10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(6):1252-1260. doi:10.7150/ijbs.28235 This issue Cite
Research Paper
Nuclear CD44 Mediated by Importin β Participated in Naïve Genes Transcriptional Regulation in C3A-iCSCs
1. Department of Cell Biology, School of Basic Medical Sciences, Peking University, Beijing, 100191, China
2. Beijing Cellonis Biotechnologies Co. Ltd, Zhongguancun Bio-Medicine Park, Beijing, People's Republic of China
3. State Key Laboratory of Organ Failure Research, Co-Innovation Center for Organ Failure Research, Guangdong Provincial Southern Medical University, Guangzhou, Guangdong Province, 510515, China
4. Medical Center of Human Reproduction, Beijing Chaoyang Hospital, Capital Medical University, Beijing, 100069, China
Received 2018-7-1; Accepted 2019-3-12; Published 2019-5-11
Abstract
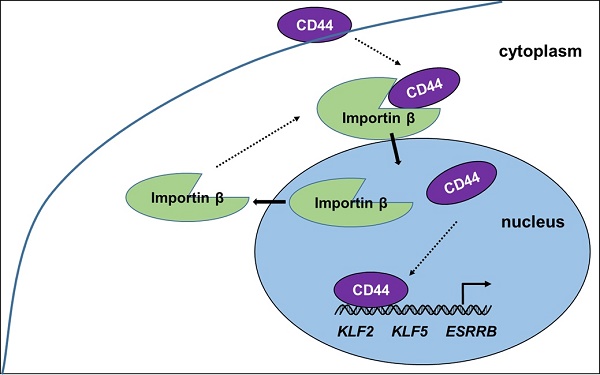
CD44 is one of biomarkers of liver cancer stem cells (CSCs). The investigation of mechanism of CD44 translocation helps to uncover new molecular pathways participated in the regulation of various cellular processes in CSCs. In the present study, we observed the translocation of CD44 from cytoplasm to nuclear in the reprogramming process of C3A cells, full-length CD44 presented in the nucleus of liver iCSCs. CD44 was bound with importin β and transportin 1 in liver iCSCs. Inhibition of importin β transport leads to reduction of CD44 in the nucleus. Translocation of CD44 is also influenced by importin α. Besides, overexpression of naïve pluripotent genes, KLF2, KLF5, DNMT3L, GBX2, ZFP42, ESRRB and DPPA4 were found in liver iCSCs. Inhibition of CD44 leads to the reduction of these naïve genes. Luciferase and chromatin immunoprecipitation (ChIP) assays further identified nuclear CD44 bound to the promoter regions of naïve genes, KLF2, KLF5, and ESRRB functioned as transcriptional activators in liver iCSCs. Our present work provides new insight into the dynamic states and functions of CD44 in iCSCs.
Keywords: CD44, importin β, liver iCSCs, naïve gene
Introduction
Cancer stem cells (CSCs) characterized as a small subpopulation of heterogeneous cells have the capacity to self-renew and differentiate, which are implicated in tumor progression and distant metastasis [1]. Liver CSCs are responsible for the high rate of recurrence and heterogeneity of tumors [2]. Therefore, therapies targeting liver CSCs hold great promise for improving survival and quality of life of liver cancer patients. There are many biomarkers including CD133, CD44, CD90 and CD24 discovered to identify and isolate liver CSCs [3-5].
The glycoprotein CD44 functions as the principal cell surface receptor for hyaluronan (HA) involved in numerous cellular processes including cell adhesion, cell migration, and epithelial-mesenchymal transitions (EMT) [6-8]. As the receptor of Wnt signaling pathway, CD44 plays a vital role in tumor stem cell niche [9]. CD44 participated in the homing of early leukemia stem cells in acute myelocytic leukemia [10]. CD44 is also identified as a negative biomarker of disturbing the forming of iPSCs during the process of fibroblast reprogramming [11]. In human colon cancer cells, CD44 was responsible for the survival of cells in suspension culture conditions. [12]. There are also some evidences that CD44 is presented in the nucleus upon engagement by HA or other ligands. It has been proved that the intracellular domain of CD44 is translocated to the nucleus following sequential cleavage, where it is believed to be involved in the regulation of gene transcription [13, 14]. In recent years, full-length CD44 has been found presented in the nucleus of gastric cancer and osteosarcoma cells [15, 16].
Proteins shuttled between cytoplasm and nucleus through nuclear pores. To traverse the barrier, proteins larger than 40kD must bind with carrier proteins or transport factors [17]. Most of transport factors belong to β-karyopherin family, known as importin and exportin [18]. Importin has two subunits, importin α and importin β. Cytoplasmic proteins containing a nuclear localization signal (NLS) are usually bound with the carrier importin β, with or without the presence of importin α [19]. Importin β-bound complex then locates to the nuclear pore complex (NPC) and releases in the nucleus following binding of importin β to Ran, a small GTP binding protein. The importin β-Ran complex is exported to the cytoplasm after it dissociates upon converting RanGTP to RanGDP [18].
