10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(12):2641-2653. doi:10.7150/ijbs.37152 This issue Cite
Research Paper
Evaluation of the effects of sequence length and microsatellite instability on single-guide RNA activity and specificity
1. Key Laboratory of Agricultural Animal Genetics, Breeding and Reproduction of Ministry of Education & Key Lab of Swine Genetics and Breeding of Ministry of Agriculture and Rural Affairs, Huazhong Agricultural University, Wuhan 430070, P. R. China;
2. Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, College of Informatics, Huazhong Agricultural University, Wuhan 430070, P. R. China;
3. The Cooperative Innovation Center for Sustainable Pig Production, Huazhong Agricultural University, Wuhan 430070, P. R. China.
*The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
Abstract
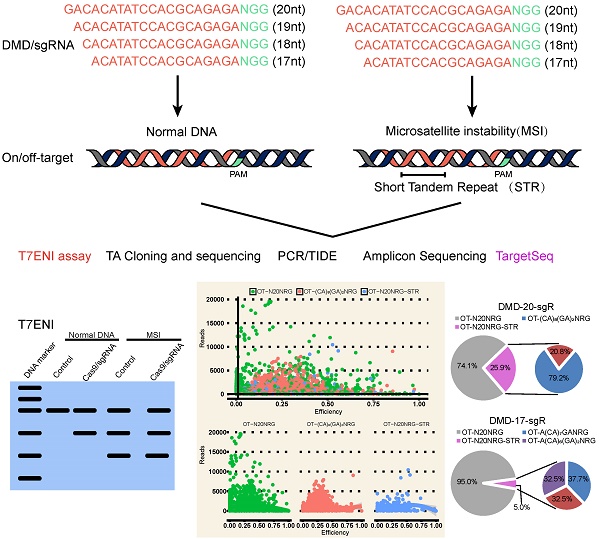
Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology is effective for genome editing and now widely used in life science research. However, the key factors determining its editing efficiency and off-target cleavage activity for single-guide RNA (sgRNA) are poorly documented. Here, we systematically evaluated the effects of sgRNA length on genome editing efficiency and specificity. Results showed that sgRNA 5′-end lengths can alter genome editing activity. Although the number of predicted off-target sites significantly increased after sgRNA length truncation, sgRNAs with different lengths were highly specific. Because only a few predicted off-targets had detectable cleavage activity as determined by Target capture sequencing (TargetSeq). Interestingly, > 20% of the predicted off-targets contained microsatellites for selected sgRNAs targeting the dystrophin gene, which can produce genomic instability and interfere with accurate assessment of off-target cleavage activity. We found that sgRNA activity and specificity can be sensitively detected by TargetSeq in combination with in silico prediction. Checking whether the on- and off-targets contain microsatellites is necessary to improve the accuracy of analyzing the efficiency of genome editing. Our research provides new features and novel strategies for the accurate assessment of CRISPR sgRNA activity and specificity.
Keywords: CRISPR/Cas9, length, microsatellite, activity, specificity

 Global reach, higher impact
Global reach, higher impact