10
Impact Factor
ISSN: 1449-2288
Int J Biol Sci 2021; 17(1):134-150. doi:10.7150/ijbs.50773 This issue Cite
Review
The Emerging Roles of Autophagy-Related MicroRNAs in Cancer
Institute of Translational Medicine, The Affiliated Hospital of Qingdao University, College of Medicine, Qingdao University, Qingdao 266021, China.
Received 2020-7-17; Accepted 2020-10-29; Published 2021-1-1
Abstract
Autophagy is a conserved catabolic process involving the degradation and recycling of damaged biomacromolecules or organelles through lysosomal-dependent pathways and plays a crucial role in maintaining cell homeostasis. Consequently, abnormal autophagy is associated with multiple diseases, such as infectious diseases, neurodegenerative diseases and cancer. Currently, autophagy is considered to be a dual regulator in cancer, functioning as a suppressor in the early stage while supporting the growth and metastasis of cancer cells in the later stage and may also produce therapeutic resistance. MicroRNAs (miRNAs) are small, non-coding RNA molecules that regulate gene expression at the post-transcriptional level by silencing targeted mRNA. MiRNAs have great regulatory potential for several fundamental biological processes, including autophagy. In recent years, an increasing number of studies have linked miRNA dysfunction to the growth, metabolism, migration, metastasis, and responses of cancer cells to therapy. Therefore, the study of autophagy-related miRNAs in cancer will provide insights into cancer biology and lead to the development of novel anti-cancer strategies. In the present review, we summarise the current knowledge of miRNA dysregulation during autophagy in cancer, focusing on the relationship between autophagy and miRNAs, and discuss their involvement in cancer biology and cancer treatment.
Keywords: Autophagy, MicroRNA, Post-translational regulation, Cancer
Introduction
Derived from Greek for 'self' (auto) and 'eating' (phagy), autophagy refers to the pathway by which cells transport cytoplasm to lysosomes for degradation [1]. Currently, there are three known forms of autophagy, including chaperone-mediated autophagy, microautophagy and macroautophagy [2, 3], which differ in their physiological functions and delivery of substances to lysosomes [3]. So-called 'autophagy' refers to macroautophagy, which is characterised by the formation of a double-layered envelope cellular structure under specific conditions that engulfs cytoplasm and organelles and transports them to lysosomes [4].
Autophagy is a conservative intracellular degradation mechanism that is maintained at low levels under normal cell conditions, while under stressful conditions, such as hunger, nutrient deficiencies, hypoxia, etc., autophagy can be rapidly upregulated [5, 6]. Then, cellular components (such as long-lived proteins, protein aggregates, pathogens and damaged organelles) are wrapped and digested into small molecules for cell metabolism and recycling [5]. However, the excessive activation of autophagy leads to cell death, which is defined as autophagic cell death and can be restrained by autophagy inhibitors [7]. Abnormalities in autophagy can cause health problems, including inflammation, neurodegenerative diseases, cardiovascular diseases and cancer [1]. Thus, identifying the molecular mechanisms of autophagic control is of crucial importance.
Non-coding RNAs, including long non-coding RNAs and circular RNAs, have been shown to participate in the regulation of autophagy [8, 9]. Furthermore, the participation of microRNAs (miRNAs) in autophagy has also been investigated [10-13]. MiRNAs are a class of non-protein-encoding RNA molecules that specifically bind to the 3'-untranslated region (3'-UTR) of target mRNAs, causing their degradation or protein translation inhibition to maintain optimal levels of the target protein [14]. Functionally, miRNAs are involved in the regulation of a variety of cellular and molecular events, including cell proliferation, differentiation, metabolism and apoptosis [15]. At present, approximately 2,000 miRNAs have been identified in humans, a number that continues to increase [16]. The anomalous expression of miRNAs is associated with a variety of diseases, such as heart failure, muscular dystrophies, type 2 diabetes, Alzheimer's disease and cancer [17], with miRNAs contributing to the initiation and progression of several stages of cancer as well as resistance to anti-cancer therapy [18]. Almost all malignant tumours exhibit differential expression of specific miRNAs between cancer and adjacent tissues [19, 20].
In the past few years, the results of a growing number of studies have emphasised the importance of miRNAs in autophagy regulation as well as the significance of autophagy in the biogenesis and function of miRNAs. In this review, we summarise recent advances in miRNA and autophagy and discuss the correlation between miRNA, autophagy and cancer.
Molecular mechanisms of autophagy
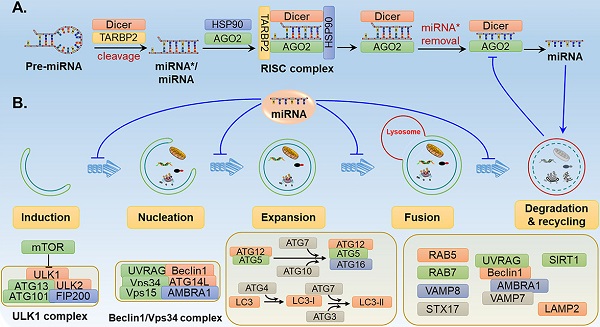
The process of autophagy involves a series of stages and sequential membrane-remodelling events, including induction, vesicle nucleation, vesicle elongation, autolysosome maturation and lysosomal fusion, and degradation and recycling [21] (Fig. 1). The molecular mechanisms of autophagy are not fully understood, but it is thought that several autophagy-related genes (ATGs) and regulatory proteins are implicated in autophagy regulation.
Induction
Target of rapamycin (TOR) kinase-containing protein complexes are the crucial autophagy regulators during the initiation stage. As shown in Fig. 1A, mammalian TOR (mTOR) is stimulated by intracellular stress and further activates the downstream serine/threonine Unc-51-like kinases 1 and 2 (ULK1 and ULK2). Subsequently, ULK1 and ULK2 form a ULK1 complex with ATG101, the family-interacting protein of 200 kDa (FIP200, the mammalian homologue of ATG17) and ATG13 [22], thereby activating the initiation and nucleation process of autophagy.
Vesicle nucleation
The second step of autophagy is vesicle nucleation. As shown in Fig. 1B, the ULK1 complex phosphorylates and activates the Beclin 1/Vps34 complex comprising Beclin 1, Vps34 (also called class III phosphatidylinositol 3-kinase, PIK3C3), ATG14L, Vps15 and the activating molecule in Beclin 1-regulated autophagy protein 1 (AMBRA1)[23]. The Beclin 1/Vps34 complex produces phosphatidylinositol 3-phosphatase (PI3P) to recruit the double-FYVE-containing protein 1 (DFCP1) that localises to the endoplasmic reticulum (ER) membrane, thereby providing a platform for expansion of the membrane and the formation of the omegasome. At present, the source of the membrane for autophagosomes has remained unclear, with potential sources including the ER membrane, the Golgi membrane, the plasma membrane and the mitochondria [24].
Expansion
Fig. 1C shows the two types of ubiquitin-like conjugation systems that participate in the elongation of autophagosomes: the ATG5-ATG12 complex conjugated with ATG16L1 and microtubule-associated protein 1 light chain 3 (MAP1LC3, also called LC3) conjugated with lipid phosphatidylethanolamine (PE) [25]. In the first system, the ubiquitin-like protein ATG12 covalently binds to ATG5 with the participation of the E1-like enzyme ATG7 and the E2-like enzyme ATG10. Next, the ATG5-ATG12 complex becomes localised to the autophagosome membrane to form a complex with ATG16, promoting the expansion of the autophagosome membrane [26]. In the second system, the archetypal form of LC3 is cleaved by ATG4B to generate the cytosolic free LC3-I form [27]. The E1-like enzyme ATG7, E2-like enzyme ATG3 and E3-like enzyme ATG12-ATG5-ATG16L1 are required for the linkage of LC3-I and PE to produce the LC3-PE complex, which is termed LC3-II [25]. The high lipophilicity of PE promotes the attachment of LC3 to both faces of autophagosome membrane, where it plays a crucial role in membrane elongation and the selective degradation of cellular materials [28].
Autolysosome maturation and lysosomal fusion
The last crucial step in the process of autophagy is the fusion of autophagosomes with lysosomes to form autolysosomes (Fig. 1D). Autophagosomes undergo several independent fusions to form amphisomes with late endosomes and subsequently forming autolysosomes with lysosomes [29]. In the first step of autophagosome-lysosome fusion, the out autophagosomal membrane fuses with lysosomal membrane. The fusion process is finished by the degradation of the inner autophagosomal membrane and the exposure of autophagosome contents to the lumen of the lysosome [30]. Several groups of proteins have been reported to be associated with the fusion process, including small GTPases, RAB proteins (RAB5 and RAB7), the UV radiation resistance-associated gene protein (UVRAG), and SNARE proteins (VAMP8 and STX17)[31, 32]. UVRAG is reported to play a role in the formation of membrane curvature, thus contributing to maturation of autolysosomes through its interaction with Beclin 1 [33].
The roles of related miRNAs during the phases of autophagy. The core proteins regulated by miRNAs are shown in the schematic diagram. (A) Under some intracellular stress, such as hypoxia or starvation, autophagy is inducted by mTOR inactivation and activation of the downstream ULK1 complex. (B) Vesicle nucleation is driven by phosphorylation and activation of the Beclin 1/Vps34 complex. PI3P produced by the Beclin 1/Vps34 pathway recruits DFCP1 to provide a framework for membrane expansion. (C) During the membrane expansion step, two types of ubiquitin-like conjugation systems (the ATG5-ATG12 complex conjugated with ATG16L1, and LC3 conjugated with PE) are involved in the elongation and formation of autophagosomes. (D) The last crucial step is the fusion of autophagosomes with lysosomes to form autolysosomes for biomacromolecule degradation and recycling. GTPases, UVRAG, SNARE and other proteins are reported to be implicated in the fusion process.
Degradation and recycling
After fusion, superfluous or malfunctioning organelles and misfolded proteins harboured by autolysosomes are degraded by lysosomal acid hydrolases such as cathepsins (CTSs) into substrates for recycling and metabolism [34] (Fig. 1D). A variety of CTSs (such as CTSB, CTSC, CTSD, CTSL, and CTSS) have been implicated in biomacromolecule degradation [35].
Interrelation between miRNAs and autophagy
Studies have demonstrated that miRNAs are crucial regulators in the autophagy process, participating in several steps of autophagy, including the upstream pathways that trigger autophagy, the subsequent developmental stages, and the later stage of degradation. Meanwhile, autophagy is also crucial in the generation and maintenance of miRNAs.
Regulation of autophagy by miRNAs
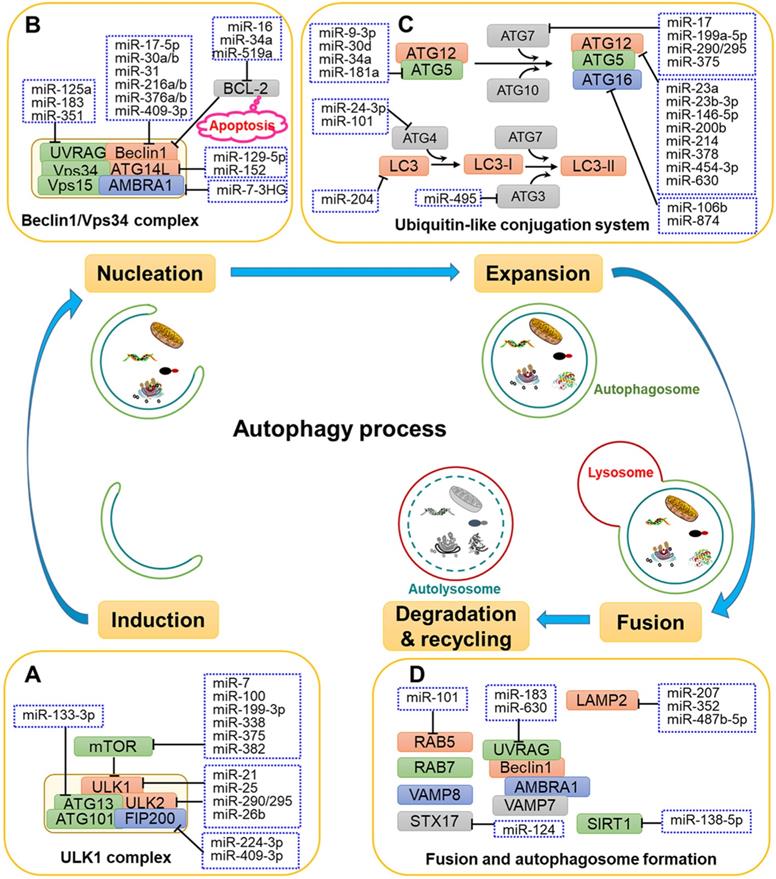
MiRNAs have been shown to affect the expression of ATGs and related regulators to influence the phases of autophagy, including induction, vesicle nucleation, phagophore expansion, autolysosome maturation, lysosomal fusion and degradation (Fig. 1 and Table 1).