The investigation of CD44 helps to understand the molecular mechanism underlying liver iCSCs. Former studies of our lab find that CD44 is overexpressed in the nucleus of C3A-iCSCs, shMBD3-iCSCs and C3A-c-Jun-iCSCs [20, 21]. This phenomenon caused our great concern. Because CD44 was found presented in the nucleus when the programming process was completed, we speculate whether entry of CD44 to the nucleus occurred in the programming process. In the present study, we observed CD44 presented at the beginning of reprogramming and translocated into nucleus in reprogramming process, indeed full-length CD44 presented in the nucleus of liver iCSCs. Furthermore, we found that CD44 was bound with importin β and transportin 1. Inhibition of importin β transport leads to CD44 reduction in the nucleus. Translocation of CD44 is also influenced by importin α. Further studies by luciferase and ChIP assays confirmed that nuclear CD44 bound to the promoter regions of naïve genes, KLF2, KLF5, and ESRRB functioned as transcriptional activators in liver iCSCs. These findings are helpful for illustrating the critical role of nuclear CD44 in liver iCSCs.
Materials and Methods
Cell culture
C3A was cultured in Eagle's minimum essential medium (Gibco) containing 10% fetal bovine serum (HyClone), 0.1mM non-essential amino acids (Gibco). Culture medium for C3A-iCSCs and H9 cells was Dulbecco's modified Eagle's medium (DMEM) / Ham's F-12 medium (Gibco) containing 20% knockout serum replacement (Gibco), 1mM L-glutamine, 0.1mM nonessential amino acids, 0.1mM 2-mercaptoethanol, and 10ng/ml recombinant human basic fibroblast growth factor (Life Technologies).
Generation of C3A-iCSCs
C3A were seeded at 6×104 cells per well in 24-well plates. C3A were infected with retroviruses vectors TetO-FUW-OSKM, comprising Oct4, Sox2, Klf4, and c-Myc (Addgene), co-transfected with retroviruses of pCMV-Gag-Pol and pCMV-VSVG. Then cells were passaged onto vitronectin-coated dishes in C3A-iCSCs culture medium supplemented with 20ng/mL doxycycline (Sigma) for 25 days. On day 26, cells were detached onto new dishes until the single cell sphere was picked up and passaged.
RNA isolation and real-time PCR
Total RNA was extracted using Trizol (Life Technologies). cDNA synthesis was performed with the M-MLV Reverse Transcriptase kit (Promega) in accordance with the manufacturer's instructions. Real-time PCR was performed with GoTaq ® qPCR Master Mix (Promega). Primer sequences are listed in Supplementary Table S1.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde. Permeabilization was performed with 0.5% Triton X-100 and blocking with 10% bovine serum albumin (Sigma). Primary antibodies included antibodies against CD44 (1:400; Proteintech), OCT4 (1:500; Santa Cruz), SOX2 (1:500; Chemicon), TRA-1-80 (1:200; Abcam), Importin β (1:1000; Abcam), Transportin 1 (1:200; Abcam). Secondary antibodies were Alexa Fluor® 488/594 goat anti-rabbit/mouse IgGs (1:500; Origene). Counter staining was performed with Hoechst 33342. Images were captured and analyzed with TCSSP8 Confocal Microscope (Leica).
RNA interference
siRNA oligonucleotides: si-KPNB1-1 (GUGCAGAGAUCCCAGUAAATT), si-KPNB1-2 (GCCCACCCUAAUAGAAUUATT), si-KPNB1-3 (GGUGGUGAAUUCCUCAAGUTT); si-KPNA2-1 (GCUGCCAGGAAACUACUUUTT), si-KPNA2-2 (GGAGGUGCCAUCCCAGCAUTT), si-KPNA2-3 (CCUAAGAGCCAUAGGGAAUTT); si-CD44 (GGACCUCUUUCAAUGACAATT). All were synthesized by GenePharma (Shanghai, China). The cells were harvested 72 hrs after transfection.
Western blot analysis
Cells were lysed in RIPA lysis buffer (50mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1 mg/ml PMSF, and 1mM proteinase inhibitor cocktails). The samples were subjected to SDS-PAGE and then transferred to PVDF membranes (Millipore). The anti-KPNB1 monoclonal antibody (1:800; Abcam), anti-KPNA2 polyclonal antibody (1:1000; Proteintech), anti-CD44 polyclonal antibody (1:1000; Proteintech), anti-laminB polyclonal antibody (1:1000; Proteintech) and anti-GAPDH monoclonal anti-body (1:2000; Santa Cruz) were applied for Western blot analysis.
