10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(12):2538-2547. doi:10.7150/ijbs.35357 This issue Cite
Research Paper
microRNA-1 Regulates NCC Migration and Differentiation by Targeting sec63
1. Jiangsu Key Laboratory of Oral Diseases, Nanjing Medical University, Nanjing 210019, China
2. MOE Key Laboratory of Model Animal for Disease Study, Model Animal Research Center, Nanjing University, Nanjing 210029, China
3. Department of Pathophysiology, Key laboratory of Cardiovascular Disease and Molecular Intervention, Nanjing Medical University, Nanjing 210029, China
* These authors contributed equally to this work.
Received 2019-3-31; Accepted 2019-7-29; Published 2019-9-7
Abstract
Background/Aims: Neural crest cells play a vital role in craniofacial development, microRNA-1 (miR-1) is essential in development and disease of the cardiac and skeletal muscle, the objective of our study is to investigate effects of miR-1 on neural crest cell in the craniofacial development and its molecular mechanism.
Methods: We knocked down miR-1 in zebrafish by miR-1 morpholino (MO) microinjection and observed phenotype of neural crest derivatives. We detected neural crest cell migration by time-lapse. Whole-mount in situ hybridization was used to monitor the expressions of genes involved in neural crest cell induction, specification, migration and differentiation. We performed a quantitative proteomics study (iTRAQ) and bioinformatics prediction to identify the targets of miR-1 and validate the relationship between miR-1 and its target gene sec63.
Results: We found defects in the tissues derived from neural crest cells: a severely reduced lower jaw and delayed appearance of pigment cells. miR-1 MO injection also disrupted neural crest cell migration. At 24 hours post fertilization (hpf), reduced expression of tfap2a, dlx2, dlx3b, ngn1 and crestin indicated that miR-1 deficiency affected neural crest cell differentiation. iTRAQ and luciferase reporter assay identified SEC63 as a direct target gene of miR-1. The defects of miR-1 deficiency could be reversed, at least in part, by specific suppression of sec63 expression.
Conclusion: miR-1 is involved in the regulation of neural crest cell development, and that it acts, at least partially, by targeting sec63 expression.
Keywords: Cranial Defect, Neural Crest Cells, microRNA, Sec63, iTRAQ
Introduction
Malformation caused by craniofacial dysplasia is a common birth defect that seriously affects human health. Most hard and soft craniofacial tissues are derived from the neural crest cells. The neural crest cells, a multipotent cell population, are induced at the neural plate border during neurulation [1]. After epithelial-mesenchymal transition, neural crest cells migrate along a specific route. Depending on extrinsic cues, they differentiate into various cell types and form structures and organs such as the nervous system, neuroendocrine cells, smooth muscle cells of the cardiac and pigment cells [2]. In the head, neural crest cells migrate from hindbrain rhombomeric regions to the pharyngeal arches where they form the facial cartilage, bone and connective tissue. Three distinguished major streams can be characterized: the mandibular, hyoid, and branchial streams [3]. Any effect on neural crest cells during development can lead to craniofacial defects [4-6].
microRNAs (miRNAs) are small, single-stranded noncoding RNAs of 18-25 nucleotides. They regulate gene expression at the posttranscriptional level by base-pairing to partially complementary sites in the 3ʹ-untranslated region (3ʹUTR) of their target genes [7, 8]. miRNAs may regulate up to 30% of genes, and genes regulated by miRNAs play an essential role in embryonic development [9]. In zebrafish, miR-1 is encoded by dre-miR-1-1 and dre-miR-1-2. The mature sequences of the resulting miRNAs are identical, i.e., 5ʹ-uggaauguaaagaaguauguau-3ʹ. In humans, miR-1 is encoded by hsa-miR-1-1 and hsa-miR-1-2, while their mature sequences are also the same, i.e., 5ʹ-uggaauguaaagaaguauguau-3ʹ. The sequences of mature miR-1 from different species, including zebrafish and humans, are identical. This degree of conservation suggests that they have similar functions in different vertebrates. miR-1 has been reported to show a tissue-specific expression pattern in zebrafish muscle, which is similar to that found in mammals [10].
The muscle-specific expression pattern of miR-1 is essential in cardiac and skeletal muscle development and disease. During cardiac development, miR-1 has been reported to control the balance between cell proliferation and differentiation [11]. In addition, miR-1 promotes myogenic differentiation and is involved in cell cycle regulation and migration processes. miR-1 also plays an essential role in Chinese sika deer-derived chondrocytes by directly targeting the 3ʹUTR of insulin-like growth factor 1 and by inhibiting the proliferation of these cartilage cells [12]. It has also been reported that miR-1 can inhibit cell proliferation and promote cell differentiation and apoptosis [13,14]. miR-1 and miR-133 form clusters on the same chromosomal loci of mouse chromosome 2 and chromosome 18. miR-1 promotes myogenesis by targeting histone deacetylase 4 [15]. Therefore, miR-1 plays a crucial regulatory role in embryonic development and disease. However, it is likely that many more functions of miR-1 are yet to be identified, and its function during neural crest development and pharyngeal cartilage formation remains unknown.
Based on the known role of miR-1 in embryonic development of vertebrates and the high degree of conservation of miR-1 during evolution, we hypothesized that miR-1 might be involved in embryonic development of neural crest-derived tissues in zebrafish through direct targeting of transcripts of proteins responsible for the differentiation of neural crest-derived tissues.
Materials and Methods
Animal maintenance
Wild-type zebrafish and the transgenic reporter strain Tg (sox10: EGFP) were raised on a 14 h/10 h light/dark cycle at 28.5 °C in the zebrafish facility of the Model Animal Research Center, Nanjing University. All work was performed with the approval of the Ethics Committee of the Stomatological School of Nanjing Medical University. All procedures were carried out according to the guidelines of the Animal Care Committee of Nanjing Medical University.