Regulation of autophagy induction by miRNAs
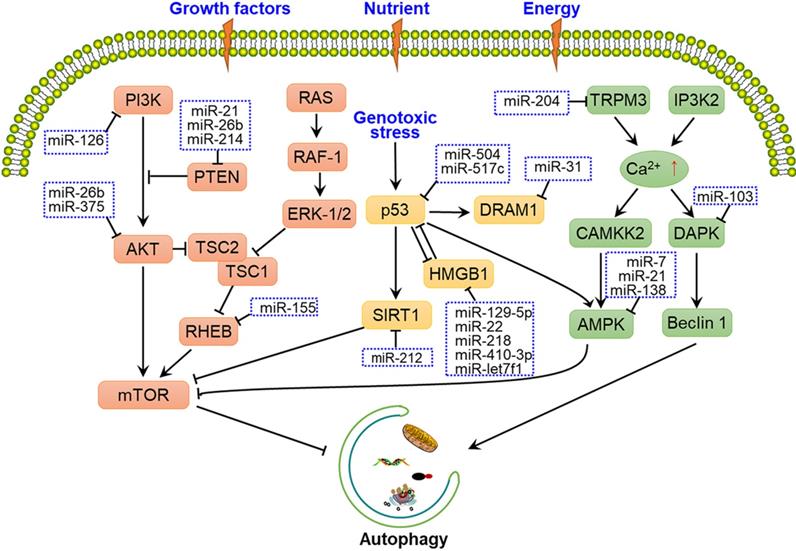
Several upstream nutrient and energy signals are involved in the regulation of autophagy induction, including the PI3K-AKT-mTOR, TP53-mTOR, and Ca2+-AMPK-mTOR pathways [36-38], with some miRNAs having been shown to alter these signals to affect phagophore induction (Fig. 2). MiR-21, miR-26b and miR-214 were shown to have a crucial role in autophagy by targeting PTEN through inhibition of the PI3K-AKT-mTOR pathway in cancer cells [39-41]. While under genotoxic stress, TP53 and HMGB1 form complex to produce the reciprocal inhibitory effect, thereby regulate the downstream SIRT1-mTOR signalling. Some miRNAs, such as miR-517c, miR-129-5p, miR-218 and miR-212 have been shown to regulate autophagy through TP53-mTOR signalling [42-45]. Some calcium-metabolizing enzymes, such as transient receptor potential melastatin 3 (TRPM3) and Drosophila inositol 1,4,5-trisphosphate kinase 2 (IP3K2) participate in the regulation of autophagy initiation through the Ca2+-AMPK-mTOR pathway. Hall et al. demonstrated that in clear cell renal cell carcinoma, miR-204 targets the TRPM3 channel, thereby promoting Ca2+ influx, the activation of CAMKK2, AMPK, and ULK1, and phagophore formation [36].
In addition to upstream signals, mTOR and ULK1 complex are directly regulated by miRNAs. For instance, miR-7, miR-99a, miR-338, miR-375 and miR-382 directly target mTOR to regulate autophagy, thereby disturbing the tumorigenic potential of cancer cells [46-50]. Moreover, miR-21, miR-25, miR-290-295 cluster and miR-26b interact with the ULK1 complex and further disrupt autophagy initiation in a number of cancers [11, 51-54]. FIP200 and ATG13 are also targeted by miRNAs, such as miR-133a-3p, miR-244-3p and miR-409-3p, thereby blocking FIP200- and ATG13-mediated autophagy [55-57] (Fig. 1A and Fig. 2).
Regulation of nucleation by miRNAs
Multiple miRNAs have been demonstrated to target the nucleation process to affect autophagy. As shown in Fig. 1B, miR-17-5p, miR-30a, miR-216, miR-376 and miR-409-3p bind to the 3'-UTR of Beclin 1 and inhibits its expression, causing the suppression of autophagy in different types of cancer cell [58, 59]. The importance of Beclin 1 is also reflected in its involvement with two cellular events, autophagy and apoptosis, which is achieved by its association with BCL-2 [60]. Some miRNAs, such as miR-16, miR-519a and miR-34a, increase apoptosis and autophagy by interacting with BCL-2 [61-63].
In addition, miRNAs targeting ATG14 and UVRAG also affect the autophagy process. For example, miR-129-5p and miR-152 adjust autophagy by targeting ATG14 and further affects the PI3K/AKT/mTOR signal pathway [64, 65]. In addition, miR-183, miR-351 and miR-125a specifically target the 3'-UTR of UVRAG, which is involved in vesicle formation [66, 67].
Regulation of membrane expansion by miRNAs
Multiple miRNAs have been reported to participate in the ATG5-ATG12-ATG16L and LC3-PE conjugation systems to regulate the elongation of autophagosomes (Fig. 1C). For example, miR-9-3p, miR-30d, miR-34a and miR-181a are demonstrated to regulate autophagy in cancer cells through targeting ATG5 [68-71]. In addition, the E1-like enzyme ATG7 is targeted by miR-7, miR-199a-5p, miR-290-295 cluster and miR-375 in several kinds of cancer cells, respectively [49, 54, 72, 73]. Moreover, miR-23a, miR-23b-3p, miR-146a-5p, miR-200b, miR-378 and miR-630 were reported to regulate autophagy by binding to ATG12 in different types of cancer cells [74-78]. Moreover, miR-874 can decrease the expression of ATG16L, thereby inhibiting autophagy and sensitizing gastric cancer (GC) cells to chemotherapy [79].
Autophagy-associated miRNAs in cancer
| Effect on autophagy | Name | Dysregulation | Autophagy-related target | Types of cancer cell line (cancer tissue) | Ref. |
|---|---|---|---|---|---|
| Inhibition | miR-7 | N.D. | LKB1-AMPK-mTOR | AsPC-1, BxPC-3 and SW1990 (pancreatic cancer) | [47] |
| miR-7-3HG | N.D. | AMBRA1 | Hela (cervical cancer) | [124] | |
| miR-9-3p | N.D. | ATG5 | TT and MZ-CRC-1 (medullary thyroid carcinoma) | [68] | |
| miR-10b | up-regulated | Bim/TFAP2C | U251, LN-308, and U373 (glioblastoma) | [153] | |
| miR-15a/16 | downregulated | BCL-2 | A549 (lung cancer) | [148] | |
| miR-17 | up-regulated | ATG7 | T98G and U373-MG (glioblastoma) | [72] | |
| miR-17-5p | downregulated | Beclin 1 | A549 (lung cancer) | [144] | |
| miR-23a | downregulated | ATG12 | WM35, WM793, 451LU, A2058 and A375 (melanoma) | [74] | |
| miR-26b | up-regulated | ULK2/AKT/PTEN | Hep-2 (laryngeal carcinoma) | [52] | |
| miR-29b | downregulated | PSME4 | AMCL1, AMCL2 (myeloma) | [154] | |
| miR-30a | downregulated | Beclin 1 | MG-63 (osteosarcoma) | [155] | |
| miR-30d | downregulated | ATG5 | HCT15, HCT116, HT-29, DLD-1 and SW480 (colon cancer) | [69] | |
| miR-34a | downregulated | ATG5 | SH-SY5Y and SK-N-SH (neuroblastoma) | [70] | |
| miR-101 | downregulated | EZH2 | HepG2 (liver cancer) | [156] | |
| miR-106b | N.D. | ATG16L1 | HCT116 (colorectal cancer) | [157] | |
| miR-124/144 | downregulated | PIM1 | DU145 and PC3 (prostate cancer) | [158] | |
| miR-133-3p | downregulated | GABARAPL1/ATG13 | BGC-823, SGC-7901, MGC-803, MKN-45, HGC-27 and AGS (gastric cancer) | [55] | |
| miR-138 | downregulated | AMPK-mTOR | A549 and Calu-3 (lung cancer) | [120] | |
| miR-140-5p | downregulated | SMAD2 | HCT116, RKO, and SW480 (colorectal cancer) | [159] | |
| miR-152 | downregulated | ATG14 | A2780/CP70, SKOV3/DDP (ovarian cancer) | [65] | |
| miR-183 | up-regulated | UVRAG | HCT116 and HT29 (colorectal cancer) | [160] | |
| miR-195 | up-regulated | GABARAPL1 | Endothelial progenitor | [131] | |
| miR-204 | N.D. | LC3 | 786-O, A498, and Caki-1 (kidney cancer) | [161] | |
| miR-212 | downregulated | SIRT1 | LNCaP (prostate cancer) | [44] | |
| miR-224-3p | up-regulated | FIP200 | HeLa, SiHa, C33A (cervical cancer) | [56] | |
| miR-290-295 cluster | up-regulated | ATG7/ULK1 | B16F1 (melanoma) | [54] | |
| miR-338 | downregulated | mTOR | Siha, HeLa, C33 A and Me180 (cervical cancer) | [48] | |
| miR-338-5p | up-regulated | PIK3C3 | SW480 and HCT116 (colorectal cancer) | [134] | |
| miR-340 | downregulated | ROCK1 | U373, U87 (glioblastoma) | [162] | |
| miR-375 | downregulated | AKT/mTOR | MKN-45 and GT3TKB (gastric cancer) | [49] | |
| miR-378 | up-regulated | ATG12 | HeLa and C-33A (cervical cancer) | [77] | |
| miR-454-3p | downregulated | ATG12 | U251, U87 and LN229 (glioma cancer) | [135] | |
| miR-502 | downregulated | RAB1B | HCT116 (colorectal cancer) | [163] | |
| miR-517c | N.D. | TP53 | U87 and U251 (glioblastoma) | [42] | |
| miR-519a | downregulated | BCL-1 | BEAS-2B (lung cancer) | [62] | |
| miR-532-3p | up-regulated | RAB3IP | AGS, MKN45, BGC823, SGC7901, MGC803, and MKN28 (gastric cancer) | [116] | |
| miR-630 | N.D. | ATG12/UVRAG | JHU-029 (squamous cell carcinoma) | [78] | |
| miR-638 | up-regulated | TP53INP2 | SK-Mel-28 and SK-Mel-147 (melanoma) | [164] | |
| miR-874 | downregulated | ATG16L1 | SGC7901, BGC823 SGC7901, BGC823 and AGS (gastric cancer) | [79] | |
| miR-1256 | downregulated | CAB39 | MKN45, MGC803, AGS, HGC27, BGC823 and SGC7901(gastric cancer) | [118] | |
| Activation | miR-18a-5p | up-regulated | EGFR | A549, H23, H1299 and H1650 (lung cancer) | [121] |
| miR-20a | up-regulated | THSB2 | SiHa and HeLa (cervical cancer) | [165] | |
| miR-21 | up-regulated | AMPK/ULK1 | A549 (lung cancer) | [11] | |
| miR-99a | downregulated | mTOR | MCF‐7 and MDA‐MB‐231 (breast cancer) | [46] | |
| miR-100 | downregulated | IGFR1/mTOR | HepG2, Huh7 (liver cancer) | [166] | |
| miR-126 | downregulated | IRS1 | H28 (sarcomatoid malignant mesothelioma) | [128] | |
| miR-133b | downregulated | PTBP1 | MKN-1, MKN-45 and KATO-III (gastric cancer) | [119] | |
| miR-145-3p | downregulated | HDAC4 | U2OS and MG-63 (osteosarcoma) | [125] | |
| miR-155 | up-regulated | RHEB/RICTOR/RPS6KB2 | CNE (nasopharyngeal cancer) and HeLa (cervical cancer) | [123] | |
| miR-155-3p | N.D. | CREBRF | U251 and T98G (glioblastoma) | [167] | |
| miR-210 | up-regulated | VEGF | RT4-D6P2T (schwannoma) | [132] | |
| miR-382 | downregulated | mTOR | Eca109 and Het-1A (esophageal squamous cell carcinoma) | [50] | |
| miR-423-3p | up-regulated | Bim | BGC823, MGC803, SGC7901, and MKN45 (gastric cancer) | [117] | |
| mR-494 | N.D. | LC3 | 769-P (renal cancer) | [129] | |
| miR-519-3p | downregulated | N.D. | HeLa (cervical cancer) | [122] | |
| miR-524-5p | downregulated | ITGA3 | TPC-1, K1, and NPA papillary (thyroid carcinoma) | [136] |
MiR-204 has been shown to regulate LC3-II in IR-induced cardiomyocyte autophagy [80]. In addition, ATG4 is reported to targeted by miR-24-3p and miR-101 in lung cancer and hepatocellular carcinoma cells, respectively [81, 82]. Moreover, the E2-like enzyme ATG3 is targeted and regulated by miR-495 under starvation conditions [83]. MiR-495 suppresses starvation-induced autophagy by inhibiting the conversion of LC3-I to LC3-II and reduces the number of autophagosomes.