Nuclear/Cytosol fractionation
Nuclear/cytosol fractionation was performed using Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime biotechnology).
Co-immunoprecipitation
Cells were lysed with IP lysis buffer (150mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50mM Tris-HCl (pH 8.0)). Add primary antibody to 500 ug protein sample and rotate at 4°C for 3-4 hrs, then incubate with agarose protein A+G beads (Santa Cruz) overnight at 4°C. Centrifuge at 3000 rpm for 3 min to collect the beads combined with proteins. Wash 3 times with 1ml PBS containing cocktails. The released proteins were analyzed by Western blotting.
Cell cycle analysis
Cells were harvested with accutase, suspended (2×106 cells / 100μl) in cold PBS, and then fixed in 70% ethanol in PBS at 4 °C for 0.5h. Cells were washed with cold PBS and stained with 0.5ml of propidium iodide (PI) staining buffer (200 mg/ml RNase A, 50 ug/ml PI) at 37°C in the dark. Signals were detected with a FACS Calibur flow cytometer (BD Biosciences).
EdU cell proliferation assay
EdU cell proliferation assay were performed using EdU HTS Kit 488 (Sigma). Cells were cultured on matrigel for 16 hrs before ethanol fixation.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed following the cross- linking chromatin immunoprecipitation protocol (http://www.abcam.cn). Real-time PCR primers used are listed in Supplementary Table S1.
Dual luciferase assay
The firefly and Renilla luciferase activities were measured by Dual-Luciferase Reporter Assay System (Promega) after 72 hrs according to the pGL3 Luciferase Reporter Vector Technical Manual (http://www. promega.com). KLF2, KLF5, and ESRRB promoter regions were amplified with specific primers and the fragments were ligated into pGL3- basic vectors (Promega). The results are showed as the ratio of Renilla to firefly luciferase activities.
Statistical analysis
The percentage of colocalization in the region of interest (ROI) was qualified; 250-300 cells analyzed per condition. All data are expressed as the mean ± SD from at least three independent experiments. The Student's two-tailed t-test with significance set at p < 0.05 was used to determine statistical significance. Error bars indicate standard deviation.
Results
Generation of C3A-iCSCs induced by OSKM
Lentivirus plasmid TetO-FUW-OSKM encoded the transcription factors Oct4, Sox2, Klf4 and c-Myc (OSKM) were used to infect the C3A cells. Doxycycline was performed to induce OSKM expression. After 25 days culture, cells were passaged onto new dishes following collagenase IV treatment. Suspended cell spheres generated and picked up were C3A-iCSCs (Figure 1A).
Generation of C3A-iCSCs induced by OSKM. A. Time schedule of C3A-iCSCs generation. B. Real-time PCR analysis of the exogenous OSKM in C3A cells, C3A-D10, C3A-D20 and C3A-iCSCs. Relative gene expression to C3A cells was calculated for C3A-D10, C3A-D20 and C3A-iCSCs. Data are presented as the means ± SD from three independent. C. Real-time PCR analysis of endogenous stem cell markers OCT4, SOX2 and NANOG in C3A cells, C3A-D10, C3A-D20, C3A-iCSCs and H9 cells. Relative gene expression to C3A cells was calculated for C3A-D10, C3A-D20, C3A-iCSCs and H9 cells. Data are presented as the means ± SD from three independent. D. Immunofluorescence staining of OCT4 / SOX2 / TRA-1-80 in C3A cells and C3A-iCSCs. Nuclei were counterstained with Hoechst 33342 (blue). Scale bar, 25μm.
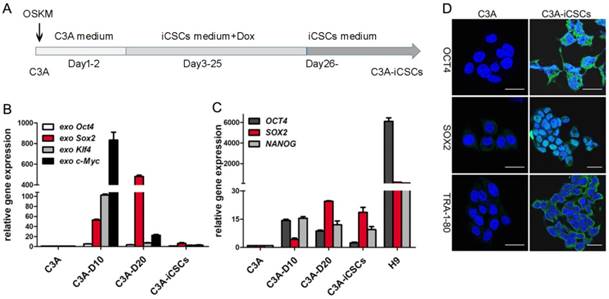
Initiation of reprogramming starts with activation of exogenous reprogramming genes, commonly OSKM. The maturation stage is considered to be the activation of endogenous expression of pluripotent core circuits, comprising Oct4, Sox2, and Nanog to achieve a state of transgene independence. The stabilization stage accompanied by the silence of exogenous genes and maintenance of endogenous pluripotent genes. [22]. We observed the exogenous expression of OSKM genes increased expression at day 10, decreased expression at day 20 and silenced expression in C3A-iCSCs (Figure 1B). Endogenous pluripotent genes showed a gradual increased expression before day 20 and maintained chiefly by SOX2 in C3A-iCSCs (Figure 1C). The expressions of pluripotent markers OCT4, SOX2, and TRA-1-80 were analyzed by immunofluorescence. OCT4 had an abundant expression dispersed in the cytoplasm and nucleus. OCT4 acted as one of core biomarkers to mark stemness [23]. Ectopic expression of OCT4 can be related to malignance and diagnosis in the neoplasm [24, 25]. SOX2 exhibited a strong positive expression in the nucleus. TRA-1-80 displayed cell membrane localization (Figure 1D). These evidences demonstrated that C3A-iCSCs possessed pluripotent capacity. H9 cells were performed as the positive control.