Cell culture
Zebrafish neural crest cells were extracted and screened as described previously [16]. Embryos (sox10: EGFP) at 11 hpf, 24 hpf, and 72 hpf were anesthetized with tricaine and then decapitated in phosphate-buffered saline (PBS). Embryos were enzymatically dissociated with trypsin-EDTA in PBS. Cells were spun down, washed, resuspended in Dulbecco's modified Eagle's medium (DMEM)-F12 (HyClone, Logan UT USA) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin, and then screened by flow cytometry.
Microinjection of MOs
The following MO antisense oligonucleotides were purchased from Gene Tools LLC (Philomath, OR, USA): miR-1 MO (5ʹ-ATACATACTTCTTTACATTCCA-3ʹ) [17], sec63 MO (5ʹ-CGTACTGAAACTGCTGTCCGGCCAT-3ʹ) [18], and negative control MO (5ʹ-CCTCTTACCTCAGTTACAATTTATA-3ʹ) [19]. One to two-cell embryos were injected with 1 nL of MO.
Whole-mount in situ hybridization
Probes including crestin, foxd3, dlx2, dlx3b, msxb, ngn1, tfap2a, snai1b and sec63 were synthesized using a DIG RNA labeling kit (Roche, Indianapolis IN USA) [20-25]. The primers listed below were designed by Primer 5.0 software. The following primers were used (forward/reverse): crestin (5ʹ-TGCCCTGGAGACGAAACA-3ʹ/5ʹ-CCCACTTCCGATCTGCTT-3ʹ);
foxd3 (5ʹ-CAAAGCATGTGTCATCTTG-3ʹ/5ʹ-TGAGAATGTCCGGCTGAT-3ʹ);
dlx2 (5ʹ-GCCAAAGAAAGTCCG-3ʹ/5ʹ-TGGCTGAAGGTGGG-3ʹ);
dlx3b (5ʹ-AGCGTATCCCACCAAGAC-3ʹ/5ʹ-ATGCGTTCAAACAGTCCA-3ʹ);
msxb (5ʹ-AAGAAGACTTACCTCCCG-3ʹ/5ʹ-TAAATAGTCCTGGCATCG-3ʹ);
ngn1 (5ʹ-CTCACAACTACATCTGGGCACT-3ʹ/5ʹ-GAGGGTTTCTTCGGGTCA-3ʹ);
tfap2a (5ʹ-GGTCACGGCATTGATACTGG-3ʹ/5ʹ-TCGCCTTGGCTGGAAACT-3ʹ);
snai1b (5ʹ-GATGCCACGCTCATTTCTT-3ʹ/5ʹ-GACCCGCACTGGTACTTCTT-3ʹ) and sec63 (5ʹ-GTACGACGACAGTGGCAACA-3ʹ/5ʹ-TATCGGAGGTGCTCCTCTTC-3ʹ). Whole-mount in situ hybridization was carried out on 4% paraformaldehyde (PFA) fixed zebrafish embryos. The prehybridization and hybridization were performed at 65°C for all riboprobes. In situ hybridization signals were examined with sheep anti-digoxigenin-AP Fab fragments. The color reaction was carried out by chromogen substrates (NBT and BCIP).
Quantitative polymerase chain reaction (qPCR)
For qPCR, total RNA was isolated from embryos using an RNA extraction kit (Takara Biotechnology, Dalian, China). cDNA was reverse-transcribed using a TAKARA PrimeScript RT reagent kit. The primer for miR-1 was designed according to Wu et al. (2016), and snRNA U6 was used as a standard for normalization. qPCR was performed according to the manufacturer's instructions, and fold change was calculated using the 2-△CT method as described previously [26]. The following primers were used (forward/reverse): U6 (5ʹ- AAAACAGCAATATGGAGCGC-3ʹ);
dre-miR-1 (5ʹ-CGCGTGGAATGTAAAGAAGTATGTA-3ʹ);
GAPDH (5ʹ-ACCACAGTCCATGCCATCAC-3ʹ/5ʹ-TCCACCACCCTGTTGCTGTA-3ʹ; and
sec63 (5ʹ-CGAGTTCACATCCCACAG-3ʹ/5ʹ-GACAGCACCATCTTCTTCC-3ʹ).
Alcian blue staining
Zebrafish embryos at 4 days post fertilization (dpf) were fixed in 4% paraformaldehyde in PBS before staining with Alcian blue (Sigma Chemical Co, St. Louis, MO, USA). Embryos were transferred to 30:70 glycerol/1% potassium hydroxide and then to 60:40 glycerol/1% potassium hydroxide before incubation for 2-3 days until they were sufficiently translucent. The key angle and length were measured as described previously [27, 28].
Time-lapse imaging
Tg (sox10: EGFP) embryos were anaesthetized and embedded in 0.8% low melting point agarose (Takara Biotechnology, Dalian, China) [29]. Confocal stack pictures of the pharyngeal arch region were taken at the indicated time points using a META Zeiss 810 confocal microscope.
Isobaric tags for relative and absolute quantitation (iTRAQ)
Each sample for iTRAQ was composed of heads of at least 100 embryos. Embryos were anaesthetized at 24 hpf in 1× PBS on ice and stripped for heads. Total proteins were extracted from the heads of embryos. For iTRAQ labeling, approximately 100 μg of proteins were reduced and alkylated by 10 Mm dithiothreitol and 55 Mm iodoacetamide. And then, each sample was digested and labeled with iTRAQ reagents. The analysis was performed by the State Key Laboratory of Reproductive Medicine at Nanjing Medical University [30].
Western blot
50 embryos were transferred to sterilized centrifuge tubes at 24 hpf. The total proteins were extracted by the ultrasonic fragmentation method. Western blot analysis was conducted according to the previous publication [31]. Primary antibody recognizing Sec63 was purchased from Santa Cruz.