Regulation of autolysosome maturation by miRNAs
The late maturation and fusion steps are regulated by multiple proteins, such as Rab5, SIRT1, UVRAG, STX17 and lysosomal associated membrane protein 2 (LAMP2) [84-86]. As shown in Fig. 1D, miR-138-5p has been shown to target the 3'-UTR of sirtuin 1 (SIRT1), further regulating the SIRT1/FoxO1/Rab7 axis and inhibiting autophagy [87]. Moreover, Huang and colleagues demonstrated that miR-124 downregulates STX17 expression by targeting the 3'-UTR to regulate retinoblastoma cell autophagy [88]. Additionally, miR-207, miR-352 and miR-487b-5p directly target LAMP2 to affect the latter stage of lysosomal-autophagy flux in cortical neuronal cells [89, 90].
Thus, the study of miRNAs as regulators of autophagy-lysosomal-associated genes is important to elucidate the molecular mechanisms underlying the development of autophagy and provides insights for clinical applications.
Regulation of miRNAs by autophagy
Autophagy is a process in which intracellular cytoplasmic substances are transported to lysosomes for degradation, and miRNAs are no exception. Considering that lysosomes contain multiple proteases and RNases, they may not only regulate RNA processes through the degradation of RNA-related proteins but could also affect RNA itself [91].
A variety of proteins are involved in the biosynthesis of miRNAs. As shown in Fig. 3A, pre-miRNAs exported from the nucleus are cleaved into double-stranded miRNA*/miRNA duplex by Dicer. Then, miRNA*/miRNA is transferred to the groove of RNA-induced silencing complex (RISC) component argonaute 2 (AGO2), and miRNA* becomes dissociated to generate mature miRNA. AGO2 can repress miRNA-targeted mRNAs through translational inhibition of those mRNAs and the promotion of mRNA decay. Gibbings et al. reported that AGO and Dicer can be degraded by autophagy [92]. Specifically, the autophagy receptor CALCOCO2 is associated with AGO2 and Dicer in a GEMIN4-dependent manner, demonstrating the crucial effects of autophagy in miRNA biosynthesis and homeostasis (Fig. 3B).
Overview of miRNAs involved in the regulation of autophagy-related signalling pathways. Several upstream nutrient and energy signals are associated with autophagy initiation, such as the PI3K-AKT-mTOR, TP53-mTOR, and Ca2+-AMPK-mTOR pathways. mTOR is the central protein in these signalling pathways, and miRNAs regulate autophagy-related signalling pathways by targeting crucial factors, such as PI3K, AKT, AMPK, and the ULK1 complex.
Schematic diagram of miRNA biosynthesis and regulation by autophagy. (A) Biosynthesis of miRNAs. In the nucleus, a pri-mRNA is transcribed under by RNA Pol Ⅰ/Ⅲ and further cleaved by Drosha and DGCR8 to generate pre-miRNA. After its nuclear export, Dicer cleaves the pre-miRNA into an miRNA*/miRNA duplex, which is then loaded to the groove of AGO2 to form the RISC complex. The miRNA then guides RISC to silence the target mRNA through mRNA cutting, transcriptional inhibition or deadenylation, while miRNA* is degraded. (B) Regulation of miRNA by autophagy. Dicer and AGO2 are associated with GEMIN4 to form the Dicer-AGO2-GEMIN4 complex. The Dicer-AGO2-GEMIN4 complex may interact with CALCOCO2 through either the ubiquitination of AGO2 or GEMIN4. Finally, Dicer and AGO2 are degraded by the autophagy-lysosome system, thereby inhibiting miRNA homeostasis. In addition, miRNA itself may be degraded by autophagy.
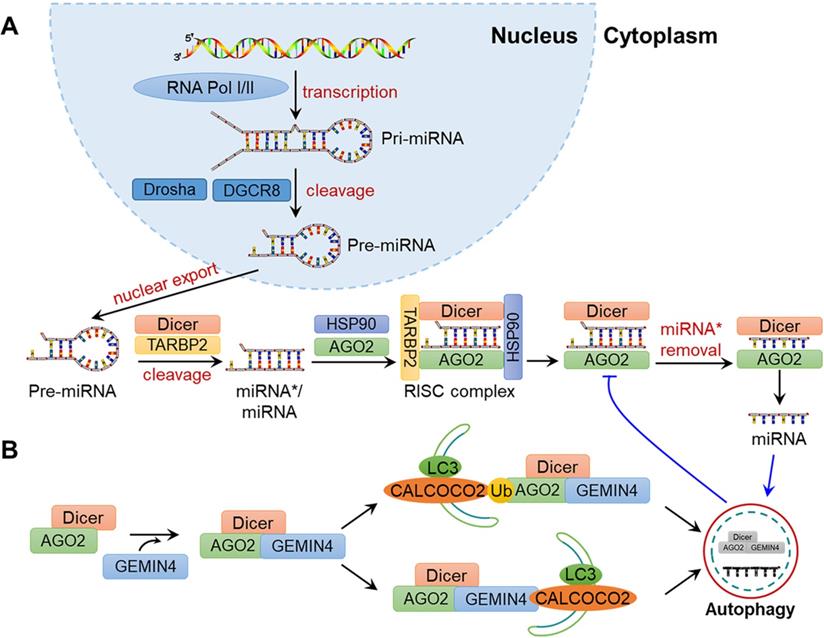
In addition, miRNAs can also be directly degraded during autophagy (Fig. 3B). For example, Lan and colleagues demonstrated that miR-224 is selectively degraded by the autophagosome-lysosome system, and the downregulation of autophagy is negatively associated with the accumulation of miR-224 in hepatocellular carcinoma (HCC)[93]. Peng et al. suggested that CDKN1B promotes the degradation of miR-6981 through SQSTM1/p62-mediated autophagy, thereby resulting in a tumour-suppressive effect [94]. On the whole, autophagy is altered under various types of cellular stress, infections and disease in mammals, which may influence the abundance, activity and function of miRNAs and provide new ideas for understanding the physiological and pathological processes in humans.
Roles of autophagy-related miRNAs in cancer
Many miRNAs have been shown to participate in various stages of autophagy by regulating the expression of autophagy-related genes and proteins. These miRNAs are also involved in different stages of cancer development and progression, affecting cancer cell survival and proliferation, metabolism, oxidative stress, metastasis and deterioration, as well as resistance to radiation and chemotherapy. Indeed, many autophagy-related miRNAs have been recognised as diagnostic and prognostic biomarkers that may be useful as clinical therapeutic targets for cancer. In this section, we review the effects of miRNAs and autophagy and their interactions with respect to cancer biology as well as their associated responses to anti-cancer therapy.
Cellular context-dependent functions of miRNAs in cancer
MiRNAs are a class of endogenous non-coding RNAs that are approximately 20-25 nucleotides in length and can recognise target mRNAs through complementary base pairing, directly silencing RISC to degrade target mRNA[95]. Recent research has revealed that miRNAs can serve as new targets for the treatment of diseases, especially in cancers.
A pioneering study by Calin and colleagues showed that miR-15 and miR-16 are deleted in chronic lymphocytic leukaemia, indicating that miRNAs may function as cancer suppressors [96]. Subsequently, countless miRNAs have been demonstrated to participate in the pathogenesis of cancer. Currently, miRNAs are known to participate in various types of cancers, including lung cancer, breast cancer, melanoma, colorectal cancer (CRC), and leukaemia [97]. The relationship between dysregulated miRNAs and tumorigenesis has been a hot topic in cancer biology investigations.
A single miRNA molecule can target tens to hundreds of different mRNAs, some of which may have opposite carcinogenic or tumour-suppressor functions. Thus, a specific miRNA may have the ability to be oncomiRNA as well as tumour-suppressive miRNA, depending on the cellular context [98]. For example, miR-7 was reported to have hundreds of targets and have cellular context-dependent activities in cancer [99]. On the one hand, miR-7 targets and downregulates tumorigenic factors in tumour-associated signalling pathways such as PA28γ, EGFR, PAK1, ACK1 and PIK3CD, demonstrating the crucial role of miR-7 in tumour suppression [100]. On the other hand, elevated levels of miR-7 have been shown to be associated with tumour aggressiveness in cervical cancer (CC) and lung adenocarcinoma (LAC) [101, 102].
Regarding members of the miR-125 family, targets include pro-apoptosis genes (P53INP1, TNFAIP3, Bmf, Puma, p53, and Bak1), anti-apoptosis genes (MUC1, Mcl-1, Bcl-w, and Bcl-2), pro-proliferation genes (c-Jun, ERBB2/3, VEGF-A, and E2F3), metastasis-inhibiting genes (STARD13, TP53INP1, and p53), metastasis promoting genes (MMP13, LIN28B, ARID3B) and pro-differentiation genes (IGF2, DICER1, LIN28A, CBFβ, and ARID3a) [103]. The balance of these oncogenes/tumour-suppressive genes in different cellular contexts leads to the oncogenic or tumour-suppressive effect of the miR-125 family in specific cancers. MiR-125b was reported to be upregulated in acute megakaryoblastic leukaemia and acute promyelocytic leukaemia, while in aggressive and indolent chronic lymphocytic leukaemia, miR-125b was observed to be downregulated [104-106].
Another example is miR-155, which in most instances is considered to function as an oncomiRNA, possessing oncogenic roles in cancers. MiR-155 has been to be overexpressed in several haematological and solid malignancies, such as Hodgkin's lymphoma, breast cancer, lung cancer, CC and thyroid carcinoma [98]. Nevertheless, miR-155 was demonstrated to play a tumour-suppressive role in some tumours. For example, Qin et al. and Li et al. confirmed that miR-155 has a tumour-suppressive effect in ovarian cancer and GC [107, 108].
The controversial properties of miRNAs in different cancers suggest that the functions of miRNAs vary with respect to the pathogenesis and progression of cancer. Therefore, the underlying mechanisms that occur under different cellular contexts, requires further investigation.
Dual roles of autophagy in cancer
Autophagy is an important biological phenomenon and is involved in regulating the balance between the synthesis, degradation and reuse of cellular material, thereby reducing the accumulation of intracellular waste and limiting abnormal mutations and genomic damage that may lead to cancer [109]. In this respect, normal autophagy can be considered to be a suppressive mechanism in the early stage of cancer. Research has shown that deletions of the tumour-suppressor gene Beclin 1 often occurs in breast, ovarian and prostate cancers, the loss of which causes a reduction of autophagy and the promotion of cellular proliferation [110, 111]. Another example is Bif-1, a protein that interacts with Beclin 1 through UVRAG and positively regulates autophagy [33]. Bif-1 knockout can lead to the inhibition of autophagy and enhance the development of cancer in mice. Taken together, these results establish the suppressive role of autophagy in cancer.
Conversely, in established tumours, autophagy can allow tumour cells to meet the high metabolic demands of continuous proliferation, thereby protecting tumour cells from metabolic stress-induced necrosis [112], which has been well illustrated for RAS-activating mutations. RAS activation leads to the upregulation of autophagy via the RAF/MEK/ERK pathway, which is associated with the regulation of mitochondrial metabolism [113]. Indeed, researchers have demonstrated increased autophagy in a variety of RAS-activated cancer cells [114, 115]. The inhibition of autophagy induces decreased glycolytic capacity, oxidative phosphorylation, and cell proliferation as well as increased apoptosis in vitro and in vivo.