Translocation of CD44 from cytoplasm to nuclear in the reprogramming process of C3A cells
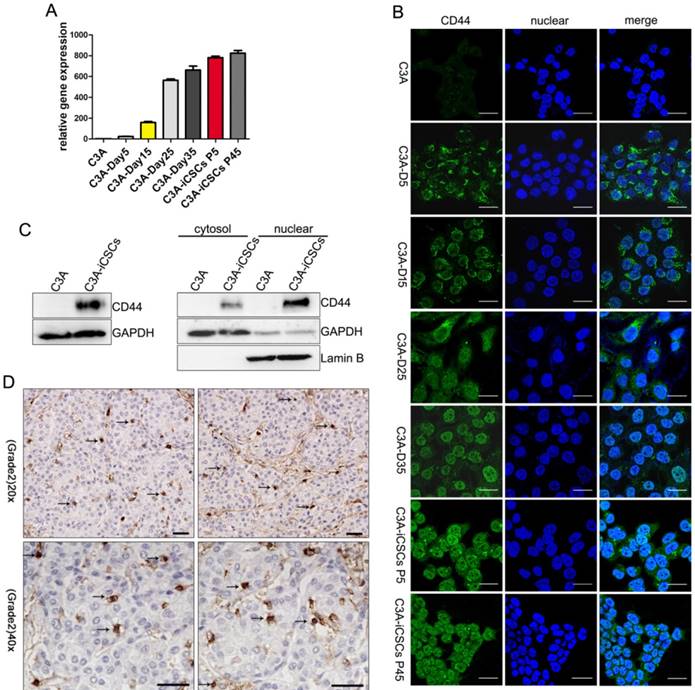
Former studies found that CD44 was overexpressed in the nucleus of C3A-iCSCs, shMBD3- iCSCs and C3A-c-Jun-iCSCs. We speculate whether entry of CD44 to the nucleus occurred in the programming process since CD44 was presented in the nucleus when the programming process was completed. To further study this phenomenon, several time points of reprogramming process were picked up to capture the expression of CD44. Analysis from real-time PCR indicated that CD44 gained a gradual increased expression in the programming process of C3A cells (Figure 2A). Data from immunofluorescence microscopy showed that CD44 presented outside of the nucleus on the day 5. Particles of CD44 were presented in the nucleus on the day 15. Increased expression of CD44 was detected diffused in the nuclear and cytoplasm on the day 25, some gathered in the nuclear. On the day 35, almost total CD44 accumulated in the nuclear (Figure 2B). Afterwards, clones began to detach from the bottom of plate. CD44 was constitutively maintained in the nucleus at the passage 5 and passage 45. These findings indicated that the translocation of CD44 from cytoplasm to nuclear occurred in the reprogramming process of C3A cells and CD44 could be stably maintained in the nucleus of C3A-iCSCs.
Western blot analysis using anti-CD44 antibody confirmed the presence of an 85-90 kDa CD44 species (Figure S1A) that corresponds to full-length CD44s in the nucleus of C3A-iCSCs (Figure 2C). Immunohistochemical staining of pathological samples of clinical hepatic carcinoma (kindly provided by Professor Gao Yi, Southern Medical University Zhujiang Hospital) confirmed that a small amount of CD44-positive markers (arrows) found in the nucleus, suggesting that nuclear CD44 was not rare to be seen in clinical liver cancer (Figure 2D).
CD44 transport was mediated by importin β and affected by importin α
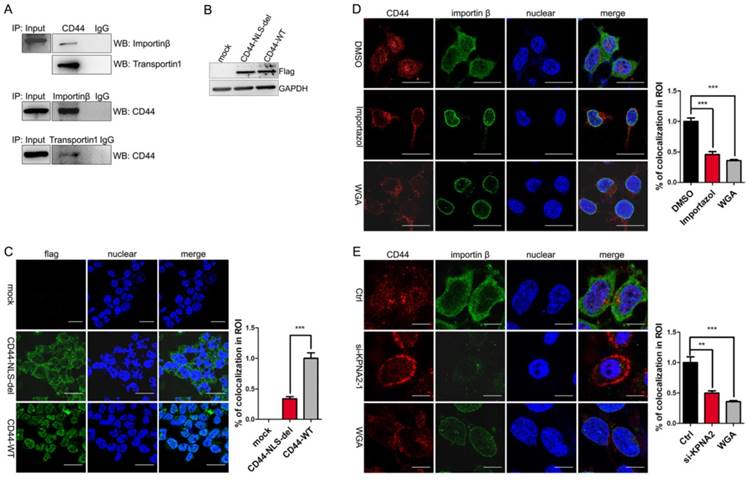
Most nuclear-cytoplasmic transportations belong to β-karyopherin family transport factors. Importin β and transportin 1 were the most studied factors. Former studies by Janiszewska M et al showed the interaction of CD44 with importin β and transportin 1 in MNNG/HOS cells [15]. We assumed that CD44 transport associated with importin β and transportin 1 in liver iCSCs. Immunofluorescence and co-immunoprecipitation assays were performed in C3A-iCSCs to identify the assumption. We confirmed that CD44 was interacted with importin β and transportin 1 in C3A-iCSCs (Figure 3A, Figure S1B-C).
