker M, Rannikko EH, Cai D. et al. KIF5C S176 Phosphorylation Regulates Microtubule Binding and Transport Efficiency in Mammalian Neurons. Front Cell Neurosci. 2016;10:57
40. Ogawa T, Saijo S, Shimizu N, Jiang X, Hirokawa N. Mechanism of Catalytic Microtubule Depolymerization via KIF2-Tubulin Transitional Conformation. Cell Rep. 2017;20:2626-38
41. Wang W, Cantos-Fernandes S, Lv Y, Kuerban H, Ahmad S, Wang C. et al. Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin. Nat Commun. 2017;8:70
42. Bell KM, Cha HK, Sindelar CV, Cochran JC. The yeast kinesin-5 Cin8 interacts with the microtubule in a noncanonical manner. J Biol Chem. 2017;12:797662
43. Subbiah S, Harrison SC. A simulated annealing approach to the search problem of protein crystallography. Acta Crystallogr A. 1989;45( Pt 5):337-42
44. Waitzman JS, Larson AG, Cochran JC, Naber N, Cooke R, Jon Kull F. et al. The loop 5 element structurally and kinetically coordinates dimers of the human kinesin-5, Eg5. Biophys J. 2011;101:2760-9
45. Maliga Z, Xing J, Cheung H, Juszczak LJ, Friedman JM, Rosenfeld SS. A pathway of structural changes produced by monastrol binding to Eg5. J Biol Chem. 2006;281:7977-82
46. Lad L, Luo L, Carson JD, Wood KW, Hartman JJ, Copeland RA. et al. Mechanism of inhibition of human KSP by ispinesib. Biochemistry. 2008;47:3576-85
47. Brier S, Lemaire D, Debonis S, Forest E, Kozielski F. Identification of the protein binding region of S-trityl-L-cysteine, a new potent inhibitor of the mitotic kinesin Eg5. Biochemistry. 2004;43:13072-82
48. Cross FR, Buchler NE, Skotheim JM. Evolution of networks and sequences in eukaryotic cell cycle control. Philos Trans R Soc Lond B Biol Sci. 2011;366:3532-44
49. Nguyen Ba AN, Moses AM. Evolution of characterized phosphorylation sites in budding yeast. Mol Biol Evol. 2010;27:2027-37
50. Koivomagi M, Valk E, Venta R, Iofik A, Lepiku M, Morgan DO. et al. Dynamics of Cdk1 substrate specificity during the cell cycle. Mol Cell. 2011;42:610-23
51. Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104-8
52. Duselder A, Fridman V, Thiede C, Wiesbaum A, Goldstein A, Klopfenstein DR. et al. Deletion of the Tail Domain of the Kinesin-5 Cin8 affects its directionality. J Biol Chem. 2015;19:620799
53. Sablin EP, Kull FJ, Cooke R, Vale RD, Fletterick RJ. Crystal structure of the motor domain of the kinesin-related motor ncd. Nature. 1996;380:555-9
54. Kull FJ, Endow SA. Kinesin: switch I & II and the motor mechanism. J Cell Sci. 2002;115:15-23
Author contact
![]() Corresponding authors: Liam J. Holt liam.holtorg and Larisa Gheber lgheberac.il
Corresponding authors: Liam J. Holt liam.holtorg and Larisa Gheber lgheberac.il

 Global reach, higher impact
Global reach, higher impact