Autophagy has important significance in the establishment and development of cancer. Thus, gaining a better understanding the cellular and functional correlation of autophagy in tumour microenvironment will help translate laboratory research into clinical applications.
Involvement of miRNAs and autophagy in cancer biology
Many autophagy-related miRNAs participate in different biological events in cancer, including cancer proliferation, cellular metabolism, angiogenesis, cancer cell migration and metastasis, as well as responses to cancer therapy. More importantly, some autophagy-related miRNAs have been regarded as cancer biomarkers and anti-cancer targets. In this section, we summarise these autophagy-related miRNAs and their roles in cancer biology and anti-cancer applications (Table 1).
Cell survival and proliferation
Autophagy is crucial for the survival and growth of cancer cells, and miRNAs have been demonstrated to regulate autophagy and control cell growth and proliferation.
For instance, Guo and colleagues observed that miR-532-3p can bind to the 3'-UTR of RAB3IP, increasing the number of autophagosomes and LC3-II expression to inhibit cell viability and growth in GC [116]. The overexpression of miR-375 has been shown inhibit cell proliferation and AKT/mTOR-dependent autophagy in GC both in vitro and in vivo, providing a potential therapeutic target for GC [49]. Additionally, miR-423-3p overexpression was shown to activate autophagy and promote proliferation in GC cell lines and animal models [117]. Thus, miR-423-3p may be a new biomarker and potential target for the treatment of GC. Moreover, Xu and colleagues reported that miR-1256 suppresses cell proliferation, thereby impairing autophagy in GC cells by targeting calcium-binding protein 39 (CAB39), an upstream regulator of the AMPK/mTOR pathway [118]. In another study, miR-133b was observed to induce autophagy and inhibit GC cell proliferation by targeting polypyrimidine tract-binding protein 1 (PTBP1) [119].
Similarly, in NSCLC, Ye et al. demonstrated that miR-138 inhibits cell proliferation by suppressing the AMPK signalling pathway and increasing mTOR phosphorylation [120]. Li et al. confirmed that miR-21 promotes the proliferation, migration and invasion of NSCLC and can induce autophagy by activating the AMPK/ULK1 pathway [11]. Moreover, miR-18a-5p is regarded to function as a regulator of autophagy by targeting EGFR, thereby inducing proliferation inhibition and apoptosis activation in NSCLC cells [121].
Autophagy-related miRNAs have also been shown to adjust cellular proliferation in CC. MiR-519d-3p, which is downregulated in CC, can inhibit cell proliferation, cause cell cycle arrest and promote apoptosis and autophagy by targeting hypoxia-inducible factor-2α (HIF-2α) [122]. In addition, Wan et al. demonstrated that miR-155 is an autophagy inducer that downregulates mTOR signalling, thereby decreasing cellular proliferation and causing cell cycle arrest in CC cells [123]. Moreover, Capizzi et al. showed that miR-7-3HG is an autophagy inhibitor that targets the 3'-UTR of AMBRA1 in CC, where inhibiting this miRNA reduced the proliferation and increased apoptosis of cancer cells [124].
Numerous studies have investigated the effects of autophagy-related miRNAs in regulating the survival and proliferation of cancer cells. For example, miR-145-3p overexpression was shown to inhibit the proliferation of osteosarcoma cells while activating apoptosis and autophagy by downregulating HDAC4 [125]. In addition, the downregulated miRNA, miR-30d, was demonstrated to bind the 3'-UTRs of ATG5, PI3K and Beclin 1, thereby negatively regulating autophagy, inhibiting the proliferation and viability of colon cancer cells [69]. MiR-26b was observed to be upregulated in laryngeal carcinoma patients [52]. Interestingly, miR-26b downregulation was shown to activate autophagy in a ULK2- and PTEN/AKT-dependent manner and could further inhibit the proliferation and induce the apoptosis of laryngeal carcinoma cells. In urothelial carcinoma, miR-331-3p was demonstrated to reduce cell proliferation by targeting nucleus accumbens-associated protein 1 (NACC1) [126].
Cell metabolism
Cancer cells undergo major changes in glucose, amino acid and fatty acid metabolism to maintain survival in a stressful microenvironment. As autophagy is an intracellular recycling process that maintains the levels of metabolites and biosynthetic intermediates under starvation or other stress conditions, it is an important mechanism for cancer cell metabolic adaptation [127]. Importantly, multiple autophagy-related miRNAs are involved in the metabolic regulation and stress responses of cancer cells. For example, Gu and colleagues reported that pancreatic cancer cells can use autophagy to meet the requirement of glycolysis [47]. Specifically, miR-7 inhibits autophagy by upregulating the LKB1-AMPK-mTOR pathway, reducing the intracellular glucose supply for glycolysis. MiR-126 was demonstrated to induce autophagy in malignant mesothelioma cells by downregulating the insulin receptor substrate-1 (IRS1) signalling pathway, further suppressing glucose uptake and leading to energy exhaustion and the AMPK-dependent phosphorylation of ULK1 [128]. Additionally, Chen et al. showed that the miR-290-295 cluster can enhance the resistance of melanoma cells to glucose starvation in an ATG7/ULK1-dependent manner, inhibiting autophagic cell death caused by glucose starvation [54].
Regarding the regulation of lipid metabolism, miR-494 can increase multilamellar bodies and lipid droplets formation that is accompanied by autophagy activation in renal cancer cells [129]. Additionally, miR-126 was demonstrated to induce autophagy by stimulating lipid droplet accumulation in an HIF-1α-dependent manner in malignant mesothelioma cells [128]. Autophagy-related miRNAs have also been shown to be involved in glutamine metabolism in cancer. Zhang and colleagues demonstrated that GC cells could supplement glutaminolysis through autophagy, whereas miR-133-3p can block this process by targeting GABA type A receptor-associated protein like 1 (GABARAPL1) and ATG13 [55].
Although these basic research studies have demonstrated that autophagy-related miRNAs are involved in the regulation of cancer cell metabolism, additional work is required to comprehensively elucidate the mechanisms underlying this metabolic regulatory network.
Angiogenesis
Autophagy is essential for endothelial cell function and angiogenesis [130]. Recently, some autophagy-related miRNAs have been shown to affect the survival, growth and spread of endothelial cells, thereby directly affecting cancer angiogenesis. For example, the inhibition of miR-195, which targets GABARAPL1, was shown to promote the proliferation and migration of endothelial progenitor cells (EPCs) as well as cancer angiogenesis under hypoxia [131]. Furthermore, miR-212 was demonstrated to negatively regulate autophagy by targeting SIRT1, thereby leading to the inhibition of angiogenesis and cell senescence in prostate cancer cells [44]. Wang and colleagues showed that miR-210 expression is associated with vascular endothelial growth factor (VEGF) levels in schwannoma cells [132]. Importantly, hypoxia was shown to induce the demethylation of miR-210 and affect HIF-1α/VEGF-mediated angiogenesis. This result led to a model in which hypoxia enhances miR-210 expression and promotes autophagy, thereby increasing angiogenesis in schwannoma. The results of these studies indicate the role of miRNAs and autophagy in regulating endothelial cell homeostasis and tumour angiogenesis in vitro, although additional research is needed to establish their relevance in vivo.
Cancer cell migration and metastasis
The link between cell autophagy and metastasis affects cancer cell mobility, invasion and metastasis, with some studies having suggested that autophagy-related miRNAs are associated with cancer migration and metastasis [133].
A number of studies have shown that miRNAs regulate migration and metastasis by inhibiting autophagy. For instance, miR-338-5p is positively associated with the stage, metastasis and poor prognosis in primary CRC by directly targeting PIK3C3 pathway, resulting in both migration and invasion in vivo [134]. In cutaneous melanoma, the downregulated miRNA, miR-23a, suppressed the invasion and migration of cells by inhibiting ATG12-mediated autophagy [74]. Moreover, miR-378 overexpression directly reduces ATG12 levels, inhibiting autophagy and resulting in enhanced migration and invasion in CC [77]. Yuan et al. observed that miR-375 inhibits autophagy in GC by targeting AKT/mTOR and is negatively associated with cellular migration capacity [49]. Similarly, miR-517c was demonstrated to inhibit autophagy and reduce glioblastoma cell migration depending on TP53 expression [42]. Additionally, miR-454-3p was shown to inhibit the migration and invasion of glioma cancer cells by targeting ATG12 [135].
In contrast, in some cases, miRNAs regulate cancer migration and metastasis by activating autophagy. For example, miR-382, promotes apoptosis and autophagy in esophageal squamous cell carcinoma (ESCC) by inhibiting mTOR and 4E-BP1 [50], and the overexpression of miR-382 was shown to suppress the migration, invasion and epithelial-mesenchymal transition (EMT) of ESCC cells. In NSCLC, miR-18a-5p was reported to promote autophagy by targeting interferon regulatory factor 2 (IRF2) to increase apoptosis while inhibiting cellular migration [121]. MiR-524-5p, a downregulated miRNA in papillary thyroid carcinoma, was shown to promote autophagy and inhibit cell viability, invasion, migration and apoptosis by targeting ITGA3 and FOXE1 in papillary thyroid cancer [136].
Autophagy-related miRNAs and their response to cancer therapy
Radiotherapy and chemotherapy are currently the most important anti-cancer strategies. However, the therapeutic resistance of cancer cells is a major challenge in cancer treatment. Autophagy induced by therapeutants has been demonstrated to be related to the response, resistance or death of cancer cells, and increasing evidence suggests that many miRNAs are involved in the regulation of autophagy processes caused by anti-cancer therapies [23].
Response to radiotherapy
Radiotherapy is a standard treatment for various cancers and involves damaging cancer cells with ionising radiation. The mechanisms of radiation therapy include the production of oxygen radicals, which damage important organelles like mitochondria, and damage to biological macromolecules like DNA [137]. Indeed, autophagy is an important factor that influences the effects of radiotherapy and cellular responses [138]. Consequently, autophagy-related miRNAs are capable of regulating the response of cancer cells to radiotherapy. In this section, we summarise the effects of autophagy-related miRNAs on cancer radiotherapy (Table 2).
For example, miR-17-5p was demonstrated to target Beclin 1 and inhibit autophagy in glioma cells [59]. In a mouse model, miRNA-17-5p overexpression sensitised the response of tumour tissues to irradiation, while miR-23b was shown to suppress irradiation-induced autophagy by targeting ATG12 in pancreatic cancer cells. Interestingly, miR-23b overexpression was observed to increase the sensitivity of pancreatic cancer cells to irradiation [139].
In contrast, in some cellular contexts, autophagy tends to reduce the sensitivity of cancer cells to radiation and can lead to resistance to radiation-induced cell death. For example, in prostate cancer cells, miR-32 was demonstrated to activate autophagy by targeting the tumour-suppressor gene DAB2 interacting protein (DAB2IP), thereby enhancing cell survival and decreasing sensitivity during radiation treatment [140]. Similarly, miR-138-5p was observed to enhance radiation-induced autophagy by targeting EIF4EBP1 in nasopharyngeal carcinoma [141]. Moreover, miR-301a/b was reported to induce autophagy and radioresistance by decreasing NARG2 expression in prostate cancer cells [142].
At present, the relationships and boundaries between protective and pro-survival autophagy, cellular apoptosis and necrosis remain unclear and requires a better understanding of the molecular mechanisms. Nonetheless, the regulation of autophagy in a cellular context-dependent manner may alter the response of cancer cells to radiotherapy.
Response to chemotherapy
A variety of chemotherapy drugs have been shown to induce the autophagy of cancer cells, and autophagy-related miRNAs have been reported to affect cancer cell susceptibility to anti-cancer drugs. Table 3 summarises the effects of autophagy-related miRNAs on cancer chemotherapy.