We further studied the relationship between CD44 and importin β. Because the nuclear translocation mediated by importin β required the guide of NLS, the CD44 NLS knockout plasmid with the flag tag (CD44-NLS-del) and the CD44 wild type plasmid with the flag tag (CD44-WT) were constructed and transduced into C3A-iCSCs respectively (Figure 3B). Results indicated that CD44-WT was presented in the nucleus, while CD44-NLS-del was blocked outside the nucleus, suggesting that the NLS domain of CD44 is essential for translocation (Figure 3C).
Importazole [26], a special inhibitor of importin β without disrupting the transportin mediated nuclear import, was performed to antagonize the entry of importin β into the nucleus. Immunofluorescence microscopy of C3A-iCSCs using an antibody of CD44 revealed the inhibition of importin β transport leaded to CD44 reduction in the nucleus. Wheat germ agglutinin (WGA), a blocker of nuclear pores [27] was used as positive control (Figure 3D). Similar results were obtained in RNA interfere assay targeting KPNB1 gene (Figure S1D-E). These findings supported the notion that CD44 transport was impaired by blocking importin β transport.
There is some evidence that the transportation of importin β were dependent upon importin α [28]. Therefore, we performed RNA interfere assay targeting KPNA2 gene to knockdown importin α in C3A-iCSCs. Real-time PCR and western blot analysis (Figure S1F) assessed the interfere effect. Immunofluorescence investigated the impact of importin α elimination on nuclear CD44. Analysis of immunofluorescence microscopy showed that nuclear CD44 were diminished when importin α were knocked down (Figure 3E). CD44 transport was affected by importin α.
Nuclear CD44 participated in naïve genes transcriptional regulation
Previously, Lee and colleagues found that triggered by ligation of OPN / H-3, CD44 could acquire more nuclear translocation [29]. Nuclear CD44 of full length integrated with STAT3 and gp130 could bind to the promoter of cyclin D1 as an transcriptional activator to impact the cell proliferation of colon cancer stem cells [30]. In our study, we found naïve pluripotent genes, KLF2, KLF5, DNMT3L, GBX2, ZFP42, ESRRB and DPPA4 were dramatically increased in C3A-iCSCs compared to C3A (Figure 4A). Inhibition of CD44 leads to the reduction of these naïve pluripotent genes (Figure 4B). These genes can be good candidates of CD44 targets. CD44 might participate in naïve genes transcriptional regulation of C3A-iCSCs. To verify this assumption, chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assay was performed in C3A cells and C3A-iCSCs. Our results confirmed that nuclear CD44 bound to promoter regions of KLF2, KLF5, and ESRRB of C3A-iCSCs (Figure 4C).
Translocation of CD44 from cytoplasm to nuclear in the reprogramming process of C3A cells. A. Real-time PCR analysis of CD44 in indicated time points of reprogramming process. Relative gene expression of CD44 to C3A cells was calculated for C3A-D5, C3A-D15, C3A-D25 C3A-D35, C3A-iCSCs P5 and C3A-iCSCs P45. Data are presented as the means ± SD from three independent. B. Immunofluorescence staining of CD44 in indicated time points of reprogramming process. Scale bar, 25μm. C. Western blot analysis of total protein (left) and cytoplasmic/nuclear protein (right) of CD44 in C3A and C3A-iCSCs. D. Immunohistochemical staining of CD44 in clinical liver cancer samples. Arrows indicated nuclear CD44-positive staining. Scale bar, 30μm.
CD44 transport was mediated by importin β and affected by importin α. A. The interaction between CD44 and importin β / transportin 1 was demonstrated by co-immunoprecipitation assay in C3A-iCSCs. B. Western blot analysis of flag after CD44-NLS-del / CD44-WT was transduced into C3A-iCSCs. C. Immunofluorescence staining analysis of flag after CD44-NLS-del / CD44-WT was transduced into C3A-iCSCs. Scale bar, 25μm. D. Immunofluorescence staining analysis of CD44 (red) and importin β (green) after Importazole was treated in C3A-iCSCs. DMSO indicated negative control, WGA indicated positive control. Scale bar, 10μm. E. Immunofluorescence staining analysis of CD44 (red) and importin β (green) after importin α was knocked down by RNA interfere assay. Scale bar, 10μm. C-E. Percentages of colocalization in ROI were qualified.