Luciferase reporter assay
The 3ʹUTR of sec63, which contains miR-1 binding site, was amplified by PCR with the following primers: F: 5ʹ-CACAACTCGAGCTGTCCTGCTGTTCCACAAAT-3ʹ and R: 5ʹ-TGAGGATCCTTGAGCCCTATTGCCATCCAC-3ʹ. The 3ʹUTR of pax7 was amplified with the following primers: F: 5ʹ-ATCGCTCGAGGAACCCGAGGTTTGTACG-3ʹ and R: 5ʹ-CACAACACAAGCGGCCGCATTCAATTAATTCTGTCTTCA-3ʹ. The partial 3ʹUTR fragment was cloned into the pMIR-REPORT Luciferase vector (Promega, Madison, WI, USA). For use as a control, the binding site was mutated and then cloned into the same vector.
HEK293T cells (5 × 104 cells per well) were seeded in 24-well plates and cultured in DMEM/F12 containing 10% FBS (Gibco, Grand Island, NY, USA) overnight. After 24 h, dre-miR-1 mimics or negative control and wild-type or mutated reporter vector were transfected into HEK293T cells (duplex as a control). After 48 h, the cell cultures were split and the luciferase activity was determined according to the manufacturer's protocol (Promega, Madison, WI, USA). The luciferase activity of each type of sample was measured at least three times [32].
Statistical analysis
Data were presented as the mean ± standard deviation. Experiments were conducted separately at least three times. Student's t-test (SPSS 19.0) was used to assess differences between groups. P value < 0.05 was considered a significant difference.
Results
miR-1 is expressed in zebrafish embryos
miR-1 is expressed in the pharyngeal region in zebrafish at 24 hpf [10]. To investigate whether miR-1 is expressed in zebrafish neural crest cells, sox10-positive (i.e., EGFP-expressing) cells were extracted and isolated by flow cytometry at 11 hpf, 24 hpf, 48 hpf, and 72 hpf from Tg (sox10: EGFP) embryos. qPCR analysis showed that miR-1 was expressed at 12 and 24hpf, and the highest level of expression was observed at 24 hpf, suggesting that miR-1 may play an important role in cranial neural crest cell migration and differentiation (Fig. S1).
Knockdown of miR-1 affects neural crest derivatives
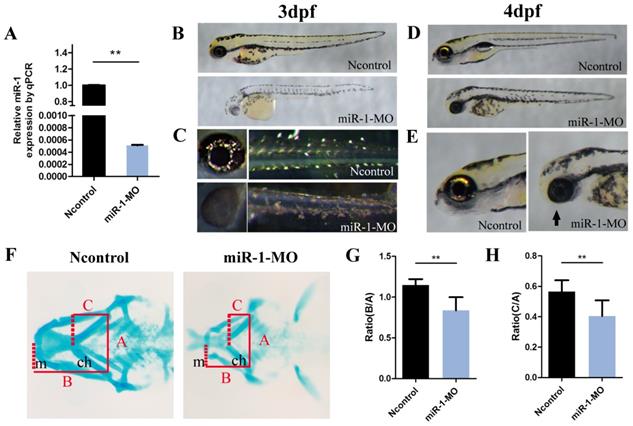
Then we used an antisense MO oligonucleotide (miR-1 MO) to inactivate mature miR-1 (Fig. 1A). First, we observed morphological defects induced by miR-1 MO. At 3 dpf, there were fewer melanophores in the yolk sac and in the body of miR-1 morphants than in the negative control MO group (Fig. 1B). There were also fewer iridophores in the eyes and yolk sac extensions in the miR-1 morphants (Fig. 1C). These results indicate that normal expression of miR-1 is required for the development of neural crest-derived chromatophores.
We also found that the mandibular length was reduced and the mouth opening was ventrally rotated (Fig. 1D, E) in the miR-1 MO-injected group. Next, we analyzed the craniofacial morphology at 4 dpf by Alcian blue staining. In miR-1 morphants, the first pair of the cartilaginous pharyngeal arches (mandibular arches) was smaller than in negative controls, and the 3rd to 5th pairs were barely detectable. Simultaneously, the ceratohyal cartilage (second pharyngeal arch) was shifted posteriorly (Fig. F). We measured the positional relationships among joints of the palatoquadrate and hyosymplectic cartilage (Meckel's cartilage) as well as the ceratohyal cartilage by analyzing three lines (Fig. 1F), which have been defined previously [27]. The statistical differences of the ratios (B:A and C:A) between the miR-1 MO-injected group and the negative control MO-injected groups are shown in Fig. 1G and H. These results indicate that knockdown of miR-1 expression affected craniofacial chondrogenesis. Therefore, we hypothesized that miR-1 might modulate the induction, migration or differentiation of cranial neural crest cells.
miR-1 deficiency does not affect neural crest induction and specification, but affects neural crest cell migration and differentiation
In order to determine the stage of neural crest development affected by miR-1 functions, we examined the expression of genes associated with specific stages of neural crest development in miR-1 morphants. A number of genes, including msxb, pax3, dlx3b, snail1b, foxd3, dlx2, ngn1, crestin and tfap2a have been shown to play important roles in the induction, specification, migration, and differentiation of neural crest cells [33,34].
Knockdown of miR-1 affects neural crest derivatives. (A) qPCR showed that miR-1 MO knocked down miR-1 efficiently (the experiments were repeated three times). Data are expressed as the mean ± SD, from at least three independent experiments, n=15, **P < 0.01. (B, C) Lateral views of live zebrafish embryos showed pigment cells at 3 dpf. Negative control-injected zebrafish exhibited neural crest-derived pigment cells: black melanophores and iridescent iridophores. The distribution and number of melanophores were reduced in miR-1 morphants. The number of iridophores was reduced in the eye and trunk. (D, E) Lateral views of embryos at 4 dpf. miR-1 morphants displayed a shorter body length with a smaller head and edema around the heart. The lower jaw was reduced in size in the miR-1 morphants (arrows) compared with the negative control embryos. (F) Alcian blue staining showed cartilage development at 4 dpf; ventral views of embryos. (G, H) Ratios B: A and C: A, as shown in (F), in the miR-1 MO-injected and negative MO-injected groups. The definition for lines A, B, and C has been described previously. microRNA-1, miR-1; morpholino, MO; dpf, days post fertilization; mc, Meckel's cartilage; pq, palatoquadrate; ch, ceratohyal cartilage; Data are expressed as the mean ± SD, from at least three independent experiments, n=15, **P < 0.01; N.S., not significant.