For instance, miR-101 and miR-199a-5p were demonstrated to intensify cisplatin-induced cell death through inhibition of autophagy in HCC cells [73, 82]. Similarly, cytoprotective autophagy was suppressed while the toxic effects of cisplatin were increased in GC cells when miR-148a-3p and miR-181a were overexpressed [71, 143]. Moreover, in lung cancer cells, miR-146a-5p, miR-17-5p, miR-200b and miR-487b-5p have been shown to inhibit autophagy and enhance cellular chemosensitivity to cisplatin, paclitaxel, docetaxel and to temozolomide, respectively [76, 90, 144, 145].
Effects of autophagy-related miRNAs on cancer radiotherapy
| Effect on autophagy | Name | Dysregulated | Autophagy-related target | Effect on radiotherapy | Tested cell line (tissue origin) | Ref. |
|---|---|---|---|---|---|---|
| Inhibition | miR-17-5p | up-regulated | Beclin 1 | radiosensitivity | U87 (glioblastoma) | [59] |
| miR-21 | up-regulated | PTEN | radiosensitivity | Hela, Siha(cervical cancer) | [168] | |
| miR-23b | downregulated | ATG12 | radiosensitivity | BxPC3, PANC-1 (pancreatic cancer) | [139] | |
| miR-31 | downregulated | Beclin 1, ATG, DRAM | radiosensitivity | Primary cultured cells (colorectal cancer) | [169] | |
| miR-93 | up-regulated | Beclin 1, ATG5, ATG4B, SQSTM1 | radiosensitivity | U87 (glioblastoma) | [170] | |
| miR-101 | downregulated | STMN1 | radioresistance | CNE-2, 5-8F (nasopharyngeal carcinoma) | [171] | |
| miR-129-5p | downregulated | HMGB1 | radiosensitivity | MCF-7, MDA-MB-231 , BT549, BT474 (breast cancer) | [43] | |
| miR-183-5p | up-regulated | ATG5 | radiosensitivity | Caco-2 (colon cancer) | [172] | |
| miR-200c | downregulated | UBQLN1 | radiosensitivity | MDA-MB-231, BT549 (breast cacer) | [173] | |
| miR-214 | downregulated | ATG12 | radiosensitivity | SW480, HCT116 (colorectal cancer) | [174] | |
| miR-216b | up-regulated | Beclin 1 | radioresistance | PANC-1 (pancreatic cancer) | [175] | |
| miR-450a-5p | downregulated | DUSP10 | radioresistance | ECA (esophageal cancer) | [176] | |
| Activation | miR-138-5p | downregulated | EIF4EBP1 | radiosensitivity | HONE1, HK1 (nasopharyngeal carcinoma) | [141] |
| miR-301a/b | up-regulated | NDRG2 | radioresistance | LNCaP (prostate cancer) | [142] | |
| miR-1246 | up-regulated | mTOR | radioresistance | A549, PC9 (lung cancer) | [177] | |
| miR-4673 | up-regulated | CDK-18 | radioresistance | SKBR3 (breast cancer) | [178] |
Effects of autophagy-related miRNAs on cancer chemotherapy
| Effect on autophagy | MiRNAs | Dysregulation | Autophagy-related target | Effect on chemotherapy | Agent | Tested cell line (tissue origin) | Ref. |
|---|---|---|---|---|---|---|---|
| Inhibition | miR-let7f1 | N.D. | HMGB1 | chemosensitivity | Cisplatin | D425, UW228 (medulloblastoma) | [179] |
| miR-30a | downregulated | Beclin 1 | chemoresistance | HeLa, MCF-7, HepG2, HepS | [180] | ||
| miR-101 | N.D. | STMN1, RAB5A, ATG4D, mTOR | chemosensitivity | HepG2 (liver cancer) | [82] | ||
| miR-146a-5p | up-regulated | ATG12 | chemoresistance | A549 (lung cancer) | [76] | ||
| miR-148a-3p | downregulated | AKAP1 | chemoresistance | BGC823 (gastric cancer) | [143] | ||
| miR-152 | downregulated | ATG14 | chemoresistance | A2780/CP70 (ovarian cancer) | [65] | ||
| miR-181a | N.D. | ATG5 | chemosensitivity | SGC7901 (gastric cancer) | [71] | ||
| miR-199a-5p | downregulated | ATG7 | chemoresistance | Huh7, HepG2 (hepatocellular carcinoma) | [73] | ||
| miR-205 | N.D. | RAB27A LAMP3 | chemoresistance | DU145, PC-3 (prostate cancer) | [181] | ||
| miR-409-3p | downregulated | FIP200 | chemosensitivity | OV-1063 (ovarian cancer) | [182] | ||
| miR-416a | up-regulated | CHOP | chemoresistance | A549, H446(lung cancer) | [183] | ||
| miR-17-5p | downregulated | Beclin 1 | chemosensitivity | Paclitaxel | A549-T24 (lung cancer) | [144] | |
| miR-216b | downregulated | Beclin 1 | chemosensitivity | A549, Calu-3 (lung cancer) | [184] | ||
| miR-218 | downregulated | HMGB1 | chemoresistance | RL95-2 (endometrial carcinoma) | [45] | ||
| miR-143 | downregulated | ATG2B | chemosensitivity | Doxorubicin | SAOS, U2OS (osteosarcoma) | [185] | |
| miR-223 | downregulated | FOXO3a | chemoresistance | HepG2, Huh7, SNU387, SNU449 (hepatocellular carcinoma) | [186] | ||
| miR-34a | downregulated | Smad4 | chemoresistance | Oxaliplatin | HT29 (colorectal cancer) | [187] | |
| miR-409-3p | downregulated | Beclin 1 | chemosensitivity | Lovo Oxa R(colon cancer) | [182] | ||
| miR-17 | up-regulated | ATG7 | chemosensitivity | Temozolomide | U373 (glioma) | [72] | |
| miR-21 | up-regulated | PTEN | chemosensitivity | Sorafenib | Huh7, HepG2 (liver cancer) | [39] | |
| miR-22 | N.D. | BTG1 | chemosensitivity | 5-Fu | SW620 (colorectal cancer) | [188] | |
| miR-25 | N.D. | ULK1 | chemosensitivity | Isoliquiritigenin | MCF-7 (breast cancer) | [53] | |
| miR-30b | N.D. | Beclin 1 | chemosensitivity | Imatinib | K562 (CML) | [189] | |
| miR-101 | N.D. | STMN1, RAB5A, ATG4D | chemosensitivity | Etoposide | MCF-7 (breast cancer) | [190] | |
| miR-143 | downregulated | GABARAPL1 | chemosensitivity | Qercetin | AGS, MKN28 (gastric cancer) | [191] | |
| miR-200b | N.D. | ATG12 | chemosensitivity | Docetaxel | SPC-A1, H1299 (lung cancer) | [145] | |
| miR-375 | N.D. | ATG7 | chemoresistance | Fulvestrant | MCF-7 (breast cancer) | [192] | |
| miR-410-3p | downregulated | HMGB1 | chemoresistance | Gemcitabine | MiaPaCa2, PANC-1 (pancreatic cancer) | [193] | |
| miR-487b-5p | up-regulated | LAMP2 | chemoresistance | Temozolomide | A549, H1299 (lung cancer) | [90] | |
| Activation | miR-125b | downregulated | Foxp3 | chemosensitivity | Cisplatin | WRO, FRO(thyroid cancer) | [194] |
| miR-425-3p | up-regulated | AKT1 | chemoresistance | A549 (lung cancer) | [146] | ||
| miR-193b | up-regulated | STMN1 | chemosensitivity | 5-Fu | KYSE450 (oesophageal cancer) | [195] | |
| miR-338-3p | up-regulated | mTOR | chemoresistance | HCT116, HT29 (colon cancer) | [147] | ||
| miR-15a/16 | N.D. | Rictor | chemosensitivity | Camptothecin | Hela (cervical carcinoma) | [148] | |
| miR-16 | downregulated | Bcl-2 | chemoresistance | Paclitaxel | A549-T24 (lung cancer) | [63] | |
| miR-30a | downregulated | Beclin 1 | chemosensitivity | Sorafenib | 786-0, A489 (renal carcinoma) | [196] |
On the other hand, miRNAs confer chemoresistance to anti-cancer agents by activating autophagy. For example, cisplatin-treated NSCLC cells were shown to become chemoresistant via miR-425-3p-facilitated autophagy activation in an AKT1-dependent manner [146]. Similarly, in colon cancer cells, miR-338-3p was shown to promote 5-Fu resistance by inhibiting mTOR and activating autophagy [147]. Moreover, miR-15a/16 was demonstrated to attenuate the phosphorylation of mTORC1 and p70S6K and to enhance camptothecin-induced autophagy in CC HeLa cells, thereby contributing to the efficacy of chemotherapy [148].
Indeed, autophagy-related miRNAs are involved in the response to chemotherapy in a variety of cancers. With the in-depth elucidation of mechanisms associated with autophagy-related miRNAs and cancer chemotherapy, additional autophagy-related miRNAs may become anti-cancer targets and prognostic biomarkers.
Conclusion and prospection
Autophagy is an evolutionary mechanism that involves the recycling of biological macromolecules and organelles [5]. MiRNAs participate in many biological processes and play a crucial role in the regulation of autophagy in cancer. The impact and number of studies on miRNAs and autophagy in cancer have continued to increase. However, autophagy and miRNA research is in its infancy, and requires further investigation, including with respect to the paradoxical effects of autophagy and miRNAs on cancer and the development of additional research methods and applications.
The role of miRNA-mediated autophagy in cancer remains controversial, and whether miRNA-regulated autophagy is a survival or death mechanism for cancer cells remains unknown [112]. In addition, miRNAs themselves appear to exert bilateral regulation in cancer [98]. Thus, the unclear role of autophagy and the dual roles of miRNAs in cancer complicate the associated regulatory mechanisms.
MiRNAs regulate gene expression before observable changes in protein levels, making autophagy-related miRNAs potential early autophagy markers that are superior to LC3 and SQSTM1 for monitoring autophagy [149]. In recent years, a variety of miRNA detection methods with high resolution have been developed. For example, Guk et al. proposed a novel strategy involving the self-circulation of molecular beacon circuits and miRNAs hybridization for miRNA detection [150]. In addition, Li and colleagues developed a novel and sensitive fluorescence polarization miRNA detection method with a detection limit of 0.001 nM [151]. Moreover, Jin et al. proposed a sensitive miRNA detection method using Chlorella virus DNA ligase that is at least 40-fold more sensitive than conventional methods and could detect individual miRNAs that differ by only one nucleotide [152]. Additionally, because studies on the effects of a specific miRNA on a single autophagy gene is often one-sided, multidisciplinary and comprehensive public databases should be used to assess the regulation of multiple genes and steps in the complex autophagy network by miRNAs.
The regulation of autophagy by miRNAs can affect the sensitivity of cancer to radiotherapy and chemotherapy. However, considering the development of cell resistance caused by autophagy during treatment, it is not appropriate to simply abandon cell survival in favour of cell death. Indeed, autophagy may be the link between cell survival and death, and these dysregulated autophagy-related miRNAs could serve as potential anti-cancer targets. Therefore, the biosafety and reliability of miRNA-based therapeutic strategies should receive widespread attention, including with respect to the exact role of miRNAs in specific autophagy steps, techniques for effectively delivering miRNA mimics in vivo, and therapeutic strategies based on miRNA off-target effects.
Autophagy-related miRNAs have great potential for use in the diagnosis, treatment, and prognosis evaluation of cancer. Thus, an in-depth exploration of autophagy and miRNA as well as related applications should be a major objective for scientists.