Nuclear CD44 participated in naïve genes transcriptional regulation. A. Real-time PCR analysis of the naïve pluripotent genes KLF2, KLF5, ZFP42, ESRRB, DNMT3L, GBX2, DPPA4 and LIFR in C3A cells and C3A-iCSCs. B. Real-time PCR analysis of CD44 and naive pluripotent genes after CD44 was silenced. C. ChIP-qPCR performed using CD44-specific antibodies in C3A cells and C3A-iCSCs. KLF2, KLF5, ESRRB represented their promoters respectively. Samples were analyzed by real-time PCR. Error bars showed the standard deviation of three independent ChIP-qPCR assays. D. Luciferase activity assay performed in C3A-iCSCs. Luciferase reporter plasmid pGL3-KLF2 / pGL3-KLF5 / pGL3-ESRRB containing their own promoter sequences was co-transfected with Renilla control vector and Ctrl / si-CD44 RNA sequences in C3A-iCSCs. Luciferase activities were measured after 72 hrs and presented as relative to the activity of Renilla luciferase. (* p<0.05, ** p<0.01, *** p<0.001, ns indicated no significant differences)
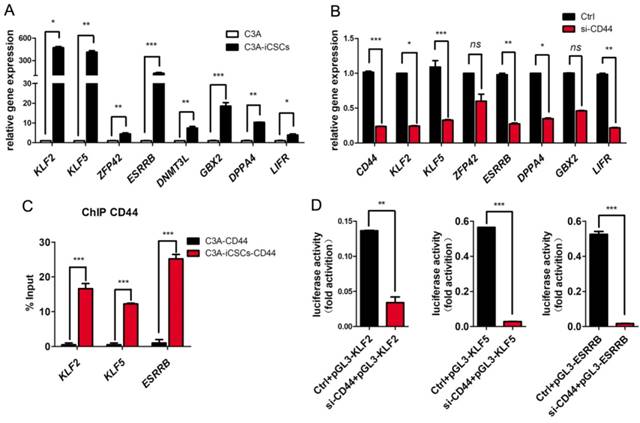
We used luciferase reporters containing the promoter sequences of KLF2, KLF5, and ESRRB of their own to reconfirm the activation of CD44 on their promoters. The luciferase activities of KLF2, KLF5, and ESRRB were decreased after CD44 was silenced respectively (Figure 3D). Nuclear CD44 binds to the promoter regions of KLF2, KLF5, and ESRRB functioned as transcriptional activators in liver iCSCs.
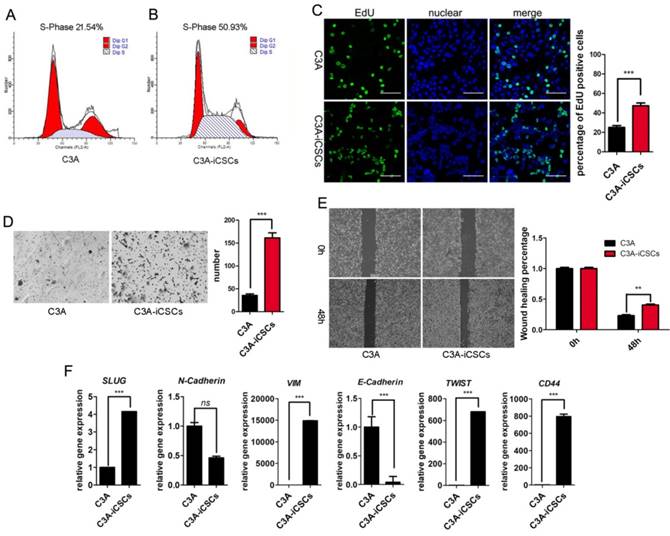
C3A-iCSCs acquired higher proliferation and migration
C3A-iCSCs displayed higher proliferation and migration properties than C3A cells. Results from cell cycle analysis showed that the total percentage of cells in S phase was 50.93% in C3A-iCSCs compared to 21.54% in C3A cells, indicating that C3A-iCSCs gained stronger proliferation (Figure 5A-B). Analysis by immunofluorescence microscopy indicated an increase of EdU incorporation in C3A-iCSCs than C3A cells (Figure 5C). Transwell assay and wound-healing assay showed that C3A-iCSCs gained more migration properties than C3A cells (Figure 5D-E). The EMT was associated with CSCs properties considered as a key program in cancer invasion and metastasis [31]. We found that mesenchymal-related markers, SLUG, VIM, TWIST1 and CD44 showed increased expressions in C3A-iCSCs, while epithelial marker E-cadherin showed a decreased expression in C3A-iCSCs (Figure 5F), suggesting that C3A-iCSCs underwent EMT associated with cancer invasion and metastasis.