Expression of selected genes in neural crest induction and specification in miR-1 MO-injected embryos at 11 hpf. The photographs show a dorsal view with the anterior to the top at 11 hpf. Expression of foxd3 (A), msxb (B), pax3 (C), dlx3b (D), and snai1b (E) in embryos is shown. The expression of foxd3, msxb, pax3, dlx3b and snai1b was similar in negative control embryos and miR-1 morphants. microRNA-1, miR-1; morpholino, MO.
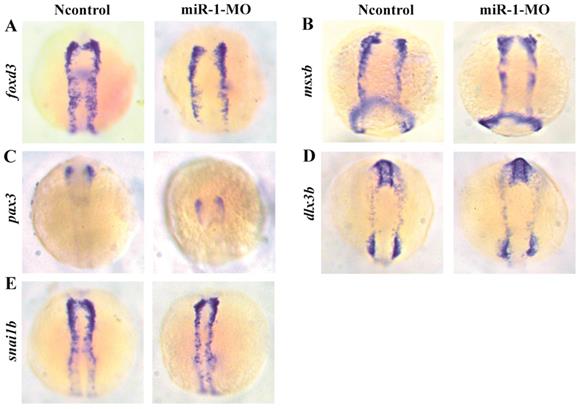
We found that the neural crest specification marker foxd3 displayed normal expression in miR-1 morphants at 11 hpf (Fig. 2A), approximately at the 3-somite stage (ss) before neural crest cell migration. Expression of the markers msxb, pax3, dlx3b, and snai1b in the miR-1 morphants was also normal (Fig. 2B, C, D, E). The normal expression indicated that miR-1 deficiency did not affect the induction and specification of neural crest cells.
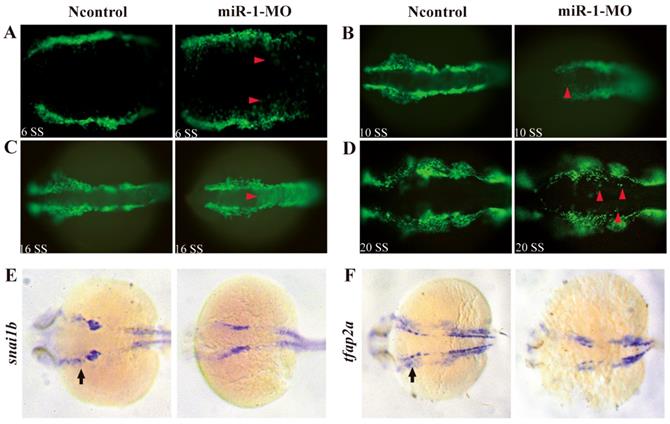
Next, we analyzed whether neural crest cell migration was altered in miR-1 morphants. miR-1 MO was injected into Tg (sox10: EGFP) embryos. Then, we mapped sox10 expression during neural crest cell migration at 6 ss, 10 ss, 16 ss and 20 ss. At 6 ss, 10 ss, 16ss and 20 ss, in miR-1 morphants, neural crest cells displayed ectopic EGFP expression in the dorsal midline, while few neural crest cells appeared in the midline in negative control embryos (Fig. 3A, B, C, D). The expression of neural crest migration marker snai1b and tfap2a was also reduced in neural crest streams in the head (Fig. 3E, F). These data implicated miR-1 regulated neural crest cell migration.
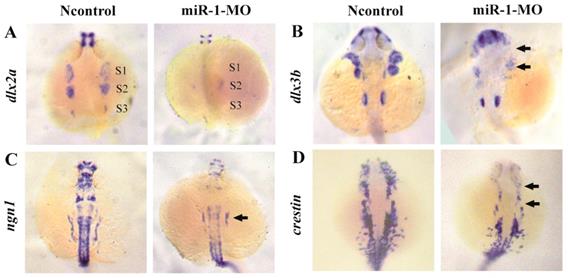
To determine whether neural crest cell differentiation was altered, we examined the expression of associated genes. tfap2a controls the differentiation of cranial neural crest cells into mature chondrocytes. At 24 hpf, tfap2a has been reported to be expressed in a highly restricted pattern in the anterior domains [35, 36]. We found that, compared with negative control embryos, tfap2a gene expression was reduced in miR-1 MO embryos, as can be most notably seen in the pharyngeal arches (Fig. 3F). At 24 hpf in miR-1 MO morphant embryos, the expression of dlx2a, a marker for the chondrogenic crest, was clearly reduced in the anterior domain of stream 1 and 2, which would give rise to the mandibular and hyoid arches, and dlx2a expression nearly disappeared in the more posterior domain of stream 3, which would migrate into the mandibular arches, the hyoid arches, and the posterior pharyngeal arches (Fig. 4A). Similarly, expression of dlx3b in the first and second arches was extremely reduced in the miR-1 MO embryos (Fig. 4B). The gene ngn1 is a critical marker for dorsal root ganglia development in zebrafish [37, 38]. Loss of miR-1 resulted in notable reduction of ngn1 expression (Fig. 4C). At 24 hpf in negative control embryos, three distinct streams of migrating cranial neural crest cells can be detected by the expression of crestin in hindbrain rhombomeres. In miR-1 MO-injected embryos, the expression of crestin was nearly absent in the anterior streams and was reduced in the posterior neural crest stream (Fig. 4D). These data implicated miR-1 regulated neural crest cell differentiation.