Abbreviations
3'-UTR: 3'-untranslated region; AGO: argonaute; AMBRA1: the activating molecule in Beclin 1-regulated autophagy protein 1; ATG: autophagy-related gene; CAB39: calcium-binding protein 39; CC: cervical cancer; CRC: colorectal cancer; CTS: cathepsin; DAB2IP: the tumour-suppressor gene DAB2 interacting protein; DFCP1: the double-FYVE-containing protein 1; EMT: epithelial-mesenchymal transition; EPC: endothelial progenitor cell; ER: endoplasmic reticulum; ESCC: esophageal squamous cell carcinoma; FIP200: the family-interacting protein of 200 kDa; GABARAPL1: GABA type A receptor-associated protein like 1; GC: gastric cancer; HCC: hepatocellular carcinoma; HIF-2α: hypoxia-inducible factor-2α; IRF2: interferon regulatory factor 2; IRS1: insulin receptor substrate-1; LAC: lung adenocarcinoma; LAMP2: lysosomal associated membrane protein 2; MAP1LC3: microtubule-associated protein 1 light chain 3; MiRNA: microRNA; mTOR: mammalian TOR; NACC1: nucleus accumbens-associated protein 1; NSCLC: non-small cell lung cancer; PE: phosphatidylethanolamine; PI(3)KC3: the class III phosphatidylinositol 3-kinase; PI3P: phosphatidylinositol 3-phosphatase; PTBP1: polypyrimidine tract-binding protein 1; PTEN: gene of phosphate and tension homology deleted on chromsome ten; RISC: the RNA-induced silencing complex; SIRT1: sirtuin 1; STX17: Syntaxin 17; THBS2: thrombospondin 2; TOR: target of rapamycin; ULK1: Unc-51-like kinases 1; ULK2: Unc-51-like kinases 2; UVRAG: the UV radiation resistance-associated gene protein; VAMP3: vesicle-associated membrane protein 3; VEGF: vascular endothelial growth factor.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Project No.: 21708021, 51803098), Natural Science Foundation of Shandong Province, China (Project No.: ZR2017BC012, ZR2018BEM032), Major Research Program of the National Natural Science Foundation of China (Project No: 91849209) and China Postdoctoral Science Foundation Funded Project (Project No.: 2018M632612).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42
2. Hussey S, Terebiznik MR, Jones NL. Autophagy: healthy eating and self-digestion for gastroenterologists. J Pediatr Gastroenterol Nutr. 2008;46:496-506
3. Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161-70
4. Klionsky DJ. Autophagy revisited: a conversation with Christian de Duve. Autophagy. 2008;4:740-3
5. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069-75
6. Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280-93
7. Clarke PG, Puyal J. Autophagic cell death exists. Autophagy. 2012;8:867-9
8. Liu CY, Zhang YH, Li RB, Zhou LY, An T, Zhang RC. et al. LncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat Commun. 2018;9:29
9. Chen X, Mao R, Su W, Yang X, Geng Q, Guo C. et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. 2019 p: 1-13
10. Tazawa H, Yano S, Yoshida R, Yamasaki Y, Sasaki T, Hashimoto Y. et al. Genetically engineered oncolytic adenovirus induces autophagic cell death through an E2F1-microRNA-7-epidermal growth factor receptor axis. Int J Cancer. 2012;131:2939-50
11. Li S, Zeng X, Ma R, Wang L. MicroRNA-21 promotes the proliferation, migration and invasion of non-small cell lung cancer A549 cells by regulating autophagy activity via AMPK/ULK1 signaling pathway. Exp Ther Med. 2018;16:2038-45
12. Li Y, Zhou D, Ren Y, Zhang Z, Guo X, Ma M. et al. Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy. 2019;15:478-92
13. Liang Y, Huo Q, Lu W, Jiang L, Gao W, Xu L. et al. Fluorescence Resonance Energy Transfer Visualization of Molecular Delivery from Polymeric Micelles. Journal of biomedical nanotechnology. 2018;14:1308-16
14. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE. et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901-6
15. Kabekkodu SP, Shukla V, Varghese VK, J DS, Chakrabarty S, Satyamoorthy K. Clustered miRNAs and their role in biological functions and diseases. Biol Rev Camb Philos Soc. 2018;93:1955-86
16. Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152-7
17. Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827-87
18. Dietrich P, Koch A, Fritz V, Hartmann A, Bosserhoff AK, Hellerbrand C. Wild type Kirsten rat sarcoma is a novel microRNA-622-regulated therapeutic target for hepatocellular carcinoma and contributes to sorafenib resistance. Gut. 2018;67:1328-41
19. Cerna K, Oppelt J, Chochola V, Musilova K, Seda V, Pavlasova G. et al. MicroRNA miR-34a downregulates FOXP1 during DNA damage response to limit BCR signalling in chronic lymphocytic leukaemia B cells. Leukemia. 2019;33:403-14
20. Karmakar S, Kaushik G, Nimmakayala R, Rachagani S, Ponnusamy MP, Batra SK. MicroRNA regulation of K-Ras in pancreatic cancer and opportunities for therapeutic intervention. Semin Cancer Biol. 2019;54:63-71
21. Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24:24-41
22. Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107-32
23. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528-42
24. Gozuacik D, Akkoc Y, Ozturk DG, Kocak M. Autophagy-Regulating microRNAs and Cancer. Front Oncol. 2017;7:65
25. Nakatogawa H. Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy. Essays Biochem. 2013;55:39-50
26. Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol. 2014;21:336-45
27. Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y. et al. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 2009;28:1341-50
28. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3-12
29. Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998;273:21883-92
30. Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 2018;14:207-15
31. Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci. 2014;39:61-71
32. Jahn R, Scheller RH. SNAREs-engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631-43
33. Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y. et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142-51
34. Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24:69-79
35. Khaket TP, Kwon TK, Kang SC. Cathepsins: Potent regulators in carcinogenesis. Pharmacol Ther. 2019;198:1-19
36. Hall DP, Cost NG, Hegde S, Kellner E, Mikhaylova O, Stratton Y. et al. TRPM3 and miR-204 establish a regulatory circuit that controls oncogenic autophagy in clear cell renal cell carcinoma. Cancer Cell. 2014;26:738-53
37. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67-93
38. Chowdhari S, Saini N. Gene expression profiling reveals the role of RIG1 like receptor signaling in p53 dependent apoptosis induced by PUVA in keratinocytes. Cell Signal. 2016;28:25-33
39. He C, Dong X, Zhai B, Jiang X, Dong D, Li B. et al. MiR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget. 2015;6:28867-81
40. Yuan T, Yang Y, Chen J, Li W, Li W, Zhang Q. et al. Regulation of PI3K signaling in T-cell acute lymphoblastic leukemia: a novel PTEN/Ikaros/miR-26b mechanism reveals a critical targetable role for PIK3CD. Leukemia. 2017;31:2355-64
41. Liu J, Chen W, Zhang H, Liu T, Zhao L. miR-214 targets the PTEN-mediated PI3K/Akt signaling pathway and regulates cell proliferation and apoptosis in ovarian cancer. Oncol Lett. 2017;14:5711-8
42. Lu Y, Xiao L, Liu Y, Wang H, Li H, Zhou Q. et al. MIR517C inhibits autophagy and the epithelial-to-mesenchymal (-like) transition phenotype in human glioblastoma through KPNA2-dependent disruption of TP53 nuclear translocation. Autophagy. 2015;11:2213-32
43. Luo J, Chen J, He L. mir-129-5p Attenuates Irradiation-Induced Autophagy and Decreases Radioresistance of Breast Cancer Cells by Targeting HMGB1. Med Sci Monit. 2015;21:4122-9
44. Ramalinga M, Roy A, Srivastava A, Bhattarai A, Harish V, Suy S. et al. MicroRNA-212 negatively regulates starvation induced autophagy in prostate cancer cells by inhibiting SIRT1 and is a modulator of angiogenesis and cellular senescence. Oncotarget. 2015;6:34446-57
45. Ran X, Yang J, Liu C, Zhou P, Xiao L, Zhang K. MiR-218 inhibits HMGB1-mediated autophagy in endometrial carcinoma cells during chemotherapy. Int J Clin Exp Pathol. 2015;8:6617-26
46. Yang Z, Han Y, Cheng K, Zhang G, Wang X. miR-99a directly targets the mTOR signalling pathway in breast cancer side population cells. Cell Prolif. 2014;47:587-95
47. Gu DN, Jiang MJ, Mei Z, Dai JJ, Dai CY, Fang C. et al. microRNA-7 impairs autophagy-derived pools of glucose to suppress pancreatic cancer progression. Cancer Lett. 2017;400:69-78
48. Lu R, Yang Z, Xu G, Yu S. miR-338 modulates proliferation and autophagy by PI3K/AKT/mTOR signaling pathway in cervical cancer. Biomed Pharmacother. 2018;105:633-44
49. Yuan KT, Li BX, Yuan YJ, Tan M, Tan JF, Dai WG. et al. Deregulation of MicroRNA-375 Inhibits Proliferation and Migration in Gastric Cancer in Association With Autophagy-Mediated AKT/mTOR Signaling Pathways. Technol Cancer Res Treat. 2018;17:1533033818806499
50. Feng J, Qi B, Guo L, Chen LY, Wei XF, Liu YZ. et al. miR-382 functions as a tumor suppressor against esophageal squamous cell carcinoma. World J Gastroenterol. 2017;23:4243-51
51. John Clotaire DZ, Zhang B, Wei N, Gao R, Zhao F, Wang Y. et al. MiR-26b inhibits autophagy by targeting ULK2 in prostate cancer cells. Biochem Biophys Res Commun. 2016;472:194-200
52. Wang S, Guo D, Li C. Downregulation of miRNA-26b inhibits cancer proliferation of laryngeal carcinoma through autophagy by targeting ULK2 and inactivation of the PTEN/AKT pathway. Oncol Rep. 2017;38:1679-87
53. Wang Z, Wang N, Liu P, Chen Q, Situ H, Xie T. et al. MicroRNA-25 regulates chemoresistance-associated autophagy in breast cancer cells, a process modulated by the natural autophagy inducer isoliquiritigenin. Oncotarget. 2014;5:7013-26
54. Chen Y, Liersch R, Detmar M. The miR-290-295 cluster suppresses autophagic cell death of melanoma cells. Sci Rep. 2012;2:808
55. Zhang X, Li Z, Xuan Z, Xu P, Wang W, Chen Z. et al. Novel role of miR-133a-3p in repressing gastric cancer growth and metastasis via blocking autophagy-mediated glutaminolysis. J Exp Clin Cancer Res. 2018;37:320
56. Fang W, Shu S, Yongmei L, Endong Z, Lirong Y, Bei S. miR-224-3p inhibits autophagy in cervical cancer cells by targeting FIP200. Sci Rep. 2016;6:33229
57. Cheng Y, Ban R, Liu W, Wang H, Li S, Yue Z. et al. MiRNA-409-3p enhances cisplatin-sensitivity of ovarian cancer cells by blocking the autophagy mediated by Fip200. Oncol Res. 2018
58. Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X. et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816-23
59. Hou W, Song L, Zhao Y, Liu Q, Zhang S. Inhibition of Beclin-1-Mediated Autophagy by MicroRNA-17-5p Enhanced the Radiosensitivity of Glioma Cells. Oncol Res. 2017;25:43-53
60. Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600-6
61. Li H, Chen L, Li JJ, Zhou Q, Huang A, Liu WW. et al. miR-519a enhances chemosensitivity and promotes autophagy in glioblastoma by targeting STAT3/Bcl2 signaling pathway. J Hematol Oncol. 2018;11:70
62. Bai W, Chen Y, Sun P, Gao A. Downregulation of B-cell lymphoma/leukemia-2 by overexpressed microRNA 34a enhanced titanium dioxide nanoparticle-induced autophagy in BEAS-2B cells. Int J Nanomedicine. 2016;11:1959-71
