10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2020; 16(11):1929-1940. doi:10.7150/ijbs.45231 This issue Cite
Review
Function and evolution of RNA N6-methyladenosine modification
Department of Pathology, Xuzhou Medical University, Xuzhou 221004, China
*These authors contributed equally to this work.
Abstract
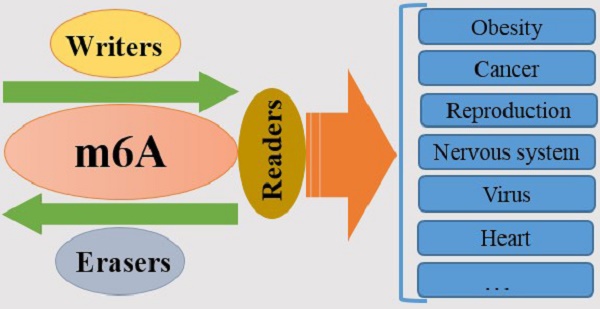
N6-methyladenosine (m6A) is identified as the most prevalent and abundant internal RNA modification, especially within eukaryotic mRNAs, which has attracted much attention in recent years since its importance for regulating gene expression and deciding cell fate. m6A modification is installed by RNA methyltransferases METTL3, METTL14 and WTAP (Writers), removed by the demethylases FTO and ALKBH5 (Erasers) and recognized by m6A binding proteins, such as YT521-B homology YTH domain-containing proteins (Readers). Accumulating evidence shows that m6A RNA methylation participates in almost all aspects of RNA processing, implying an association with important bioprocesses. In this review, we mainly summarize and discuss the functional relevance and importance of m6A modification in cellular processes.
Keywords: m6A modification, methylation, RNA, function

 Global reach, higher impact
Global reach, higher impact