Discussion
In this study, we elucidated the mechanism upon CD44 transport to the nucleus and the critical role of nuclear CD44 in transcriptional regulation to naïve genes in liver iCSCs. We observed the translocation of CD44 from cytoplasm to nuclear in the reprogramming process of C3A cells, full-length CD44 presented in the nucleus of liver iCSCs. CD44 is bound with importin β and transportin 1. Inhibition of importin β transport leads to CD44 reduction in the nucleus. Translocation of CD44 is also influenced by importin α. Nuclear CD44 binds to the promoter regions of naïve genes, KLF2, KLF5, and ESRRB functions as a transcriptional activator in liver iCSCs.
C3A-iCSCs acquired higher proliferation and migration. A-B. Flow cytometric analysis of the cell cycle of C3A cells and C3A-iCSCs. C. Nuclear EdU staining of C3A and C3A-iCSCs. Scale bar, 100 μm. Percentages of EdU-positive cells were calculated, n=5. D. Transwell assay performed in C3A and C3A-iCSCs. Cells were starved 24 hrs before seeding into tranwell chamber; images were captured 48 hrs later after seeding. Cells of lower layer of chamber were counted, n=5. E. Wound-healing assay performed in C3A and C3A-iCSCs. Quantities analysis of wound-healing assay were performed, n=5. F. Real-time PCR analysis of the EMT-related genes in C3A cells and C3A-iCSCs. (* p<0.05, *** p<0.001, ns indicated no significant differences)
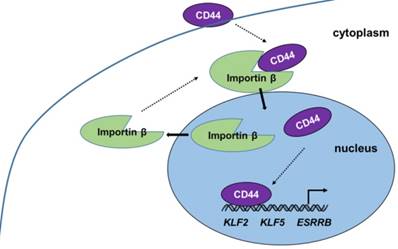
Proposed model of CD44 transport from cytoplasm to nuclear. CD44 transport was mediated by importin β. Nuclear CD44 participated in transcriptional regulation of KLF2, KLF5, and ESRRB genes in C3A-iCSCs.
It has been proved that CD44 intracellular domain could be hydrolyzed and released from the cell membrane to obtain the nuclear translocation following sequential cleavage, since it possessed NLS sequences. [32]. In recent years, there are reports the presence of full-length CD44 in the nucleus. Former study by Lee et al. [30] have shown nuclear localization of the full-length CD44 in the H1299 gastric cancer cell line. Janiszewska et al. demonstrated that full-length CD44 appeared in the nucleus in MNNG/HOS cells [15]. In our study, we performed western blot in C3A-iCSCs, noting the presence of CD44 of full-length in the nucleus of liver iCSCs.
In the reprogramming process of human parental fibroblasts, CD44 is gradually lost throughout the process and absent in fully reprogrammed cells which is identified as the negative surface marker [11]. In our study, CD44 is presented from cytoplasm to nuclear in the reprogramming process of C3A cells. Cancer cells are hard to be reprogrammed. C3A-iCSCs may belong to partially reprogrammed cells. However, the role of CD44 in the reprogramming process of C3A cells remains to be further studied.
As it is informed that CD44 is one of markers of EMT associated with cancer invasion and metastasis [33]. Cells need to undergo an early EMT state to start reprogramming [34]. Nuclear CD44 was responsible for the survival of suspended sphere state of colon CSCs. In our study, we observed the suspension spheres of C3A-iCSCs when CD44 presented in the nucleus totally. This transition might be related to the function of CD44. Nevertheless, the specific mechanism remains to be elucidated.
Inhibition of transportin 1 blocked the entry of CD44 into the nucleus in MNNG/HOS cells [15]. In our study, we found that CD44 protein bound to importin β and transportin 1. Inhibition of importin β transport resulted in significant reduction of CD44 in the nucleus. Nuclear transport of CD44 was mediated by importin β in liver iCSCs.
In this study, we find that nuclear CD44 functions as a transcription factor involved in the transcriptional regulation of promoter regions of KLF2, KLF5 and ESRRB in C3A-iCSCs. It has been demonstrated that CD44 participates in the transcriptional regulation of genes. JL Lee found the nuclear CD44 bound to the promoters of c-Myc and Twist1, leading to changes in cell fate through transcriptional reprogramming [12]. CD44 was involved in the transcriptional regulation of SOX2 and c-Myc associated with the stemness of liver iCSCs.