63. Chatterjee A, Chattopadhyay D, Chakrabarti G. MiR-16 targets Bcl-2 in paclitaxel-resistant lung cancer cells and overexpression of miR-16 along with miR-17 causes unprecedented sensitivity by simultaneously modulating autophagy and apoptosis. Cell Signal. 2015;27:189-203
64. Zhang H, Zhang X, Zhang J. MiR-129-5p inhibits autophagy and apoptosis of H9c2 cells induced by hydrogen peroxide via the PI3K/AKT/mTOR signaling pathway by targeting ATG14. Biochem Biophys Res Commun. 2018;506:272-7
65. He J, Yu JJ, Xu Q, Wang L, Zheng JZ, Liu LZ. et al. Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy. 2015;11:373-84
66. Kim Y, Kang YS, Lee NY, Kim KY, Hwang YJ, Kim HW. et al. Uvrag targeting by Mir125a and Mir351 modulates autophagy associated with Ewsr1 deficiency. Autophagy. 2015;11:796-811
67. Yuan Y, Zhang Y, Han L, Sun S, Shu Y. miR-183 inhibits autophagy and apoptosis in gastric cancer cells by targeting ultraviolet radiation resistance-associated gene. Int J Mol Med. 2018;42:3562-70
68. Gundara JS, Zhao J, Gill AJ, Lee JC, Delbridge L, Robinson BG. et al. Noncoding RNA blockade of autophagy is therapeutic in medullary thyroid cancer. Cancer Med. 2015;4:174-82
69. Zhang R, Xu J, Zhao J, Bai J. Mir-30d suppresses cell proliferation of colon cancer cells by inhibiting cell autophagy and promoting cell apoptosis. Tumour Biol. 2017;39:1010428317703984
70. Cheng X, Xu Q, Zhang Y, Shen M, Zhang S, Mao F. et al. miR-34a inhibits progression of neuroblastoma by targeting autophagy-related gene 5. Eur J Pharmacol. 2019;850:53-63
71. Zhao J, Nie Y, Wang H, Lin Y. MiR-181a suppresses autophagy and sensitizes gastric cancer cells to cisplatin. Gene. 2016;576:828-33
72. Comincini S, Allavena G, Palumbo S, Morini M, Durando F, Angeletti F. et al. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 2013;14:574-86
73. Xu N, Zhang J, Shen C, Luo Y, Xia L, Xue F. et al. Cisplatin-induced downregulation of miR-199a-5p increases drug resistance by activating autophagy in HCC cell. Biochem Biophys Res Commun. 2012;423:826-31
74. Guo W, Wang H, Yang Y, Guo S, Zhang W, Liu Y. et al. Down-regulated miR-23a Contributes to the Metastasis of Cutaneous Melanoma by Promoting Autophagy. Theranostics. 2017;7:2231-49
75. An Y, Zhang Z, Shang Y, Jiang X, Dong J, Yu P. et al. miR-23b-3p regulates the chemoresistance of gastric cancer cells by targeting ATG12 and HMGB2. Cell Death Dis. 2015;6:e1766
76. Yuwen DL, Sheng BB, Liu J, Wenyu W, Shu YQ. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2017;21:2650-8
77. Tan D, Zhou C, Han S, Hou X, Kang S, Zhang Y. MicroRNA-378 enhances migration and invasion in cervical cancer by directly targeting autophagy-related protein 12. Mol Med Rep. 2018;17:6319-26
78. Huang Y, Guerrero-Preston R, Ratovitski EA. Phospho-DeltaNp63alpha-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle. 2012;11:1247-59
79. Huang H, Tang J, Zhang L, Bu Y, Zhang X. miR-874 regulates multiple-drug resistance in gastric cancer by targeting ATG16L1. Int J Oncol. 2018;53:2769-79
80. Xiao J, Zhu X, He B, Zhang Y, Kang B, Wang Z. et al. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J Biomed Sci. 2011;18:35
81. Pan B, Chen Y, Song H, Xu Y, Wang R, Chen L. Mir-24-3p downregulation contributes to VP16-DDP resistance in small-cell lung cancer by targeting ATG4A. Oncotarget. 2015;6:317-31
82. Xu Y, An Y, Wang Y, Zhang C, Zhang H, Huang C. et al. miR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;29:2019-24
83. Li W, Yang Y, Hou X, Zhuang H, Wu Z, Li Z. et al. MicroRNA-495 regulates starvation-induced autophagy by targeting ATG3. FEBS Lett. 2016;590:726-38
84. Hyttinen JM, Niittykoski M, Salminen A, Kaarniranta K. Maturation of autophagosomes and endosomes: a key role for Rab7. Biochim Biophys Acta. 2013;1833:503-10
85. Takats S, Nagy P, Varga A, Pircs K, Karpati M, Varga K. et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201:531-9
86. Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256-69
87. Tian S, Guo X, Yu C, Sun C, Jiang J. miR-138-5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget. 2017;8:11071-82
88. Huang J, Yang Y, Fang F, Liu K. MALAT1 modulates the autophagy of retinoblastoma cell through miR-124-mediated stx17 regulation. J Cell Biochem. 2018;119:3853-63
89. Tao J, Liu W, Shang G, Zheng Y, Huang J, Lin R. et al. MiR-207/352 regulate lysosomal-associated membrane proteins and enzymes following ischemic stroke. Neuroscience. 2015;305:1-14
90. Bao L, Lv L, Feng J, Chen Y, Wang X, Han S. et al. miR-487b-5p Regulates Temozolomide Resistance of Lung Cancer Cells Through LAMP2-Medicated Autophagy. DNA Cell Biol. 2016;35:385-92
91. Gibbings D, Mostowy S, Voinnet O. Autophagy selectively regulates miRNA homeostasis. Autophagy. 2013;9:781-3
92. Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012;14:1314-21
93. Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF. et al. Autophagy-preferential degradation of MIR224 participates in hepatocellular carcinoma tumorigenesis. Autophagy. 2014;10:1687-9
94. Peng M, Wang J, Tian Z, Zhang D, Jin H, Liu C. et al. Autophagy-mediated Mir6981 degradation exhibits CDKN1B promotion of PHLPP1 protein translation. Autophagy. 2019;15:1523-38
95. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-97
96. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524-9
97. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203-22
98. Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016;76:3666-70
99. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609-12
100. Kalinowski FC, Brown RA, Ganda C, Giles KM, Epis MR, Horsham J. et al. microRNA-7: a tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol. 2014;54:312-7
101. Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290-7
102. Chou YT, Lin HH, Lien YC, Wang YH, Hong CF, Kao YR. et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010;70:8822-31
103. Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6
104. Klusmann JH, Li Z, Bohmer K, Maroz A, Koch ML, Emmrich S. et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010;24:478-90
105. Zhang H, Luo XQ, Feng DD, Zhang XJ, Wu J, Zheng YS. et al. Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol Cancer. 2011;10:108
106. Tili E, Michaille JJ, Luo Z, Volinia S, Rassenti LZ, Kipps TJ. et al. The down-regulation of miR-125b in chronic lymphocytic leukemias leads to metabolic adaptation of cells to a transformed state. Blood. 2012;120:2631-8
107. Qin W, Ren Q, Liu T, Huang Y, Wang J. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013;587:1434-9
108. Li CL, Nie H, Wang M, Su LP, Li JF, Yu YY. et al. microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep. 2012;27:1960-6
109. Eisenberg-Lerner A, Kimchi A. The paradox of autophagy and its implication in cancer etiology and therapy. Apoptosis. 2009;14:376-91
110. Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A. et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809-20
111. Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E. et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59-65
112. White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401-10
113. Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124-31
114. Perera RM, Stoykova S, Nicolay BN, Ross KN, Fitamant J, Boukhali M. et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015;524:361-5
115. Guo JY, White E. Autophagy is required for mitochondrial function, lipid metabolism, growth, and fate of KRAS(G12D)-driven lung tumors. Autophagy. 2013;9:1636-8
116. Guo W, Chen Z, Chen Z, Yu J, Liu H, Li T. et al. Promotion of Cell Proliferation through Inhibition of Cell Autophagy Signalling Pathway by Rab3IP is Restrained by MicroRNA-532-3p in Gastric Cancer. J Cancer. 2018;9:4363-73
117. Kong P, Zhu X, Geng Q, Xia L, Sun X, Chen Y. et al. The microRNA-423-3p-Bim Axis Promotes Cancer Progression and Activates Oncogenic Autophagy in Gastric Cancer. Mol Ther. 2017;25:1027-37
118. Xu Z, Li Z, Wang W, Xia Y, He Z, Li B. et al. MIR-1265 regulates cellular proliferation and apoptosis by targeting calcium binding protein 39 in gastric cancer and, thereby, impairing oncogenic autophagy. Cancer Lett. 2019;449:226-36
119. Sugiyama T, Taniguchi K, Matsuhashi N, Tajirika T, Futamura M, Takai T. et al. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci. 2016;107:1767-75
120. Ye Z, Fang B, Pan J, Zhang N, Huang J, Xie C. et al. miR-138 suppresses the proliferation, metastasis and autophagy of non-small cell lung cancer by targeting Sirt1. Oncol Rep. 2017;37:3244-52
121. Liang C, Zhang X, Wang HM, Liu XM, Zhang XJ, Zheng B. et al. MicroRNA-18a-5p functions as an oncogene by directly targeting IRF2 in lung cancer. Cell Death Dis. 2017;8:e2764
122. Jiang L, Shi S, Shi Q, Zhang H, Xia Y, Zhong T. MicroRNA-519d-3p Inhibits Proliferation and Promotes Apoptosis by Targeting HIF-2alpha in Cervical Cancer Under Hypoxic Conditions. Oncol Res. 2018;26:1055-62
123. Wan G, Xie W, Liu Z, Xu W, Lao Y, Huang N. et al. Hypoxia-induced MIR155 is a potent autophagy inducer by targeting multiple players in the MTOR pathway. Autophagy. 2014;10:70-9
124. Capizzi M, Strappazzon F, Cianfanelli V, Papaleo E, Cecconi F. MIR7-3HG, a MYC-dependent modulator of cell proliferation, inhibits autophagy by a regulatory loop involving AMBRA1. Autophagy. 2017;13:554-66
125. Wu G, Yu W, Zhang M, Yin R, Wu Y, Liu Q. MicroRNA-145-3p suppresses proliferation and promotes apotosis and autophagy of osteosarcoma cell by targeting HDAC4. Artif Cells Nanomed Biotechnol. 2018;46:579-86
126. Morita K, Fujii T, Itami H, Uchiyama T, Nakai T, Hatakeyama K. et al. NACC1, as a Target of MicroRNA-331-3p, Regulates Cell Proliferation in Urothelial Carcinoma Cells. Cancers (Basel). 2018 10
127. Cheong H. Integrating autophagy and metabolism in cancer. Arch Pharm Res. 2015;38:358-71
128. Tomasetti M, Monaco F, Manzella N, Rohlena J, Rohlenova K, Staffolani S. et al. MicroRNA-126 induces autophagy by altering cell metabolism in malignant mesothelioma. Oncotarget. 2016;7:36338-52
129. Dutta P, Haller E, Sharp A, Nanjundan M. MIR494 reduces renal cancer cell survival coinciding with increased lipid droplets and mitochondrial changes. BMC Cancer. 2016;16:33
130. Schaaf MB, Houbaert D, Mece O, Agostinis P. Autophagy in endothelial cells and tumor angiogenesis. Cell Death Differ. 2019;26:665-79
131. Mo J, Zhang D, Yang R. MicroRNA-195 regulates proliferation, migration, angiogenesis and autophagy of endothelial progenitor cells by targeting GABARAPL1. Biosci Rep. 2016 36
132. Wang Z, Deng M, Liu Z, Wu S. Hypoxia-induced miR-210 promoter demethylation enhances proliferation, autophagy and angiogenesis of schwannoma cells. Oncol Rep. 2017;37:3010-8
133. Mowers EE, Sharifi MN, Macleod KF. Functions of autophagy in the tumor microenvironment and cancer metastasis. FEBS J. 2018;285:1751-66
134. Chu CA, Lee CT, Lee JC, Wang YW, Huang CT, Lan SH. et al. MiR-338-5p promotes metastasis of colorectal cancer by inhibition of phosphatidylinositol 3-kinase, catalytic subunit type 3-mediated autophagy pathway. EBioMedicine. 2019;43:270-81