miR-1 directly regulates sec63 expression
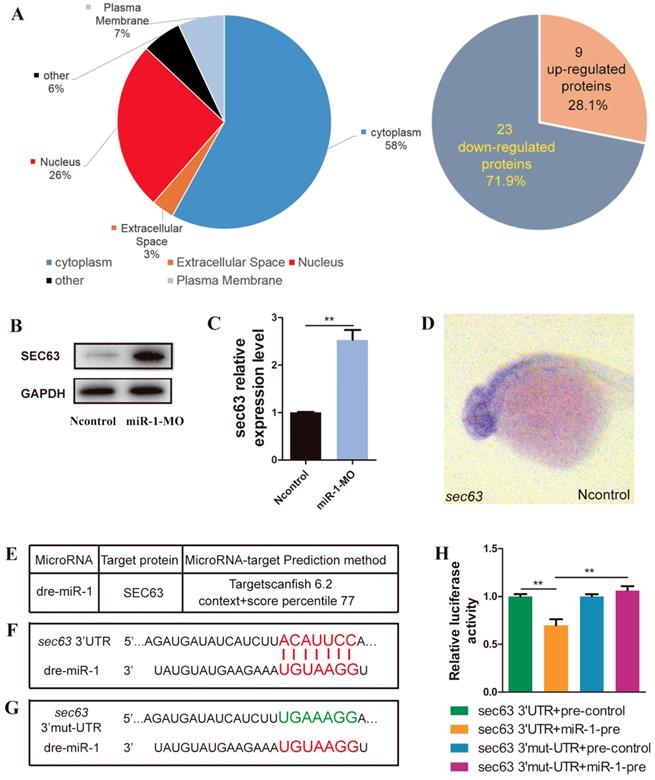
We analyzed the differential expression of proteins between miR-1 morphants and negative control embryos by iTRAQ analysis. Compared with negative control embryos, 32 proteins were significantly altered after knockdown of miR-1, including upregulation of 9 proteins and downregulation of 23 proteins (Fig. 5A, Fig. S2). Our results revealed that SEC63 was among the upregulated proteins after miR-1 knockdown, and we chose SEC63 for further analysis because of the reasons below. We found a significant difference in the level of sec63 Mrna and protein between the miR-1 morphants and negative control embryos at 24 hpf (Fig. 5B, C). We established that the gene sec63 was expressed in the very domain where the neural crest cells are found (Fig. 5D). Moreover, using the TargetScan search program (Release 6.2), miR-1 was found to be complementary to a RNA sequence from the sec63 3ʹUTR. We used a luciferase assay to test the functional interaction between miR-1 and the sec63 3ʹ-UTR in vitro (Fig. 5E, F, G). The results showed that the miR-1 precursor significantly repressed the expression of luciferase-sec63-3ʹUTR but not luciferase-sec63 3ʹ-mut-UTR (Fig. 5H). Together, these data indicate that miR-1 directly targeted the 3ʹUTR of sec63, thereby modulating the level of sec63 mRNA and protein.
miR-1 morphants displayed defective migration of neural crest cells. Live embryo imaging in miR-1 morphants and negative control Tg (sox10: EGFP) embryos was used to analyze the movement of neural crest cells, viewed dorsally. (A-D) Representative images are shown from the time-lapse photography from 6 segment stage (ss) to 20 ss. At 6ss, 10 ss, 16 ss and 20 ss, neural crest cells in miR-1 morphants displayed ectopic EGFP expression (red arrowheads) in the dorsal midline (A, B, C and D). At 24 hpf, expression of snai1b (E) and tfap2a (F) in negative control and miR-1 morphants is shown. At this time, increasing numbers of the migrating lateral line primordium (arrow) expressed tfap2a and snail1b. The expression of neural crest migration markers (snai1b, tfap2a) was reduced (arrows) (E, F). microRNA-1, miR-1; morpholino, MO; ss, segment stage.
miR-1 MO-injected embryos displayed defective neural crest cells differentiation. Embryos were viewed dorsally, anterior to the top, at 24 hpf. Expression of dlx2a (A), dlx3b (B), ngn1 (C), and crestin (D) is shown. Expression of the pharyngeal homing marker gene dlx2a was reduced in the neural crest stream (S1, S2, S3) in miR-1 MO-injected embryos (A). General features of dorso-ventral patterning appeared affected as shown (arrows) by decreased expression of dlx3b (B). ngn1, a marker of neuron differentiation, was downregulated in miR-1 morphants (arrows) (C). The expression of neural crest marker crestin was reduced in the cranial region and the branchial primordia (arrows) (D). microRNA-1, miR-1; morpholino, MO. Taken together, our data suggest that miR-1 loss of function interfered with the development of normal neural crest derivatives through disruption of neural crest cell migration and differentiation.
miR-1 directly targeted sec63. (A) Subcellular localization of potentially targeted proteins differentially expressed in miR-1 morphants and negative control embryos identified by gene ontology annotations. 9 proteins were upregulated and 23 proteins were downregulated in miR-1 morphants. (B) Protein expression of Sec63 in the control and miR-1 MO-injected groups. (C) A candidate protein for upregulation, sec63, was determined by iTRAQ analysis. Validation of increased sec63 expression in miR-1 morphants by qPCR in vivo at 24 hpf. Data are expressed as the mean ± SD, from at least three independent experiments, n=15, **P < 0.01. (D) Expression of sec63 in embryos at 24 hpf. Embryos were viewed laterally, with the anterior to the left. sec63-3ʹUTR binding site of miR-1 was predicted using TargetScan (E, F). (G) Mutant 3ʹUTR of sec63 in dual luciferase reporter plasmids. Overexpression of miR-1 (miR-1-pre) reduced sec63-3ʹUTR luciferase activity in vitro, but not the luciferase activity of mutated sec63-3ʹUTR. Three independent experiments were performed, and each experiment was carried out in duplicate (H).