Our present work provides new insight into the progress and function of CD44 in iCSCs. CD44 is shown to be transported from the cytoplasm to the nucleus by importin β, where it induces naïve genes transcription in liver iCSCs. (Figure 6) The investigation of mechanism of CD44 translocation can help uncover new molecular pathways thus it participates in the regulation of various cellular processes in CSCs.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (81572313) and Science and Technology Planning Project of Guangdong Province, China (2015B020229002).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P. et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973-8
2. Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123:1911-8
3. Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P. et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153-66
4. Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ. et al. Aldehyde dehydrogenase discriminates the CD133 liver cancer stem cell populations. Mol Cancer Res. 2008;6:1146-53
5. Klonisch T, Wiechec E, Hombach-Klonisch S, Ande SR, Wesselborg S, Schulze-Osthoff K. et al. Cancer stem cell markers in common cancers - therapeutic implications. Trends Mol Med. 2008;14:450-60
6. Goldstein LA, Zhou DF, Picker LJ, Minty CN, Bargatze RF, Ding JF. et al. A human lymphocyte homing receptor, the hermes antigen, is related to cartilage proteoglycan core and link proteins. Cell. 1989;56:1063-72
7. Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J. et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121:1064-74
8. Li W, Ma H, Zhang J, Zhu L, Wang C, Yang Y. Author Correction: Unraveling the roles of CD44/CD24 and ALDH1 as cancer stem cell markers in tumorigenesis and metastasis. Sci Rep. 2018;8:4276
9. Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843-50
10. Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167-74
11. Quintanilla RH Jr, Asprer JS, Vaz C, Tanavde V, Lakshmipathy U. CD44 is a negative cell surface marker for pluripotent stem cell identification during human fibroblast reprogramming. PLoS One. 2014;9:e85419
12. Su YJ, Lai HM, Chang YW, Chen GY, Lee JL. Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J. 2011;30:3186-99
13. Okamoto I, Kawano Y, Murakami D, Sasayama T, Araki N, Miki T. et al. Proteolytic release of CD44 intracellular domain and its role in the CD44 signaling pathway. J Cell Biol. 2001;155:755-62
14. Gao R, Li D, Xun J, Zhou W, Li J, Wang J. et al. CD44ICD promotes breast cancer stemness via PFKFB4-mediated glucose metabolism. Theranostics. 2018;8:6248-62
15. Janiszewska M, De Vito C, Le Bitoux MA, Fusco C, Stamenkovic I. Transportin regulates nuclear import of CD44. J Biol Chem. 2010;285:30548-57
16. Lee JL, Wang MJ, Chen JY. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. J Cell Biol. 2009;185:949-57
17. Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195-208
18. Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647-71
19. Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441-51
20. Han S, Guo J, Liu Y, Zhang Z, He Q, Li P. et al. Knock out CD44 in reprogrammed liver cancer cell C3A increases CSCs stemness and promotes differentiation. Oncotarget. 2015;6:44452-65
21. Li R, He Q, Han S, Zhang M, Liu J, Su M. et al. MBD3 inhibits formation of liver cancer stem cells. Oncotarget. 2017;8:6067-78
22. Hawkins K, Joy S, McKay T. Cell signalling pathways underlying induced pluripotent stem cell reprogramming. World J Stem Cells. 2014;6:620-8
23. Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26:3068-74
24. Atlasi Y, Mowla SJ, Ziaee SA, Bahrami AR. OCT4, an embryonic stem cell marker, is highly expressed in bladder cancer. Int J Cancer. 2007;120:1598-602
25. Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465-77
26. Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara-Bingen M. et al. Importazole, a small molecule inhibitor of the transport receptor importin-beta. ACS Chem Biol. 2011;6:700-8
27. Finlay DR, Newmeyer DD, Price TM, Forbes DJ. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J Cell Biol. 1987;104:189-200
28. Liang P, Zhang H, Wang G, Li S, Cong S, Luo Y. et al. KPNB1, XPO7 and IPO8 mediate the translocation ofNF-kappaB/p65 into the nucleus. Traffic. 2013;14:1132-43
29. Lee JL, Wang MJ, Sudhir PR, Chen JY. CD44 engagement promotes matrix-derived survival through the CD44-Src-integrin axis in lipid rafts. Mol Cell Biol. 2008;28:5710-23
30. Lee JL, Wang MJ, Chen JY. Acetylation and activation of STAT3 mediated by nuclear translocation of CD44. Journal of Cell Biology. 2009;185:949-57
31. Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704-15
32. Nagano O, Saya H. Mechanism and biological significance of CD44 cleavage. Cancer Sci. 2004;95:930-5
33. Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740-6
34. Liu X, Sun H, Qi J, Wang L, He S, Liu J. et al. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat Cell Biol. 2013;15:829-38
Author contact
![]() Corresponding author: Li Shen; E-mail: shenliedu.cn
Corresponding author: Li Shen; E-mail: shenliedu.cn

 Global reach, higher impact
Global reach, higher impact