135. Shao N, Xue L, Wang R, Luo K, Zhi F, Lan Q. miR-454-3p Is an Exosomal Biomarker and Functions as a Tumor Suppressor in Glioma. Mol Cancer Ther. 2019;18:459-69
136. Liu H, Chen X, Lin T, Chen X, Yan J, Jiang S. MicroRNA-524-5p suppresses the progression of papillary thyroid carcinoma cells via targeting on FOXE1 and ITGA3 in cell autophagy and cycling pathways. J Cell Physiol. 2019;234:18382-91
137. Schaue D, McBride WH. Opportunities and challenges of radiotherapy for treating cancer. Nat Rev Clin Oncol. 2015;12:527-40
138. Tam SY, Wu VW, Law HK. Influence of autophagy on the efficacy of radiotherapy. Radiat Oncol. 2017;12:57
139. Wang P, Zhang J, Zhang L, Zhu Z, Fan J, Chen L. et al. MicroRNA 23b regulates autophagy associated with radioresistance of pancreatic cancer cells. Gastroenterology. 2013;145:1133-43 e12
140. Liao H, Xiao Y, Hu Y, Xiao Y, Yin Z, Liu L. microRNA-32 induces radioresistance by targeting DAB2IP and regulating autophagy in prostate cancer cells. Oncol Lett. 2015;10:2055-62
141. Gao W, Lam JW, Li JZ, Chen SQ, Tsang RK, Chan JY. et al. MicroRNA-138-5p controls sensitivity of nasopharyngeal carcinoma to radiation by targeting EIF4EBP1. Oncol Rep. 2017;37:913-20
142. Wang W, Liu M, Guan Y, Wu Q. Hypoxia-Responsive Mir-301a and Mir-301b Promote Radioresistance of Prostate Cancer Cells via Downregulating NDRG2. Med Sci Monit. 2016;22:2126-32
143. Li B, Wang W, Li Z, Chen Z, Zhi X, Xu J. et al. MicroRNA-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017;410:212-27
144. Chatterjee A, Chattopadhyay D, Chakrabarti G. miR-17-5p downregulation contributes to paclitaxel resistance of lung cancer cells through altering beclin1 expression. PLoS One. 2014;9:e95716
145. Pan B, Feng B, Chen Y, Huang G, Wang R, Chen L. et al. MiR-200b regulates autophagy associated with chemoresistance in human lung adenocarcinoma. Oncotarget. 2015;6:32805-20
146. Ma Y, Yuwen D, Chen J, Zheng B, Gao J, Fan M. et al. Exosomal Transfer Of Cisplatin-Induced miR-425-3p Confers Cisplatin Resistance In NSCLC Through Activating Autophagy. Int J Nanomedicine. 2019;14:8121-32
147. Han J, Li J, Tang K, Zhang H, Guo B, Hou N. et al. miR-338-3p confers 5-fluorouracil resistance in p53 mutant colon cancer cells by targeting the mammalian target of rapamycin. Exp Cell Res. 2017;360:328-36
148. Huang N, Wu J, Qiu W, Lyu Q, He J, Xie W. et al. MiR-15a and miR-16 induce autophagy and enhance chemosensitivity of Camptothecin. Cancer Biol Ther. 2015;16:941-8
149. Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58-63
150. Guk K, Hwang SG, Lim J, Son HY, Choi Y, Huh YM. et al. Fluorescence amplified sensing platforms enabling miRNA detection by self-circulation of a molecular beacon circuit. Chem Commun (Camb). 2019;55:3457-60
151. Li X, Huang N, Zhang L, Zhao J, Zhao S. A T7 exonuclease assisted dual-cycle signal amplification assay of miRNA using nanospheres-enhanced fluorescence polarization. Talanta. 2019;202:297-302
152. Jin J, Vaud S, Zhelkovsky AM, Posfai J, McReynolds LA. Sensitive and specific miRNA detection method using SplintR Ligase. Nucleic acids research. 2016;44:e116
153. Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J. et al. Human glioma growth is controlled by microRNA-10b. Cancer Res. 2011;71:3563-72
154. Jagannathan S, Vad N, Vallabhapurapu S, Vallabhapurapu S, Anderson KC, Driscoll JJ. MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib. Leukemia. 2015;29:727-38
155. Xu R, Liu S, Chen H, Lao L. MicroRNA-30a downregulation contributes to chemoresistance of osteosarcoma cells through activating Beclin-1-mediated autophagy. Oncol Rep. 2016;35:1757-63
156. Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A. et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60:590-8
157. Zhai Z, Wu F, Chuang AY, Kwon JH. miR-106b fine tunes ATG16L1 expression and autophagic activity in intestinal epithelial HCT116 cells. Inflamm Bowel Dis. 2013;19:2295-301
158. Gu H, Liu M, Ding C, Wang X, Wang R, Wu X. et al. Hypoxia-responsive miR-124 and miR-144 reduce hypoxia-induced autophagy and enhance radiosensitivity of prostate cancer cells via suppressing PIM1. Cancer Med. 2016;5:1174-82
159. Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget. 2015;6:19735-46
160. Huangfu L, Liang H, Wang G, Su X, Li L, Du Z. et al. miR-183 regulates autophagy and apoptosis in colorectal cancer through targeting of UVRAG. Oncotarget. 2016;7:4735-45
161. Mikhaylova O, Stratton Y, Hall D, Kellner E, Ehmer B, Drew AF. et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21:532-46
162. Huang D, Qiu S, Ge R, He L, Li M, Li Y. et al. miR-340 suppresses glioblastoma multiforme. Oncotarget. 2015;6:9257-70
163. Zhai H, Song B, Xu X, Zhu W, Ju J. Inhibition of autophagy and tumor growth in colon cancer by miR-502. Oncogene. 2013;32:1570-9
164. Bhattacharya A, Schmitz U, Raatz Y, Schonherr M, Kottek T, Schauer M. et al. miR-638 promotes melanoma metastasis and protects melanoma cells from apoptosis and autophagy. Oncotarget. 2015;6:2966-80
165. Zhou Q, Dong J, Luo R, Zhou X, Wang J, Chen F. MicroRNA-20a regulates cell proliferation, apoptosis and autophagy by targeting thrombospondin 2 in cervical cancer. Eur J Pharmacol. 2019;844:102-9
166. Ge YY, Shi Q, Zheng ZY, Gong J, Zeng C, Yang J. et al. MicroRNA-100 promotes the autophagy of hepatocellular carcinoma cells by inhibiting the expression of mTOR and IGF-1R. Oncotarget. 2014;5:6218-28
167. Xue H, Yuan G, Guo X, Liu Q, Zhang J, Gao X. et al. A novel tumor-promoting mechanism of IL6 and the therapeutic efficacy of tocilizumab: Hypoxia-induced IL6 is a potent autophagy initiator in glioblastoma via the p-STAT3-MIR155-3p-CREBRF pathway. Autophagy. 2016;12:1129-52
168. Song L, Liu S, Zhang L, Yao H, Gao F, Xu D. et al. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1alpha feedback loop and the Akt-mTOR signaling pathway. Tumour Biol. 2016;37:12161-8
169. Yang X, Xu X, Zhu J, Zhang S, Wu Y, Wu Y. et al. miR-31 affects colorectal cancer cells by inhibiting autophagy in cancer-associated fibroblasts. Oncotarget. 2016;7:79617-28
170. Huang T, Wan X, Alvarez AA, James CD, Song X, Yang Y. et al. MIR93 (microRNA -93) regulates tumorigenicity and therapy response of glioblastoma by targeting autophagy. Autophagy. 2019;15:1100-11
171. Sun Q, Liu T, Zhang T, Du S, Xie GX, Lin X. et al. MiR-101 sensitizes human nasopharyngeal carcinoma cells to radiation by targeting stathmin 1. Mol Med Rep. 2015;11:3330-6
172. Zheng S, Zhong YF, Tan DM, Xu Y, Chen HX, Wang D. miR-183-5p enhances the radioresistance of colorectal cancer by directly targeting ATG5. J Biosci. 2019 44
173. Sun Q, Liu T, Yuan Y, Guo Z, Xie G, Du S. et al. MiR-200c inhibits autophagy and enhances radiosensitivity in breast cancer cells by targeting UBQLN1. Int J Cancer. 2015;136:1003-12
174. Hu JL, He GY, Lan XL, Zeng ZC, Guan J, Ding Y. et al. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis. 2018;7:16
175. Zhang X, Shi H, Lin S, Ba M, Cui S. MicroRNA-216a enhances the radiosensitivity of pancreatic cancer cells by inhibiting beclin-1-mediated autophagy. Oncol Rep. 2015;34:1557-64
176. Chen H, Yao X, Di X, Zhang Y, Zhu H, Liu S. et al. MiR-450a-5p inhibits autophagy and enhances radiosensitivity by targeting dual-specificity phosphatase 10 in esophageal squamous cell carcinoma. Cancer Lett. 2020;483:114-26
177. Fan L, Wang J, Cao Q, Ding X, Li B. Aberrant miR-1246 expression promotes radioresistance in non-small cell lung cancer: a potential prognostic biomarker and radiotherapy sensitization target. Am J Cancer Res. 2020;10:314-35
178. Dokumcu K, Simonian M, Farahani RM. miR4673 improves fitness profile of neoplastic cells by induction of autophagy. Cell Death Dis. 2018;9:1068
179. Pannuru P, Dontula R, Khan AA, Herbert E, Ozer H, Chetty C. et al. miR-let-7f-1 regulates SPARC mediated cisplatin resistance in medulloblastoma cells. Cell Signal. 2014;26:2193-201
180. Zou Z, Wu L, Ding H, Wang Y, Zhang Y, Chen X. et al. MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J Biol Chem. 2012;287:4148-56
181. Pennati M, Lopergolo A, Profumo V, De Cesare M, Sbarra S, Valdagni R. et al. miR-205 impairs the autophagic flux and enhances cisplatin cytotoxicity in castration-resistant prostate cancer cells. Biochem Pharmacol. 2014;87:579-97
182. Tan S, Shi H, Ba M, Lin S, Tang H, Zeng X. et al. miR-409-3p sensitizes colon cancer cells to oxaliplatin by inhibiting Beclin-1-mediated autophagy. Int J Mol Med. 2016;37:1030-8
183. Tan W, Liao Y, Qiu Y, Liu H, Tan D, Wu T. et al. miRNA 146a promotes chemotherapy resistance in lung cancer cells by targeting DNA damage inducible transcript 3 (CHOP). Cancer Lett. 2018;428:55-68
184. Chen K, Shi W. Autophagy regulates resistance of non-small cell lung cancer cells to paclitaxel. Tumour Biol. 2016;37:10539-44
185. Zhou J, Wu S, Chen Y, Zhao J, Zhang K, Wang J. et al. microRNA-143 is associated with the survival of ALDH1+CD133+ osteosarcoma cells and the chemoresistance of osteosarcoma. Exp Biol Med (Maywood). 2015;240:867-75
186. Zhou Y, Chen E, Tang Y, Mao J, Shen J, Zheng X. et al. miR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepatocellular carcinoma cells. Cell Death Dis. 2019;10:843
187. Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC, Yao L. et al. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-beta/Smad4 pathway. World J Gastroenterol. 2017;23:1816-27
188. Zhang H, Tang J, Li C, Kong J, Wang J, Wu Y. et al. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015;356:781-90
189. Yu Y, Yang L, Zhao M, Zhu S, Kang R, Vernon P. et al. Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia. 2012;26:1752-60
190. Frankel LB, Wen J, Lees M, Hoyer-Hansen M, Farkas T, Krogh A. et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30:4628-41
191. Du F, Feng Y, Fang J, Yang M. MicroRNA-143 enhances chemosensitivity of Quercetin through autophagy inhibition via target GABARAPL1 in gastric cancer cells. Biomed Pharmacother. 2015;74:169-77
192. Liu L, Shen W, Zhu Z, Lin J, Fang Q, Ruan Y. et al. Combined inhibition of EGFR and c-ABL suppresses the growth of fulvestrant-resistant breast cancer cells through miR-375-autophagy axis. Biochem Biophys Res Commun. 2018;498:559-65
193. Xiong J, Wang D, Wei A, Ke N, Wang Y, Tang J. et al. MicroRNA-410-3p attenuates gemcitabine resistance in pancreatic ductal adenocarcinoma by inhibiting HMGB1-mediated autophagy. Oncotarget. 2017;8:107500-12
194. Wang S, Wu J, Ren J, Vlantis AC, Li MY, Liu SYW. et al. MicroRNA-125b Interacts with Foxp3 to Induce Autophagy in Thyroid Cancer. Mol Ther. 2018;26:2295-303
195. Nyhan MJ, O'Donovan TR, Boersma AW, Wiemer EA, McKenna SL. MiR-193b promotes autophagy and non-apoptotic cell death in oesophageal cancer cells. BMC Cancer. 2016;16:101
196. Zheng B, Zhu H, Gu D, Pan X, Qian L, Xue B. et al. MiRNA-30a-mediated autophagy inhibition sensitizes renal cell carcinoma cells to sorafenib. Biochem Biophys Res Commun. 2015;459:234-9
Author contact
![]() Corresponding authors: E-mail: shanchan2019com; wangk696com.
Corresponding authors: E-mail: shanchan2019com; wangk696com.

 Global reach, higher impact
Global reach, higher impact