Reducing sec63 partly restored the neural crest derivative defects in miR-1 morphants
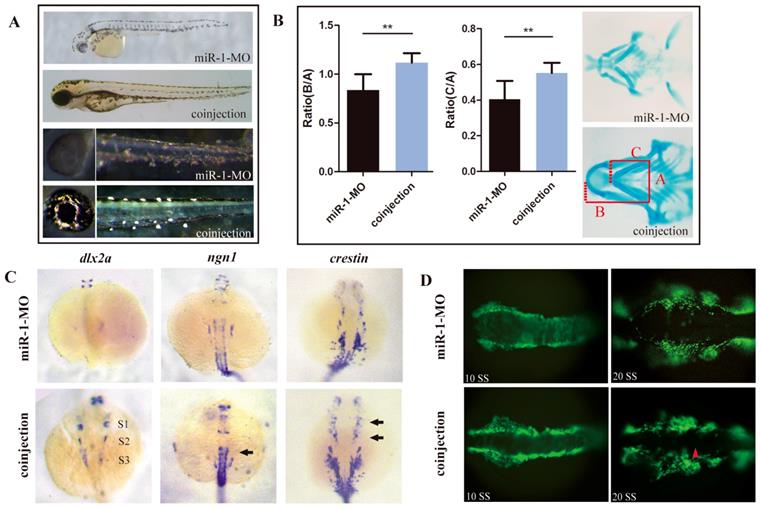
We hypothesized that upregulation of sec63 was one of the likely causes for the defects of neural crest derivatives in miR-1-deficient embryos. To test this possibility, we knocked down sec63 using a MO oligonucleotide in the miR-1 morphants. Coinjection of miR-1 and sec63 MOs greatly reversed the delayed appearance of pigment cells (Fig. 6A). Alcian blue staining demonstrated that the defects in craniofacial cartilage were also corrected in coinjected zebrafish at 4 dpf (Fig. 6B). The reduced expression of dlx2, ngn1 and crestin was partly corrected at 24 hpf (Fig. 6C). Compared with miR-1 morphants, the number of neural crest cells showing ectopic expression of EGFP during the migration was reduced (Fig. 6D). We concluded that miR-1 regulates neural crest cell migration and derivation in zebrafish by targeting sec63.
Reducing sec63 partly restored the neural crest derivative defects in miR-1 morphants. (A) The defects in melanophores and iridophores were reversed in embryos co-injected with both miR-1 MO and sec63 MO. (B) The craniofacial cartilage were corrected in embryos co-injected with miR-1-MO and sec63-MO. Data are expressed as the mean ± SD, from at least three independent experiments, n=15, **P < 0.01. (C) The expression of dlx2a, ngn1 and crestin were partly rescued at 24 hpf in embryos co-injected with both miR-1 MO and sec63 MO (S1, S2, S3, arrows). (D) The number of ectopic neural crest cells was reduced. microRNA-1, miR-1; morpholino, MO.
Discussion
In this study, we discovered a novel role for miR-1 in neural crest cell s, pharyngeal cartilage, and lower jaw development in zebrafish embryos. Inactivation of miR-1 by antisense MO oligonucleotides led to marked phenotypic differences between miR-1 morphants and negative controls. The majority of the defects in the miR-1 morphants were in cells and tissues derived from the neural crest, including the heart, pigment cells, and craniofacial cartilage, confirming the essential role for miR-1 in neural crest cell development. miR-1 inactivation did not affect neural crest cell induction and specification because there was no major reduction in the number of premigratory neural crest cells at 11 hpf. Neural crest cells displayed ectopic localization in miR-1 morphants at 10 ss and 20 ss, representing early migration and late migration, respectively. At both 10 ss and 16 ss, the movement of cranial neural crest cells was delayed in the miR-1 morphants. Based on our findings, we conclude that the primary role of miR-1 in neural crest cells development is to regulate neural crest cell directional migration and differentiation. In addition, it is possible that miR-1 plays a role in neural crest cell apoptosis and proliferation, but this will require further study.
Several direct targets of miR-1 have been determined in vivo, including Hand2 and the Notch ligand delta in Drosophila. In addition, miR-1 regulates EDN1 in hepatocellular carcinoma progression [11,14,39]. Sera [40] predicted that pax7 might be the direct of miR-1. We carried out luciferase assay to test the functional interaction between miR-1 and the pax7 3ʹ-UTR in vitro (Fig. S3A). The results revealed that the miR-1 precursor significantly repressed the expression of luciferase-pax7-3ʹUTR but not luciferase-pax7 3ʹ-mut-UTR (Fig. S3B). However, coinjection of pax7 and miR-1 MOs could not reverse the delayed appearance of pigment cells and the defects in craniofacial cartilage. In order to gain insight into the molecular mechanism of the regulatory effect of miR-1 in neural crest development, we searched different databases for matched sequences between miR-1 and potential target genes in iTRAQ results. After confirmation of function by luciferase assays, we determined zebrafish sec63 as a direct target of miR-1.
SEC63 is a member of the endoplasmic reticulum (ER) translocon [41]. sec63 encodes an integral membrane protein of the ER that is highly conserved from yeast to humans. SEC63 is required in both post-translational and co-translational pathways [42]. Mutations in sec63 cause polycystic liver disease in humans, and even zebrafish sec63 mutants develop liver pathology. In zebrafish embryos, sec63 is expressed at all stages examined and in all tissues, and it is broadly expressed in liver and pancreas. Disruption of sec63 in zebrafish leads to abnormalities in myelinating glia in both the central and peripheral nervous system [43]. The function of sec63 during neural crest development and craniofacial cartilage formation has not been studied yet.
SEC63 is part of post-translational quality control and processing of polycystin-1 (PC1) and polycystin-2 (PC2) [44]. The polycystin complex has been implicated in MAPK/ERK and cAMP pathways [45]. Function of the PCs has been found in different cellular and tissue, is their role in regulation of cell migration and tissue morphogenesis [46]. When the expression of PCs was changed, it could lead to edema and cardiac defects in embryos [46]. And we also observe the edema and cardiac defects in miR-1 MO groups. Then we supposed that Sec63 was predicted target of miR-1 and responsible for proper neural crest cell migration and differentiation.
