10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2019; 15(13):2897-2910. doi:10.7150/ijbs.30499 This issue Cite
Research Paper
Sphingolipidomic Analysis of C. elegans reveals Development- and Environment-dependent Metabolic Features
1. Faculty of Health Sciences, University of Macau, Taipa, Macau SAR 999078, China
2. Centre of Reproduction, Development and Ageing, University of Macau, Taipa, Macau SAR 999078, China
#These Authors contributed equally to this work.
Abstract
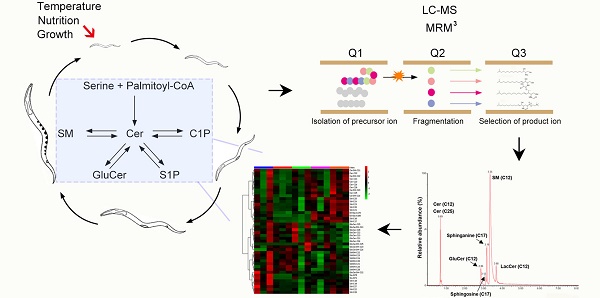
Sphingolipids (SLs) serve as structural and signaling molecules in regulating various cellular events and growth. Given that SLs contain various bioactive species possessing distinct roles, quantitative analysis of sphingolipidome is essential for elucidating their differential requirement during development. Herein we developed a comprehensive sphingolipidomic profiling approach using liquid chromatography-mass spectrometry coupled with multiple reaction monitoring mode (LC-MS-MRM). SL profiling of C. elegans revealed organism-specific, development-dependent and environment-driven metabolic features. We showed for the first time the presence of a series of sphingoid bases in C. elegans sphingolipid profiles, although only C17-sphingoid base is used for generating complex SLs. Moreover, we successfully resolved growth-, temperature- and nutrition-dependent SL profiles at both individual metabolite-level and network-level. Sphingolipidomic analysis uncovered significant SL composition changes throughout development, with SMs/GluCers ratios dramatically increasing from larva to adult stage whereas total sphingolipid levels exhibiting opposing trends. We also identified a temperature-dependent alteration in SMs/GluCers ratios, suggesting an organism-specific strategy for environmental adaptation. Finally, we found serine-biased GluCer increases between serine- versus alanine-supplemented worms. Our study builds a “reference” resource for future SL analysis in the worm, provides insights into natural variability and plasticity of eukaryotic multicellular sphingolipid composition and is highly valuable for investigating their functional significance.
Keywords: Caenorhabditis elegans, sphingolipidomics, development, temperature, nutrition, LC/MS/MRM.

 Global reach, higher impact
Global reach, higher impact