To determine whether sec63 expression is regulated by miR-1, we carried out rescue experiments. Embryos co-injected with both miR-1 MO and sec63 MO displayed relatively normal body length, head size, heart, and the appearance of pigment cells was also normal. Significantly, the lower jaw extended more anteriorly compared with miR-1 morphant embryos. And the Meckel's cartilage could extend anteriorly beyond (but close to) the eyes. Whole-mount hybridization analyses demonstrated that the expression was similar to that in negative control embryos at 24 hpf. Migration of neural crest cells in miR-1 MO and sec63 MO morphants was relatively normal. This partial effect of sec63 MO in miR-1 MO morphants confirms that sec63 is a target gene of miR-1. These results confirmed the effect of miR-1 on sec63 that was observed in the iTRAQ experiments. In addition, luciferase reporter assays confirmed that sec63 is a direct target of miR-1.
Our findings provide the first evidence that miR-1 regulates sec63 by directly targeting the 3ʹUTR of sec63. Taken together, our study suggests a molecular mechanism in which miR-1 participates in the control of sec63 expression and regulates neural crest migration and formation of Meckel's cartilage during embryonic development in zebrafish. These findings will hopefully help us to better understand craniofacial development and promote effective therapy and treatment for craniofacial malformations.
Supplementary Material
Supplementary figures.
Acknowledgements
Author contributions
Jun-Qing Ma, Jing-Jing Ben, contributed to conception, design and critically revised the manuscript; Dong-Yue Wang, Ya-Juan Weng, contributed to conception, data acquisition, analysis, drafted and critically revised the manuscript; Shu-yu Guo, contributed to data analysis and critically revised the manuscript; Jie-li Ni, Lei Yu, Yang Zhang, contributed to data acquisition and analysis; Qing-Shun Zhao, contributed to conception and design. All authors gave final approval and agree to be accountable for all aspects of the work.
Sources of Funding
This work was supported by the National Natural Science Foundation of China (81371179 and 81771029), the Natural Science Foundation of Jiangsu Province (BK20150048), and Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (2014-037).
Transparency document
The Transparency document associated with this article can be found, in online version.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Rogers CD, Jayasena CS, Nie S, Bronner ME. Neural crest specification: tissues, signals, and transcription factors. Wiley Interdiscip Rev Dev Biol. 2012;1:52-68
2. Dorsky RI, Moon RT, Raible DW. Environmental signals and cell fate specification in premigratory neural crest. Bioessays. 2000;22:708-716
3. Kimmel CB, Miller CT, Keynes RJ. Neural crest patterning and the evolution of the jaw. J Anat. 2001;1:105-120
4. Mayor R, Theveneau E. The neural crest. Development. 2013;140:2247-2251
5. Steventon B, Carmona-Fontaine C, Mayor R. Genetic network during neural crest induction: From cell specification to cell survival. Semin Cell Dev Biol. 2005;16:647-654
6. Santagati F, Rijli FM. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci. 2003;4:806-818
7. Tian S, Tian J, Chen X, Li L, Liu Y, Wang Y, Sun Y, Zhang C, Lou G. Identification of Subpathway Signatures For Ovarian Cancer Prognosis by Integrated Analyses of High-Throughput miRNA and mRNA Expression. Cell Physiol Biochem. 2017;44:1325-1336
8. Worku T, Bhattarai D, Ayers D, Wang K, Wang C, Rehman ZU, Talpur HS, Yang L. Long Non-Coding RNAs: the New Horizon of Gene Regulation in Ovarian Cancer. Cell Physiol Biochem. 2017;44:948-966
9. Ying SY, Lin SL. Current perspectives in intronic micro RNAs (miRNAs). J Biomed Sci. 2006;13:5-15
10. Wienholds E, Kloosterman WP, Miska E, Alvarez-Aaavedra E, Berezikov E, de bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310-311
11. Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214-220
12. Hu W, Meng X, Lu T, Wu L, Li T, Li M, Tian Y. MicroRNA-1 inhibits the proliferation of Chinese sika deer-derived cartilage cells by binding to the 3ʹ-untranslated region of IGF-1. Mol Med Rep. 2013;8:523-528
13. Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394-33405
14. Lu JW, Liao CY, Yang WY, Lin YM, Jin SL, Wang HD, Yuh CH. Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PLoS One. 2014;9:e85318
15. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228-233
16. Ramachandran KV, Hennessey JA, Barnett AS, Yin X, Stadt HA, Foster E, Shah RA, Yazawa M, Dolmetsch RE, Kirby ML. et al. Calcium influx through L-type CaV1.2 Ca2+ channels regulates mandibular development. J Clin Invest. 2013;123:1638-1646
17. Mishima Y, Abreu-Goodger C, Staton AA, Stahihut C, Shou C, Cheng C, Gerstein M, Enright AJ, Giraldez AJ. Zebrafish miR-1 and miR-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23:619-632
18. Tietz Bogert PS, Huang BQ, Gradilone SA, Masyuk TV, Moulder GL, Ekker SC, Larusso NF. The zebrafish as a model to study polycystic liver disease. Zebrafish. 2013;10:211-217
19. Sundaramoorthi H, Khandekar G, Kim S, Jagadeeswaran P. Knockdown of αIIb by RNA degradation by delivering deoxyoligonucleotides piggybacked with control vivo-MOs into zebrafish thrombocytes. Blood Cells Mol Dis. 2015;54:78-83
20. Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J Neurosci. 1994;14:3475-3486
21. Angelo S, Lohr J, Lee KH, Ticho BS, Breitbart RE, Hill S, Yost HJ, Srivastava D. Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech Dev. 2000;95:231-237
22. Dutton KA, Pauliny A, Lopes SS, Elworthy S, Carney TJ, Rauch J, Geisler R, Haffter P, Kelsh RN. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development. 2001;128:4113-4125
23. Luo R, An M, Arduini BL, Henion PD. Specific pan-neural crest expression of zebrafish Crestin throughout embryonic development. Dev Dyn. 2001;220:169-174
24. Odenthal J, Nüsslein-Volhard C. Fork head domain genes in zebrafish. Dev Genes Evol. 1998;208:245-258
25. Stewart RA, Arduini BL, Berghmans S, George RE, Kanki JP, Henion PD, Look AT. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev Biol. 2006;292:174-188
26. Wu M, Wu D, Wang C, Guo Z, Li B, Zuo Z. Hexabromocyclododecane exposure induces cardiac hypertrophy and arrhythmia by inhibiting miR-1 expression via up-regulation of the homeobox gene Nkx2.5. J Hazard Mater. 2016;302:304-313
27. Goudevenou K, Martin P, Yeh YJ, Jones P, Sablitzky F. Def6 is required for convergent extension movements during zebrafish gastrulation downstream of Wnt5b signaling. PLoS One. 2011;6:e26548
28. Rooryck C, Diaz-Font A, Osborn DP, Chabchoub E, Hernandez-Hernandez V, Shamseldin H, Kenny J, Waters A, Jenkins D, Kaissi AA. et al. Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nat Genet. 2011;43:197-203
29. Ning G, Liu X, Dai M, Meng A, Wang Q. MicroRNA-92a upholds Bmp signaling by targeting noggin3 during pharyngeal cartilage formation. Dev Cell. 2013;24:283-295
30. Gao W, Xu J, Wang F, Zhang L, Peng R, Shu Y, Wu J, Tang Q, Zhu Y. Plasma membrane proteomic analysis of human Gastric Cancer tissues: revealing flotillin 1 as a marker for Gastric Cancer. BMC Cancer. 2015;15:367
31. Yu S, Li C, Biswas L, Hu X, Liu F, Reilly J, Liu X, Liu Y, Huang Y, Lu Z. et al. CERKL gene knockout disturbs photoreceptor outer segment phagocytosis and causes rod-cone dystrophy in zebrafish. Hum Mol Genet. 2017;26:2335-2345
32. Zarelli VE, Dawid IB. Inhibition of neural crest formation by Kctd15 involves regulation of transcription factor AP-2. Proc Natl Acad Sci U S A. 2013;110:2870-2875
33. Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291-299
34. Gammill LS, Bronner-Fraser M. Neural crest specification: migrating into genomics. Nat Rev Neurosci. 2003;4:795-805
35. Barrallo-Gimeno A, Holzschuh J, Driever W, Knapik EW. Neural crest survival differentiation in zebrafish depends on mont blanc/tfap2a gene function. Development. 2004;131:1463-1477
36. Knight RD, Nair S, Nelson SS, Afshar A, Javidan Y, Geisler R, Rauch GJ, Schilling TF. Lockjaw encodes a zebrafish tfap2a required for early neural crest development. Development. 2003;130:5755-5768
37. Cornell RA, Eisen JS. Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development. 2002;129:2639-2648
38. Andermann P, Ungos J, Raible DW. Neurogenin1 defines zebrafish cranial sensory ganglia precursors. Dev Biol. 2002;251:45-58
39. Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102:18986-18991
40. Sera SR, Zur Nieden NI. mircroRNA regulation of skeletal development. Curr Osteoporos Rep. 2017;15:353-366
41. Lang S, Benedix J, Fedeles SV, Schorr S, Schirra C, Schauble N, Jalal C, Greiner M, Hassdenteufel S, Tatzelt J. et al. Different effects of Sec61α, Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J Cell Sci. 2012;125:1958-1969
42. Davila S, Furu L, Gharavi AG, Tian X, Onoe T, Qian Q, Li A, Cai Y, Kamath PS, King BF. et al. Mutations in SEC63 cause autosomal dominant polycystic liver disease. Nat Genet. 2004;36:575-577
43. Monk KR, Voas MG, Franzini-Armstrong C, Hakkinen IS, Talbot WS. Mutation of sec63 in zebrafish causes defects in myelinated axons and liver pathology. Dis Model Mech. 2013;6:135-145
44. Wills ES, Roepman R, Drenth JP. Polycystic liver disease: ductal plate malformation and the primary cilium. Trends Mol Med. 2014;20:261-270
45. Ma M, Tian X, Igarashi P, Pazour GJ, Somlo S. Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic disease. Nat Genet. 2013;45:1004-1012
46. Outeda P, Huso DL, Fisher SA, Halushka MK, Kim H, Qian F, Germino GG, Wantnick T. Polycystin signaling is required for directed endothelial cell migration and lymphatic development. Cell Rep. 2015;7:634-644
Author contact
![]() Corresponding authors: Jun-Qing Ma. Address: Jiangsu Key Laboratory of Oral Diseases, Nanjing Medical University, 136 Hanzhong Road, Nanjing, 210029, China. Tel/Fax. +86 025-85031976. Email: jmaedu.cn. Jing-Jing Ben. Address: Department of Pathophysiology, Key laboratory of Cardiovascular Disease and Molecular Intervention, Nanjing Medical University, 136 Hanzhong Road, Nanjing, 210029, China. Tel/Fax. +86 025-86862886 Email: bjjedu.cn
Corresponding authors: Jun-Qing Ma. Address: Jiangsu Key Laboratory of Oral Diseases, Nanjing Medical University, 136 Hanzhong Road, Nanjing, 210029, China. Tel/Fax. +86 025-85031976. Email: jmaedu.cn. Jing-Jing Ben. Address: Department of Pathophysiology, Key laboratory of Cardiovascular Disease and Molecular Intervention, Nanjing Medical University, 136 Hanzhong Road, Nanjing, 210029, China. Tel/Fax. +86 025-86862886 Email: bjjedu.cn

 Global reach, higher impact
Global reach, higher impact